Abstract
Mammalian Musashi1 (Msi1) is an RNA-binding protein that regulates the translation of target mRNAs, and participates in the maintenance of cell ‘stemness’ and tumorigenesis. Msi1 reportedly binds to the 3′-untranslated region of mRNA of Numb, which encodes Notch inhibitor, and impedes initiation of its translation by competing with eIF4G for PABP binding, resulting in triggering of Notch signaling. Here, the mechanism by which Msi1 recognizes the target RNA sequence using its Ribonucleoprotein (RNP)-type RNA-binding domains (RBDs), RBD1 and RBD2 has been revealed on identification of the minimal binding RNA for each RBD and determination of the three-dimensional structure of the RBD1:RNA complex. Unique interactions were found for the recognition of the target sequence by Msi1 RBD1: adenine is sandwiched by two phenylalanines and guanine is stacked on the tryptophan in the loop between β1 and α1. The minimal recognition sequences that we have defined for Msi1 RBD1 and RBD2 have actually been found in many Msi1 target mRNAs reported to date. The present study provides molecular clues for understanding the biology involving Musashi family proteins.
INTRODUCTION
The Musashi (Msi) family comprises a group of RNA-binding proteins that function as translational regulators of target mRNAs and that play critical roles in stem cell maintenance and self-renewal capability (1,2). Msi and Msi-like proteins have been discovered in various multicellular animals, such as Drosophila (3), Caenorhabditis elegans (4), newt (5), mouse (6,7) and human (8) (64 genes in the NCBI database, Feb 2010). In mammals, two members, Msi1 and Msi2 have been identified to date (2,6,7). Both mouse Msi1 and Msi2 have two tandemly connected ribonucleoprotein-type RNA-binding domains (RBDs), also known as RRMs (RNA recognition motifs), RBD1 and RBD2, in its N-terminal region, which are followed by a putative PABP [poly(A)-binding protein] binding region (9). Mouse Msi1 and Msi2 consist of 362 and 346 amino acid residues, respectively. Their sequence identity is 69%, whereas that of the region containing the two RBDs (20–191 in Msi1 and 21–192 in Msi2) is 86%.
RBDs are one of the families of well-known single-stranded polynucleotide binding domains that have a β1α1β2β3α2β4 topology, forming a four-stranded anti-parallel β sheet packed against two α-helices (10,11). In the β-sheet, there are two highly conserved sequence stretches, RNP1 (in the β3 strand) and RNP2 (in the β1 strand). Many of the so-called classical RBDs contain three aromatic amino acid residues in RNP1 and RNP2 that play important roles in nucleic acid recognition. We previously determined the structures of RBD1 and RBD2 of mouse Msi1, and showed that each RBD has three solvent-exposed phenylalanines in RNP1 and RNP2 (12,13). Subsequent RNA binding and molecular dynamics studies involving nuclear magnetic resonance (NMR) have revealed that these phenylalanines are important for direct RNA binding (12,13). Substitution of these phenylalanines of RBD1 with alanines results in loss of translational repression and the RNA-binding activity of Msi1 (14), which is consistent with the results of our NMR experiments.
The binding sequences of mouse Msi1 have been identified as (G/A)UnAGU (n = 1–3) by means of an in vitro selection method (14). Five target mRNAs containing this sequence in their 3′-UTRs have been identified and studied: numb (14), p21WAF−1 (15), c-mos (16) and doublecortin (dcx) (17) and Adenomatous Polyposis Coli (APC) (18). Activation of the Notch signaling pathway positively regulates the self-renewal of various stem cells including neural stem cells (19,20) and cancer development (21). The numb mRNA encodes a protein that suppresses the Notch signaling pathway, whereby Numb promotes the differentiation of neural stem cells (NSCs). Msi1 has been proposed to bind to the 3′-UTR of numb mRNA, and then to compete with translation initiation factor eIF4G for PABP binding (9). This will lead to translational repression of numb mRNA, thereby maintaining the stem cell state. The Wnt signaling pathway plays an important role in the maintenance of pluripotency, as well as in the process of somatic cell reprogramming (22). p21WAF−1 mRNA encodes a cyclin-dependent kinase inhibitor, which negatively regulates Wnt gene expression (23). Msi1 binds to the 3′-UTR of p21WAF−1 mRNA and represses its translation. This results in upregulation of the cell cycle via the Wnt signaling pathway, whereby cell proliferation and multipotency are maintained (15). dcx mRNA, which encodes a protein that is related to the migration of newborn neurons and neural development (24,25), was identified as another target of Msi1 by means of an in vitro virus (IVV) method (17). The Msi1 binding sequence was again found in the 3′-UTR of dcx mRNA. Recently, more genes have been shown to contain Msi1 binding sequences (26). Collectively, the products of these genes of Msi1 targets were shown to be involved in cell cycle regulation, cell proliferation, self-renewal and apoptosis.
Strong expression of Msi1 has also been observed in proliferative cell populations in tumor tissues, such as medulloblastomas (27,28), gliomas (29,30), astrocytomas (31), retinoblastomas (32) and colorectal adenomas (33). Recent reports indicate that a causal role of elevated Msi2 expression in the progression of chronic myeloid leukemia (CML) from a slow growing chronic form to an aggressive blast crisis form through the translational suppression of numb mRNA and subsequent activation of Notch pathway (34–36). These reports emphasize the importance of examining the cooperation and division of roles of Msi1 and Msi2 proteins to fully understand the physiological functions of the Msi family and its relation with diseases.
Another line of evidence suggested that Msi1 can influence stem cell maintenance and differentiation by controlling the subcellular localization of proteins involved in miRNA biogenesis, as well as by regulating the translation of its target mRNA. It was shown that Msi1 acts in concert with Lin28 to regulate post-transcriptional microRNA (miRNA) biogenesis in the cropping step, which occurs in the nucleus (37).
As mentioned above, our knowledge about the target mRNAs and regulatory pathways of Msi family members is increasing. To obtain a clear understanding of the mechanism of action in translational repression and other obscure functions, and of the specificity determinants of their RBDs, we have undertaken NMR-based binding and structural studies on the two RBDs from Msi1. NMR titration experiments involving 6-nt RNA oligomers derived from the target sequence revealed the binding properties of RBD1 and RBD2. The consensus sequences for the binding of RBD1 and RBD2 were identified as r(GUAG) and r(UAG), respectively. The three-dimensional solution structure of Msi1 RBD1:r(GUAGU) revealed the characteristics of the interactions. The adenine and guanine residues at the third and fourth positions of r(GUAG) are stacked on the conserved F23 and F65 in the RNP2- and RNP1-sequences, respectively. Interestingly, the third adenine is sandwiched by another phenylalanine, F96, which resides immediately after the 4th β-strand. In addition, the first guanine residue stacks onto W29 in the loop between β1 and α1. These stacking interactions are unique to the recognition of its target RNA by Msi1 RBD1. F96 and W29 turned out to be limited to, but conserved among, members of the Msi protein family (Msi1 and Msi2). Thus, the structure we have defined shows the mechanism underlying the architecture of RNA recognition by Msi1 RBD1. Msi1 RBD2 partly contains the same recognition platform as that in Msi1 RBD1. This architecture is highly conserved among Msi proteins. Thus, this study provides a molecular basis for rationally predicting the interaction of each member of the Msi protein family with its de novo target RNA sequence.
MATERIALS AND METHODS
Preparation of Msi1 and RNA
DNA encoding RBD1 and RBD2 of Msi1 (residues 20–103 and 109–191, respectively) was amplified by polymerase chain reaction (PCR) from the mouse Msi1 gene by the conventional polymerase chain reaction, and then subcloned into pET15b (Novagene). The proteins were expressed with an extra His6-tail and a thrombin cleavage site at the N-terminus in Escherichia coli, BL21(DE3) pLysS, at 37°C in minimal medium (M9) containing 1 g/l [15N] ammonium chloride, 3 g/l [13C] glucose (for 15N and 13C-labeled proteins), vitamins, mononucleotides, metals, 100 mg/l carbenicillin and 34 mg/l chloramphenicol (38). Cells were grown to OD600 ∼ 0.5 and induced with 1 mM iso-propylthio-β-D-galactopyranoside (IPTG). Cells were harvested after 4 h by centrifugation. Cell pellets were resuspended in lysis buffer [1 M NaCl, 10 mM benzamidine, 1 mM PMSF, 50 mM Tris–HCl (pH 8.0)], disrupted by mild sonication and then centrifuged at 20 000 g. The soluble fraction containing the fusion protein was purified by Ni2+-affinity chromatography (Hi-Trap chelating HP column, GE Healthcare). The purest fractions were pooled and dialyzed against 20 mM Tris–HCl buffer (pH 7.5) containing 5 mM DTT and 1 M NaCl (MWCO 3000) in order to remove RNA impurities, and then further dialyzed against the same solution but containing no NaCl. The dialyzed protein solution was loaded onto the cation exchange column (HiTrap SP HP column, GE Healthcare) and further purified. The purified protein was dialyzed against NMR buffer [5 mM DTT, 20 mM MES (pH 6.0)] and concentrated by ultrafiltration [Amicon Ultra (MWCO 3500), Millipore]. The concentrations of the Msi1 RBD1 and RBD2 solutions were determined by ultraviolet (UV) spectroscopy. No detectable impurities were found on 15% polyacrylamide gel electrophoresis (PAGE) analysis. RNA oligomers (Table 1) were purchased from Hokkaido System Science Inc.
Table 1.
RNA sequences used for the NMR titration experiments in this study
| RNA oligomer | Sequence |
|---|---|
| numb15 | UAGGUAGUAGUUUUA |
| numb6-1 | UAGGUA |
| numb6-2 | GGUAGU |
| numb6-3 | UAGUAG |
| numb6-4 | GUAGUU |
| numb6-5 | AGUUUU |
| numb6-6 | GUUUUA |
| numb5 | GUAGU |
NMR spectroscopy
NMR spectra were recorded at 298 K on Bruker Avance 600, Avance 800 and Avance II 900 spectrometers, all of which are equipped with a triple resonance (1H/13C/15N) cryoprobe. Data were processed with NMRPipe (39), and analyzed with Kujira (40) and Sparky (http://www.cgl.ucsf.edu/home/sparky).
NMR titration experiments on Msi1 RBD1 or RBD2 with a series of RNA oligomers: r(UAGGUAGUAGUUUUA) (numb12), r(UAGGUA) (numb6-1), r(GGUAGU) (numb6-2), r(UAGUAG) (numb6-3), r(GUAGUU) (numb6-4), r(AGUUUU) (numb6-5), r(GUUUUA) (numb6-6) and r(GUAGU) (numb5) were performed by recording 1H-15N HSQC spectra at 298 K. Increasing amounts of the unlabeled RNA oligomers were added to 50 µM 15N-labeled proteins, to obtain molar ratios of 1:0, 1:0.1, 1:0.2, 1:0.5, 1:0.7, 1:1.0, and 1:1.2.
NMR spectra of the Msi1:numb5 complex were recorded using a sample comprising 300 µM 13C, 15N-labeled Msi1 protein and 300 µM unlabeled numb5 dissolved in the NMR buffer containing 95% H2O/5% D2O. 15N and 13C resonances of the protein portion were assigned by standard double and triple resonance NMR experiments (41–43). Sequence-specific backbone assignment of Msi1 RBD1 (Cα, Cβ, C′, N, HN) was made based on HNCO, HNCA, HN(CA)CO, HN(CO)CA, CBCA(CO)NH and HNCACB. The side chains of Msi1 RBD1 were assigned based on HBHA(CO)NH, HC(CCO)NH, C(CCO)NH, H(C)CH-TOCSY, H(C)CH-COSY, CCH-TOCSY, CCH-COSY, 1H-13C NOESY-HSQC and 1H-15N NOESY-HSQC (80 and 200 ms mixing times). RNA assignments were made based on 13C, 15N-[f1,f2]-filtered NOESY (200 ms mixing times), 13C, 15N-[f2]-filtered TOCSY (43 ms mixing time) and DQF-COSY. Inter-molecular nuclear Overhauser effects (NOEs) were recorded using 1H-13C NOESY-HSQC, 1H-15N NOESY-HSQC (80 and 200 ms mixing times), and 13C, 15N-[f2]-filtered NOESY (150 and 200 ms mixing times). The assignments are deposited in the BMRB data bank under accession number 150218.
Structure calculation
The 3D structure of the Msi1 RBD1:r(GUAGU) (numb5) complex was determined by combining the automated NOESY cross-peak assignments and the structure calculations with torsion angle dynamics implemented in the program CYANA 2.1. Protein backbone ϕ, ψ torsion angle restraints were determined by chemical shift database analysis, using the program TALOS (44). Restraints for the χ1 dihedral angles were obtained by analyzing the pattern of inter- and intra-residual nuclear overhauser effect (NOE) intensities (45). Automatic assignment of the intra-protein NOEs of RBD1 in complex with numb5 was carried out with CYANA. The inter-molecular protein–RNA inter-molecular and intra-RNA NOEs were manually assigned, using 2D filtered-NOESY spectra. Protein–RNA inter-molecular and intra-RNA NOEs from the 2D filtered-NOESY spectrum with a mixing time of 80 ms were divided into two groups with upper distance bounds of 3.5 and 5.0 Å, respectively according to their intensities. The upper distance bound of 5.5 Å was applied for the inter-molecular NOEs that could only be identified from the 2D filtered-NOESY spectrum with a mixing time of 150 ms. In total, 58 inter-molecular NOEs between Msi1 RBD1 and numb5 were used for the structure calculations. RNA sugar puckering of Gua1, Ura2, Ade3 and Gua4 was determined from TOCSY and DQF-COSY spectra (46). All the RNA χ torsion angles were determined from the intra-residual NOE intensities (46).
The structure calculations started from 200 randomized conformers and involved the standard CYANA simulated annealing schedule, with 40 000 torsion angle dynamics steps per conformer. The 40 conformers with the lowest final CYANA target function values were further refined with the AMBER9 program, using an AMBER 2003 force field and a generalized Born solvation model, as described previously (47). The 20 conformers that were most consistent with the experimental restraints were selected as final structures. PROCHECK-NMR (48) and MOLMOL (49) were used to validate and to visualize the final structures, respectively. The coordinates for the ensembles of the 20 conformers of the Msi1 RBD1 in its numb5 bound form was deposited in the RCSB Protein Data Bank.
RESULTS
RNA binding of two connected RBDs, RBD1–2, may cause multiple registration
The signals in the 1H-15N HSQC spectrum for each individual [15N]-labeled RBD were dispersed and exhibited uniform peak intensities, which are characteristics of a well-folded polypeptide. We previously determined the structure of individual RBD1 and RBD2 in their free states (12,13). A protein construct containing the two RBDs (RBD1–2, 20–191) also gave well-resolved 1H-15N HSQC spectra in the free state (Supplementary Figure S1). The spectrum of RBD1-2 turned out to be almost the summation of the spectra of both RBDs. Some differences are found for the signals that are assigned to the linker region that connects the two RBDs. This linker region does not exist in the individual RBD constructs. These observations indicate that there is no inter-domain interaction between RBD1 and RBD2. This was also supported by paramagnetic relaxation enhancement (PRE) experiments. PRE is a technique that provides longer distance information (∼15 Å) than commonly used nuclear Overhauser effect spectroscopy (NOESY) (∼5 Å) (50). PRE experiments were carried out to obtain the inter-domain distances between RBD1 and RBD2 within the RBD1–2. Msi1 RBD1–2 V118C mutant, which was spin-labeled with (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate (MTSL) at C118 in RBD2 (C118 locates in the loop between β1 and α1) was used. No inter-domain PREs, however, were observed. This indicates that the two domains within RBD1–2 are not arranged in any particular orientation or they are apart in the free state (Supplementary Figure S2). We then titrated the target RNA, r(UAGGUAGUAGUUUUA) [numb15; (14)], against RBD1–2 and monitored the signal perturbations in the 1H-15N HSQC spectra. First, the sample solution became turbid when the molar ratio reached [RBD1-2]:[numb15] ∼ 1.0:0.5. Under these conditions, the signals were significantly line-broadened. As in the titration experiments, we have noticed that in the range of [RBD1–2]:[numb15] = 1.0:0.5 to 1.0:2.0, the chemical shifts of most of the signals changed linearly in one direction. However, some signals started to change in different directions when the molar ratio exceeded [RBD1-2]:[numb15] ∼ 1.0:1.0 (Supplementary Figure S1). The origin of this behavior is not clear. We suppose that it could be due to multiple registration of numb15 onto RBD1-2, that may cause the formation of oligomerized complexes under the present NMR conditions. An alternative possibility is that an initial encounter complex is formed in the initial phase, followed by the formation of some more specific complex later on.
Identification of the consensus RNA sequences for the binding of Msi1 RBD1 and RBD2
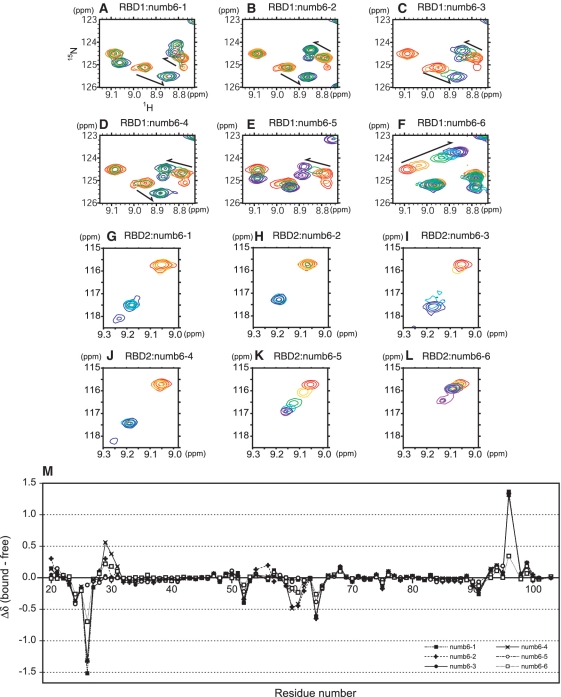
To avoid analysis of such inhomogeneous samples, we made protein constructs, each of which contained only the RBD1 or RBD2 portion of Msi1, and to identify the RNA sequences that bind to RBD1 and RBD2, we prepared six 6-nt RNA oligomers: r(UAGGUA) (numb6-1), r(GGUAGU) (numb6-2), r(UAGUAG) (numb6-3), r(GUAGUU) (numb6-4), r(AGUUUU) (numb6-5) and r(GUUUUA) (numb6-6), all of which were derived from r(UAGGUAGUAGUUUUA) (numb15), as described in Table 1. NMR titration experiments were then performed on each of the 15N-labeled RBDs (50 µM), a series of 2D 1H−15N HSQC spectra being measured with the addition of each of the RNA oligomers. All 6-nt RNA oligomers had an extensive effect on the 1H-15N HSQC spectra upon titration. These changes reached a plateau when an equimolar quantity of RNA was added to the RBDs (Figure 1). This indicates that both RBD1 and RBD2 interact with each of the RNA oligomers in a 1:1 stoichiometric manner. It should be noted that the binding of RBD1 with numb6-2 or numb6-4 (Figure 1B and D), and that of RBD2 with numb6-1, numb6-2 or numb6-4 (Figure 1G, H and J) exhibited slow exchange on the NMR timescale, while the other pairs exhibited either fast (RBD1: Figure 1E and F; RBD2: Figure 1K and L) or intermediate exchange (RBD1: Figure 1A and C; RBD2: Figure 1I). In general, the binding of two molecules exhibiting slow exchange on the NMR time scale is considered to be higher affinity than the binding of two molecules showing either fast or intermediate exchange. Thus, these results indicate that RBD1 binds to numb6-2 and numb6-4 with higher affinity than to the other four RNA oligomers. Figure 1M presents the chemical shift changes of the HN signals of the Msi1 RBD1 residues upon the addition of each of the RNA oligomers in a 1:1 molar ratio. In this figure, the values for the chemical shift changes upon the addition of numb6-2 and numb6-4 superimpose very well on those of all the residues of Msi1 RBD1.
Figure 1.
Binding of 6-nt RNA oligomers to individual RBDs of Msi1. Overlays of the 15N-HSQC spectra sections of RBD1 (red), and RBD1 in the presence of 0.2 (orange), 0.5 (green), 1.0 (blue) and 1.2 (purple) equivalents of 6-nt RNA oligomers: (A) r(UAGGUA) (numb6-1); (B) r(GGUAGU) (numb6-2); (C) r(UAGUAG) (numb6-3); (D) r(GUAGUU) (numb6-4); (E) r(AGUUUU) (numb6-5) and (F) r(GUUUUA) (numb6-6). Overlays of the 15N-HSQC spectra sections of RBD2 (red) and RBD2 in the presence of 0.2 (orange), 0.5 (green), 1.0 (blue) and 1.2 (purple) equivalents of 6-nt RNA oligomers: (G) numb6-1; (H) numb6-2; (I) numb6-3; (J) numb6-4; (K) numb6-5 and (L) numb6-6. (M) The chemical shift changes, Δδ, were obtained by subtracting the chemical shifts of Msi1 backbone 1HN for the free protein from the chemical shifts of Msi1 backbone 1HN in the complex (Msi1:RNA oligomer = 1:1).
A series of 6-nt RNA titration experiments indicated that Msi1 RBD1 binds r(GUAGU)-containing RNA oligomers with high affinity. In order to determine the effect of the extra guanine on the 5′ side of numb6-2 and uracil on the 3′ side of numb6-4 on the affinity, we performed an experiment on competition between numb6-2 and numb6-4 in RBD1 binding by means of chemical shift perturbation. The addition of numb6-2 (12.5 nmol) and numb6-4 (12.5 nmol) simultaneously to [15N]-labeled RBD1 (12.5 nmol) resulted in two sets of protein signals (Supplementary Figure S3). One set overlapped the protein signals for the RBD1:numb6-2 complex and the other with those for the RBD1:numb6-4 complex. The intensities of the signals originating from the RBD1:numb6-2 and RBD1:numb6-4 complexes were equivalent, suggesting that the magnitudes of their dissociation constants are comparable. Therefore, we chose r(GUAGU) as a minimal RNA oligomer for use in further structural studies.
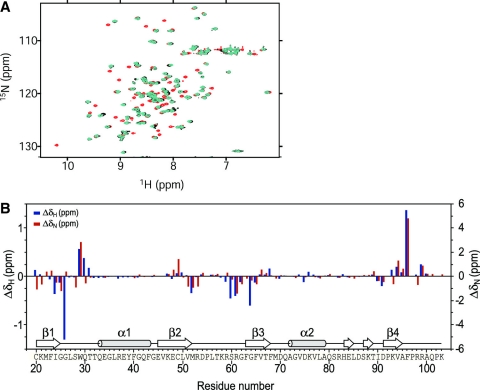
Next, we have synthesized a 5-nt RNA oligomer numb5, r(GUAGU), and performed NMR titration experiments in order to determine whether or not this sequence is of sufficient length for structural determination. During the numb5 titration, many of the residues of [15N]-labeled RBD1 exhibited two distinct cross peaks in 1H-15N HSQC spectra, suggesting that numb5 binding exhibits a slow exchange regime on the chemical shift timescale. No free protein signals were observed when the molar ratio reached RBD1:numb5 = 1:1, just like in the cases when numb6-4 and numb6-2 were titrated against RBD1. Almost all protein signals of the RBD1:numb5 complex and those of the RBD1:numb6-2 and RBD1:numb6-4 complexes overlapped each other (Figure 2A). In fact, the 1H and 15N chemical shift differences of [15N]-labeled RBD1 in a complex with numb5 (Figure 2B) and those with either numb6-2 or numb6-4 turned out to be within the range of 0.05–0.3 ppm, respectively. Altogether, these data demonstrated that numb5 could be used for the further structural study to obtain important insights into the nature of the RNA binding by Msi1 RBD1.
Figure 2.
r(GUAGU) (numb5) and r(GUAGUU) (numb6-4) bind equally well to RBD1 of Msi1. (A) Overlay of the 15N-HSQC spectra of RBD1 (red), RBD1 in the presence of a 1.0 equivalent of numb5 (cyan) and RBD1 in the presence of a 1.0 equivalent of numb6-4 (black). (B) The chemical shift changes, ΔδH (ΔδN), were obtained by subtracting the chemical shifts of Msi1 backbone 1HN (15N) for the free protein from the chemical shifts of Msi1 backbone 1HN (15N) in the complex.
As for the RNA oligomers that bind to RBD2, numb6-1, numb6-2 and numb6-4 exhibited a slow exchange regime on the chemical shift timescale, indicating that they exhibit stronger affinity than the other three. Each of the numb6-1, numb6-2 and numb6-4 contains UAG and GUA sequences in common, whereby they can be considered as candidates for the Msi1 RBD2 recognition sequences. We further analyzed the amino acid residues of Msi1 RBD2 that are involved in RNA binding, and their conservation with those of Msi1 RBD1. As a result, we were able to define UAG as the recognition sequence for Msi1 RBD2, with a possible involvement of an arbitrary nucleotide in the fourth position. The binding mode of Msi1 RBD2 and UAG will be discussed in the ‘Discussion’ section.
Structure determination
In the previous section, we showed that Msi1 RBD1 binds r(GUAGU)-containing RNA oligomers with high affinity. Here, we determined the solution structure of the complex between Msi1 RBD1 (20–103) and Gua1-Ura2-Ade3-Gua4-Ura5 (numb5) using the NMR method. We have 1716 intra-protein, 16 intra-RNA and 58 inter-molecular protein–RNA upper distance limits. Intra-protein NOEs were collected from 15N-edited NOESY-HSQC and 13C-edited NOESY-HSQC spectra. Intra-RNA NOEs were obtained from 13C, 15N-[f1,f2]-filtered NOESY spectra, whereas 13C, 15N-[f2]-filtered NOESY spectra contained both intra-RNA and inter-molecular protein–RNA NOEs. We were able to isolate just the inter-molecular protein–RNA NOEs by careful comparison between these two sets of filtered NOESY spectra. Inter- and intra-molecular distance restraints were derived from the intensities of the obtained NOEs. Inspection of the J-coupling of H1′–H2′ cross-peaks in the DQF-COSY spectra showed that the Gua1, Ura2, Ade3 and Gua4 sugars adopt the C2′-endo conformation [JH1′-H2′ ∼11 Hz; (46)].
The structure calculation and refinement were carried out using program CYANA (51) and Amber9 (52). Among the 200 independently calculated structures, the 40 conformers with the lowest CYANA target functions were refined by restrained energy minimization using solvation simulation with program Amber9 (Table 2). The 20 conformers that were most consistent with the experimental restraints were selected as the final structures (Figure 3A and B).
Table 2.
Structural statistics for the Msi1 RBD1:numb5 complex
| Restraints for final structure calculations | |
|---|---|
| Upper distance limits | |
| Total | 1790 |
| Intra-protein | 1716 |
| short-range, |i−j|≤1 | 835 |
| medium-range, 1<|i−j|<5 | 281 |
| long-range, | i−j |≥5 | 600 |
| Intra-RNA | 16 |
| Inter-molecular | 58 |
| Number of dihedral angle restraints | |
| χ angles | 4 |
| Number of hydrogen bond restraints | 0 |
| Structure statistics (20 structures) | |
| Number of NOE violations >0.2Å | 0 |
| Number of dihedral angle violations >5° | 0 |
| Average CYANA target function (Å): | 0.18 |
| Average AMBER energy (kcal·mol−1): | −4059.47 |
| Average AMBER total restraint violation (kcal·mol−1) | 4.27 |
| RMS deviation from the mean coordinate structure | |
| Backbone heavy atoms (Å) | |
| Protein (21–97) | 0.33 ± 0.09 |
| All heavy atoms (Å) | |
| Protein (21–97) | 0.96 ± 0.11 |
| Protein (21–97) and RNA | 0.94 ± 0.11 |
| Ramachandran statistics (%) | |
| Residues in most favored regions | 87.6 |
| Residues in additional allowed regions | 11.6 |
| Residues in generously allowed regions | 0.3 |
| Residues in disallowed regions | 0.5 |
Figure 3.
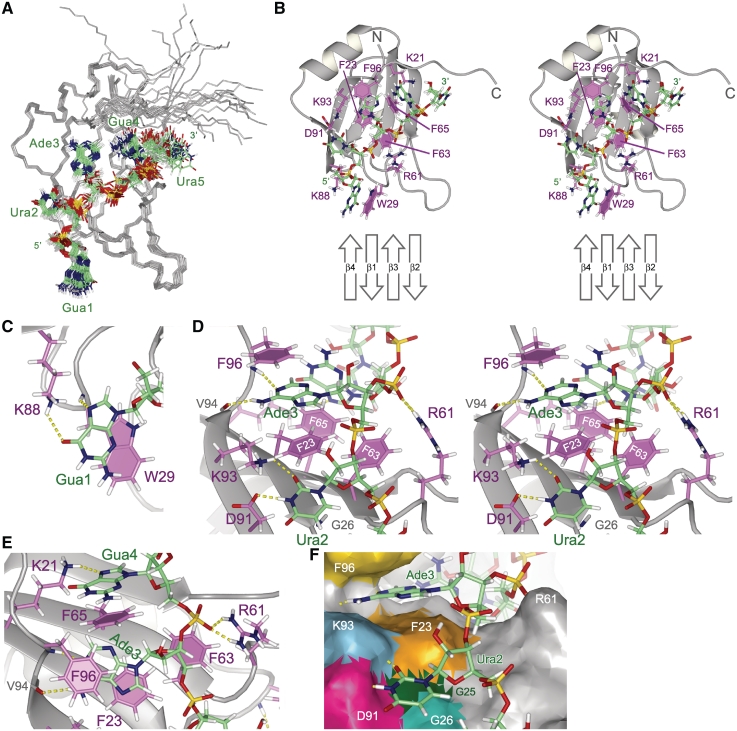
Structure of Msi1 RBD1 in a complex with r(GUAGU) (numb5). (A) Superpositioning of the 20 lowest energy conformers of the RBD1:numb5 complex. The protein backbone (residues 20-103) is colored gray. RNA is shown as a stick model: hydrogen (pale gray), carbon (green), nitrogen (blue), oxygen (red) and phosphorus (yellow). (B) The lowest energy conformer is presented and viewed from the same direction as in (A). Protein side chains contributing to the RNA binding are shown as a stick model: hydrogen (pale gray), carbon (magenta), nitrogen (blue) and oxygen (red); and aromatic rings are filled. RNA is represented as in (A). A schematic drawing of the RNA-binding β-sheet is presented at the bottom. (C) Recognition of Gua1. The Gua1 base stacks onto the W29 indole ring; and hydrogen bonds, Gua1 N7-W29 HN and Gua1 O6- K88 Hζ, are formed. (D) Recognition of Ade3. The Ade3 base is sandwiched between the aromatic rings of F23 and F96; and hydrogen bonds, Ura2 N3-D91 Oδ, Ura2 O2-K93 Hζ, Ade3 H6-V94 O and Ade3 N1-F96 HN, are formed. (E) Recognition of Gua4. The Gua4 base stacks onto the F65 aromatic ring, and Gua4 N7 hydrogen bonds with K21 Hζ. R61 Hη may form a salt bridge with the 5′ phosphate group of Gua4. (F) The Ura2 base stacks in the pocket. The solvent accessible surface of RBD1 is viewed from a similar direction to as in (D). D91, K93, F23 and F63 form a rim, and G25 and G26 form the bottom of the pocket. Hydrogen bonds are indicated by dotted yellow lines. (B) and (D) are stereo diagrams.
Structure description of the Msi1 RBD1: numb5 complex and the base-specific RNA recognition by Msi1 RBD1
The structure of Msi1 RBD1 in the complexed form adopts a βαββαβ topology: β1 (21–25), α1 (33–42), β2 (47–52), β3 (63–67), α2 (72–79) and β4 (91–94), with additional short β-strands, sβ (84–85, 88–89). These secondary structure elements form a four-stranded anti-parallel β-sheet, which is backed up by two α-helices on one side. The additional short β-strands form a β-turn located between α2 and β4. The overall protein structure turned out to be identical to that in the RNA free state (13). In the complex, r(GUAGU) (numb5) lies across the four-stranded anti-parallel β-sheet, the 5′-end of numb5 (Gua1) being located between two loops (loop β1 -α1 and loop α2 -β4), with the 3′-end (Ura5) on β2 (Figure 3B). Four nucleotide residues, Gua1, Ura2, Ade3 and Gua4, turned out to participate in the specific interaction with RBD1. These structural features will be described in detail below.
The base of Gua1 stacks onto the indole ring of W29 and N7 of Gua1 seems to form a hydrogen bond with the amide proton of W29 (Figure 3C). The latter is consistent with the fact that the signal of the amide proton of W29 shows a large downfield shift upon numb5 binding (Figure 2B). The ring current effect of the nearby Gua1 base may also contribute to this large downfield shift. Additionally, Gua1 O6 and K88 Hζ are located close to each other, the distance between them being appropriate for the formation of hydrogen bond. This is supported by the inter-molecular NOEs between Gua1 H8 and K88 Hδ. The backbone amide proton signals of S60 and R61 were upfield shifted upon numb5 binding (Figure 2B). This ring current effect of the indole of W29, which is close to these amide protons, may be responsible for this observation.
The structure of the complex indicated that Ura2 is recognized through two hydrogen bonds: One between O2 of the Ura2 base and K93 Hζ, and the other between the imino proton of the Ura2 base and D91 Oδ (Figure 3D). These interactions result in the placement of the ring of Ura2 proximal to the backbone amide proton of G26 (Figure 3D). The structure obtained suggests the upfield shift of the G26 HN chemical shift can be induced through the ring current of the Ura2 ring. Indeed, we observed a 1.3 ppm upfield shift of the G26 HN chemical shift as compared with the chemical shift of G26 HN in the RBD1 free form (Figure 2B).
The base of Ade3 is sandwiched between the rings of F23 (RNP2) and F96 (C-terminal region flanking β4) (Figure 3D and E). This structural feature is supported by three inter-molecular NOEs: F96 HN-Ade3 H2, F96 Hε-Ade3 H8, and F96 Hδ-Ade3 H3. No NOE was observed between the F23 and F96 rings; this agrees with the obtained structure as these rings are located on opposite sides of Ade3. In addition, the structure suggests two possible inter-molecular hydrogen bonds: one between N1 of the Ade3 base and the backbone NH of F96, and the other between Ade3 NH2 (or H6) and V94 CO (Figure 3E). The former is supported by the 1.4 ppm downfield shift of F96 HN upon complex formation (Figure 2B). Additionally, the strong NOE observed between Ade3 H4′ and F63 Hδ Hε suggests that the ribose ring of Ade3 is located near the F63 aromatic ring (Figure 3D).
The base of Gua4 stacks onto the aromatic ring of F65, which is one of the three conserved phenylalanines in RNP1 (Figure 3E). Among the intra-residue NOEs involving Gua4 H8, the NOE between H8 and sugar H1′ exhibited the strongest intensity, whereas the intensities of the NOEs between H8 and H2′, H8 and H3′, and H8 and H5′/H5″ were either very weak or unobservable. This indicates that Gua4 takes on the syn conformation around the glycosidic bond (χ angle). This conformation is also supported by the observation of inter-molecular NOEs between Gua4 H8 and the protons of RBD1 residues such as M52 Hε, L50 Hδ and F65 Hδ/Hε/Hζ; since the inter-molecular distances between the corresponding protons would be too great if the χ angle was in the anti-conformation. The structure also suggests that K21 Hζ forms a hydrogen bond with Gua4 N7. This is mostly due to the observed NOEs between K21 Hδ and Hβ and Hδ of F65, that is located just under Ade3 (Figure 3E).
Finally, inspection of the 20 final structures of the Msi1 RBD1:numb5 complex showed that Ura5 is located near β2 with poor convergence. This is due to the fact that no intra- and inter-molecular NOEs involving Ura5 were detected. Therefore, we concluded that Ura5 is not actively recognized by Msi1 RBD1.
DISCUSSION
Features of the Msi1 RBD1:numb5 complex
First, in this study, we identified the minimal binding RNA sequences for the individual RBD1 and RBD2 of Msi1. Then, we determined the three-dimensional structure of the Msi1 RBD1:r(GUAGU) (numb5) complex. The structure illustrated that Msi1 RBD1 lacks the interaction with Ura5 at the 3′-end; therefore, we concluded that Msi1 RBD1 requires just four bases, GUAG, for the recognition. Msi1 RBD1 utilizes its β-sheet surface to bind GUAG, just like most classical RBDs whose structures in the complex state with a cognate RNA or DNA have been determined (11). The structure of the Msi1 RBD1:numb5 complex shares several attributes with most Ribonucleoprotein (RNP)-type RBD:nucleic acids complexes, and some features that are likely to be specific to the Msi1 RBD1:RNA complex. We will discuss the following points: first, common interactions between Msi1 RBD1 and numb5 that are shared by other RNP-type RBDs and cognate RNAs, and then unique interactions that are observed in the numb5 recognition by Msi1 RBD1.
In Msi1 RBD1, F23 of RNP2 (on β1) and F65 of RNP1 (on β3) are used to interact with Ade3 and Gua4, respectively. These phenylalanines are conserved among many RNP-type RBDs (10), such as those of hnRNP A1, hnRNP D0 and Hrp1 (Figure 4). In all of these RBDs, the base moiety of the nucleotide residues stacks onto the aromatic ring of the conserved phenylalanines. Actually, F18 of hnRNP A1, F162 of Hrp1 and F185 of hnRNP D0, all of which correspond to F23 of Msi1, bind to adenine of a cognate RNA (for hnRNP D0 and Hrp1) or DNA (for hnRNP A1). In addition to this aromatic stacking interaction between the adenine and phenylalanine residues of RNP2, hydrogen bonds are formed between Ade3 and amino acid residues on β4 (F96 HN- Ade3 N1 in Msi1, R88 CO-Ade203 H6 in hnRNP A1 and I234 HN-Ade6 N1 in Hrp1). In the case of the Msi1:numb5 complex, the backbone amide proton of F96 and the carbonyl oxygen of V94 are hydrogen bonded to N1 and the amino group of Ade3. Similarly, the amide protons of V90 in hnRNPA1, M258 in hnRNP D0 and I234 in Hrp1 form hydrogen bonds with adenine N1 of the cognate RNA; and R232 CO of Hrp1 forms a hydrogen bond with adenine H6 of the cognate RNA.
Figure 4.
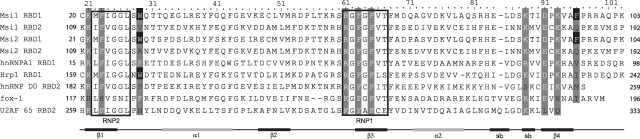
Amino acid sequence alignment of the RBDs of Msi family proteins and other proteins; Mus Musculus Msi1 RBD1 and RBD2, M. Musculus Msi2 RBD1 and RBD2, Homo sapiens hnRNP A1 RBD1 (56), Saccharomyces cerevisiae Hrp1 RBD1 (54), H. sapiens Fox-1 (53), H. sapiens hnRNP D0 RBD2 (57) and H. sapiens U2AF65 (58) are listed. The residues of Msi1 RBD1 that are suggested to interact with r(GUAGU) (numb5), and the residues at the equivalent positions in the listed RBDs are highlighted in gray. W29 and F96 of Msi1 RBD1 are highly conserved in the Msi family members (highlighted in black). The positions of the secondary structure elements (β, β-strand; α, α-helix; sb, short β-strands that form a β-turn) determined for Msi1 RBD1 are shown at the bottom.
The aromatic ring of F65 in Msi1 interacts with the base of Gua4 by means of aromatic stacking. The equivalent residues are F59 in hnRNP A1 and F227 in hnRNP D0, both of which interact with guanine bases through aromatic stacking. Interestingly, the corresponding guanine in each of these complexes takes on the syn conformation as to the χ angle. In addition, in all of these complexes, the Gua4 base is hydrogen-bonded with the side chains of amino acid residues on β1 (K21 in Msi1, K15 in hnRNP A1 and K183 in hnRNP D0). K21 Hζ (on β1) in Msi1 approaches to N7 of the Gua4 base and forms a hydrogen bond (Figure 4). Analogous hydrogen bonds are formed in hnRNP A1 and hnRNP D0, each in the complex form, where K15 and K183 correspond to K21 of Msi1, respectively (Figure 4). This lysine on β1 is exclusively conserved among RBD1 and RBD2 of the Msi family throughout different species (Supplementary Figure S6). In some RBDs, such as Fox-1, the corresponding amino acid residue is arginine (Figure 4), which can also form a hydrogen bond with a guanine residue of a cognate RNA (53). It seems that the basic amino acid (lysine or arginine) on β1 at this position is important for support and stabilization of the guanine–phenylalanine aromatic stacking.
Inspection of the RBD1:r(GUAGU) complex structure revealed that when Gua4 in the current syn conformation is converted to anti, serious steric clashes are caused with the surrounding amino acid residues. Steric clashes are also caused when Gua4 is replaced by an adenosine in the anti conformation. Thus, the syn conformation is needed at the fourth position to avoid the steric clashes. A guanosine can adopt the syn conformation more easily than an adenosine. This could be a reason why a guanosine is preferred over an adenosine at the fourth position. The inspection also revealed that an adenosine in the syn conformation can be accommodated without a steric clash in replacement of Gua4, although amino protons of the adenosine are located close to the side chain of Lys21. Thus, the steric hindrance involving amino protons of an adenine alone does not account for the preference of a guanosine at the fourth position. This also supports the idea that a guanosine is preferred at the fourth position due to its easiness to take on the syn conformation over an adenosine.
Figure 3F shows a pocket on the molecular surface of Msi1 RBD1 in which Ura2 fits. This pocket comprises D91, K93 and F23, forming a rim; and G25 and G26, forming the bottom of the basin. All of these residues are highly conserved in the Msi1 family (Figure 4). F23, G25 and G26 are part of RNP2 (β1) and F63 is in RNP1 (β3). It is important that the residues at the positions of G25 and G26 have no side chains. This requirement allows the side chains of D91 and K93 on β4 to approach the base of Ura2 from the horizontal direction and form hydrogen bonds (Figure 3D). These charged amino acid residues on the β4 are conserved in many RBDs (Figure 4). For example, the residues at these positions in RBD2 of hnRNP A1 and RBD1 of Hrp1 are aspartate and lysine; and those in RBD1 of hnRNP A1 and RBD2 of hnRNP D0 are glutamate and lysine (Figure 4). The pocket described above is also formed in these RBDs.
R61 of Msi1 RBD1, which is located at the N-terminus of the RNP1 motif, reaches out to the phosphate backbone of the nucleic acid sequence and undergoes an electrostatic interaction (Figure 3D and Supplementary Figure S3A). In many RNP-type RBDs whose structures in complexes with cognate RNAs have been determined, the residue at the equivalent position is either arginine or lysine, all of which interact electrostatically with the phosphate backbone (Supplementary Figure S4A and C). So far, we have discussed the interactions found in the Msi1 RBD1:numb5 complex that are commonly found in complexes between other RNP-type RBDs and their cognate nucleic acids.
Next, we will focus on the interactions that are unique to the Msi1 RBD1:numb5 complex. The indole ring of W29, which is located in the β1 -α1 loop, and the purine ring of Gua1 interact by means of aromatic stacking (Figure 3C and Supplementary Figure S3B). First of all, tryptophan is rarely found at this position in RNP-type RBDs: 10% in 1200 RBDs (recorded in Pfam database on February 2010). Hrp1, whose structure has been determined in the complex state with its cognate RNA, also has a tryptophan at the corresponding position (54). However, Hrp1 uses this tryptophan to interact with the base of an adenine by means of the aromatic stacking (Supplementary Figure S4D). The difference in base-recognition between Msi1 and Hrp1 originates from the hydrogen bonds that support aromatic stacking in each case. In the Msi1:numb5 complex, Gua1 N7 and O6 are hydrogen-bonded with K88 Hζ; while in the Hrp1 and cognate RNA complex, Ade4 N7 and H6 are hydrogen-bonded with W168 HN and N167 Oδ1, respectively (Supplementary Figure S4D). The common architecture for the purine ring recognition at the β1–α1 loop between these two complexes is purine–indole aromatic stacking, and the hydrogen bond between N7 of purine and the amide of a tryptophan (Supplementary Figure S3B and S3D). However, these two proteins recognize different purine rings, because the hydrogen bond networks are different in these complexes.
Another unique feature found in the structure of the Msi1 RBD1:numb5 complex is as follows. The interaction between F23 of Msi1 RBD1 and Ade3 of numb5 has already been described as typical aromatic stacking among RNP-type RBDs. However, additional and apparently specific interactions that involve Ade3 have also been found. The aromatic ring of F96 stacks on top of the purine base of Ade3, which is stacked on the aromatic ring of F23 (Figure 3D). Thus, a F96-Ade3-F23 sandwich structure is formed. A similar sandwich structure was observed previously for the complex of an atypical RBD and its target nucleic acid, which has a rather specially modified base. CBP20, a component of the nuclear cap-binding protein complex, sandwiches the 7-methylguanosine cap structure of the eukaryotic 5′-terminus between two tyrosines, Y43 on the β-sheet and Y20 in the N-terminal unstructured region (55). In this case, however, the direction of the RNA bound to the β-sheet surface was different to that of RNA bound to common RNP-type RBDs including RBD of Msi1. In the canonical mode of RNA binding, RNA crosses diagonally in the 5′–3′ direction on the β-sheet surface, in the order of β1 (RNP1), β3 (RNP2) and β2 of RBD. On the other hand, the cap structure, having a unique 5′-5′ connection with the tri-phosphate bridge, lies in the opposite direction on the β-sheet surface to CBP20. As far as we know, this is the first report that a sandwich structure could be used for the recognition of a nucleic acid by a typical RNP-type RBD.
Residues of RBD2 suggested to be involved in RNA binding
RBD1 and RBD2 of mouse Msi1 exhibit high sequence homology, the sequence identity and similarity being 45 and 65%, respectively. Our NMR titration experiments have suggested that RBD2 recognizes either r(UAG) or r(GUA). Among the residues of RBD1 that are involved in RNA binding, K21, F23, R61, F63, F65, K88 and K93 are conserved in RBD2 (K110, F112, R150, F152, F154, K177 and K182, respectively). Since all of these amino acid residues, except for K88 are involved in binding to the r(UAG) moiety in the RBD1:r(GUAGU) (numb5) complex, it seems likely that RBD2 also uses the same set of amino acid residues to bind the r(UAG). In fact, experimental results on the chemical shift perturbation support this. For example, in the case of RBD1, Ade3 stacks onto F23 (Figure 3D) and accordingly, the following residue, I24, exhibits large negative chemical shift perturbation due to the ring current effect (Figure 2B). The residue of RBD2 that corresponds to F23 of RBD1 is F112 (Figure 4). Therefore, stacking of Ade3 onto F112 is expected. Large negative chemical shift perturbation observed for V113 that follows F112 (Supplementary Figure S5) supports this expected stacking. Thus, it is strongly suggested that RBD2 uses the same set of amino acid residues as RBD1 for binding and that the recognition sequence of RBD2 is r(UAG).
The residues that are not conserved in these two domains but are involved in RNA binding in RBD1 are W29 (Gua1 binding), D91 (Ura2 binding) and F96 (Ade3 binding), which are substituted by valine, glutamate and glutamine in RBD2, respectively. Among them, as described in the text, W29 of Msi1 RBD1 is important for the stacking interaction with the first guanine, Gua1, of r(GUAGU) (numb5). The corresponding residue in Msi1 RBD2 is V118, which cannot undergo a stacking interaction due to its chemical structure, thus V118 seems not to interact with Gua1. This is supported by the experimental result that V118, N119 and T120 of RBD2 exhibited much smaller chemical shift perturbation than the corresponding W29, Q30 and T31 of RBD1, upon addition of any of the RNA oligomers used in this study (Figure 2B and Supplementary Figure S5). Thus, Gua1 of r(GUAGU) is not recognized by RBD2. This reveals that the recognition sequence of RBD2 is not r(GUA). Thus, we can conclude that the recognition sequence of RBD2 is r(UAG).
E180 of RBD2, which corresponds to D91 in RBD1, probably interacts with uracil (Ura2) in the same manner. F96 of RBD1 stacks onto Ade3, which is stacked onto F23, to form an F23–Ade3–F96 sandwich. It is not likely that Q185 of RBD2, which corresponds to F96 of RBD1, undergoes a similar stacking interaction, although the possibility of a stacking interaction cannot be excluded completely because of the sp2 feature of the glutamine side chain. Msi1 RBD2 reportedly exhibits lower RNA-binding affinity than RBD1 (13). We assume that this lower binding affinity is due to the lack of tryptophan–guanine stacking and/or phenylalanine–adenine–phenylalanine sandwich stacking interactions. In summary, RBD2 recognizes three nucleotides, r(UAG), with a possibility of involvement of an arbitrary nucleotide in the fourth position.
Our chemical shift perturbation experiments showed that the binding between Msi1 RBD2 and numb6-3exhibits an intermediate exchange regime on the NMR timescale. Since numb6-3 has two sequential r(UAG) sequences, this phenomena can be explained by the multiple registration of Msi1 RBD2 for numb6-3 binding.
The possible target RNAs of Msi1 possessing r(GUAG) and r(UAG)
The target RNA of Drosophila Msi and mouse Msi1 was originally identified as r(GUU…UAG)n or r(GUU…UG)n (n = 2, 3) and (G/A)UnAGU (n = 1–3), respectively by means of in vitro selection, SELEX (14,59). From this information, Drosophila ttk69 and mouse numb, each of which encodes an important factor that plays a key role in the differentiation regulation of neural stem cells, were identified (14,59). In the present study, we used NMR spectroscopy to define the minimal RNA-binding sequences of mouse Msi1 RBDs. It was found that r(GUAG) and r(UAG) are the minimal recognition sequences for Msi1 RBD1 and RBD2, respectively.
Here, we have reanalyzed the sequence of Drosophila ttk69 mRNA, and searched for r(GUAG) and r(UAG). It turned out that there are several sequence stretches that contain these minimal recognition sequences connected by variable linker regions (2–4 nt) (Supplementary Figure S6). We also compared the amino acid sequences of the Msi proteins from different species (Supplementary Figure S6). It turned out that mouse Msi1 and Drosophila Msi exhibit high sequence homology: 55% identity and 73% similarity for RBDs; and that the residues of Msi1 RBDs that participate in the RNA binding are highly conserved in Drosophila Msi (Supplementary Figure S6). These results imply that Drosophila Msi and mouse Msi1 recognize their target RNA sequences in a similar, if not the same, manner.
Next, we searched for the minimal recognition sequences, r(GUAG) and r(UAG), in the 3′-UTR of numb mRNAs originating from other vertebrate species. Again, we were able to find these minimal recognition sequences located within 3–4 nt. The Supplementary Figure S6 demonstrates that all the residues of Msi1 RBDs that participate in the binding of the minimal recognition sequences are conserved in Msi1 from other vertebrates. These results strongly suggest that the molecular mechanisms by which the Msi proteins regulate translation of numb mRNAs are conserved widely among vertebrates.
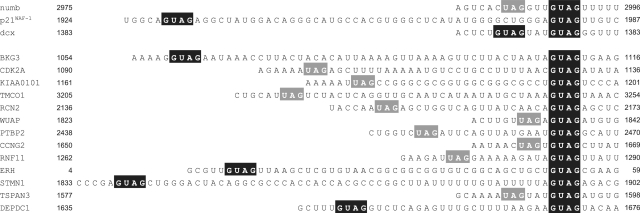
Many vertebrate mRNAs reportedly contain Msi1 binding sites in their 3′-UTRs (17,26). Next, we searched for r(GUAG) and r(UAG) within these 3′-UTRs. Indeed, we were able to list the portions of the 3′-UTRs that contain these minimal recognition sequences (Figure 5). r(GUAG) and r(UAG) turned out to be connected via variable linkers (1–50 nt long).
Figure 5.
A list of 3′-UTR regions of the putative Msi1 targets; H. sapiens numb (accession code NM_001005744.1), H. sapiens p21WAF-1 (NM_078467.1), M. musculus dcx (NM_001110223.1), H. sapiens BKG3 (NM_006806.4), H. sapiens CCNG2 (NM_004354.2), H. sapiens CDK2A (NM_001195132.1), H. sapiens DEPDC1 (NM_001114120.1), H. sapiens ERH (NM_004450.2), H. sapiens KIAA0101 (NM_001029909.1), H. sapiens PTBP2 (NM_021190.2), H. sapiens RCN2 (NM_002902.2), H. sapiens RNF11 (NM_014372.4), H. sapiens STMN1 (NM_001145454.1), H. sapiens TMCO1 (NM_019026.3), H. sapiens TSPAN3 (NM_001168412.1) and H. sapiens WUAP (NM_004906.3). Binding sequences for Msi1 RBD1 (GUAG) and RBD2 (UAG) are highlighted in black and gray, respectively.
Thus, the present study has revealed that r(GUAG) and r(UAG) are the minimal recognition sequences for Msi proteins and are extensively conserved within the target mRNAs of Msi proteins.
Biological implications for RNA recognition by the Msi family
In mammals, two members of the Msi family, not only Msi1, but also Msi2 are co-expressed in neural precursor cells, including central nervous system (CNS) stem cells. Neurosphere formation is reportedly inhibited when the Msi1 and Msi2 genes are knocked out simultaneously, however, single knockout does not bring about such disturbance (60). Two recent reports identified Msi2 as a key regulator in the progression of CML from the chronic phase to the blast crisis phase (34,35). According to these reports, Numb is downregulated in blast crisis CML and exogenous expression of Numb inhibits leukemogenesis. Consequently, upregulated Msi2 in blast crisis CML negatively regulates the expression of Numb. High expression of Msi2 was also found in leukemic cells of acute myelogenous leukemia (AML) patients, and elevated Msi2 expression was shown to be associated with a poor prognosis in both AML and CML. Exogenous expression of Msi2 enhanced the development of aggressive immature leukemia induced by BCR-ABL. These reports emphasize the importance of the Msi2-Numb pathway in regulating the development of aggressive myeloid leukemia.
Next, we examined if the present study could provide information on the RNA recognition by Msi2. We compared the amino acid sequences of Msi2 and Msi1. The RBD1–2 regions of Msi1 and Msi2 of both mouse and human exhibit high sequence homology: 85% identity and 91% similarity. The residues that are important for Msi1 RBD1 to recognize r(GUAG) turned out to be completely conserved in Msi2 RBD1 (Figure 4). As described in the previous section, we also identified the residues of Msi1 RBD2 that are possibly involved in r(UAG) recognition. These residues are also fully conserved in Msi2 RBD2 (Figure 4). These findings suggest that Msi1 and Msi2 may recognize the same RNA sequences, which explains the results of double knockout experiments regarding the neurosphere formation. Consequently, it is tempting to assume that Msi1 and Msi2 target for the same mRNAs and play crucial roles in controlling self-renewal, proliferation and leukemogenesis by either suppressing (2,34,35) or promoting (16) translation. Our next questions would be: what are the functions of Msi1 and Msi2, and how are they regulated? Although the RBD1–2 regions of Msi1 and Msi2 are highly conserved, the C-terminal regions of these two proteins are quite different in length and amino acid sequence: 56% identity and 63% similarity. We therefore hypothesize that their C-terminal regions may play important roles in protein–protein interactions and/or autoregulation. Indeed, Msi1 has a putative PABP binding site following RBD2 (9). This region is not conserved in Msi1 and Msi2. It is also possible that Msi1 and Msi2 are regulated by post-translational modifications. Future biological and structural studies on Msi1 and Msi2 will facilitate not only understanding of the functions of Msi family proteins but also reveal therapeutic strategies against aggressive forms of myeloid leukemia.
ACCESSION NUMBERS
The coordinates for the ensembles of the 20 conformers of the Msi1 RBD1 in its numb5 bound form was deposited in the RCSB Protein Data Bank, under the accession codes 2RS2
The inter-molecular NOE assignments are deposited in the BMRB data bank under accession number 150218.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6 and Supplementary References [61–63].
FUNDING
Ministry of Education, Science, Sports and Culture of Japan (Grants-in-Aid for Scientific Research: 22121517, 23657072 to M.K., 20570111, 23570146 to T.N. and 21370047 to M.K. and T.N.); Yokohama City University (T2203 to M.K. and T.N.); Advanced Medical Research Center of Yokohama City University [Research and Development Project II (10) and S2210 to M.K. and T.N.]; Yokohama Academic Foundation (to T.N.); Japan Science and Technology (SENTAN) (to M.K.); Naito Foundation (to M.K.); Sumitomo Denko Foundation (to M.K.); Iwatani Foundation (to M.K.). Funding for open access charge: MEXT Grants-in-Aid for Scientific Research (21370047).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Okano H, Imai T, Okabe M. Musashi: a translational regulator of cell fate. J. Cell Sci. 2002;115:1355–1359. doi: 10.1242/jcs.115.7.1355. [DOI] [PubMed] [Google Scholar]
- 2.Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Exp. Cell Res. 2005;306:349–356. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura M, Okano H, Blendy JA, Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:67–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- 4.Yoda A, Sawa H, Okano H. MSI-1, a neural RNA-binding protein, is involved in male mating behaviour in Caenorhabditis elegans. Genes Cells. 2000;5:885–895. doi: 10.1046/j.1365-2443.2000.00378.x. [DOI] [PubMed] [Google Scholar]
- 5.Kaneko J, Chiba C. Immunohistochemical analysis of Musashi-1 expression during retinal regeneration of adult newt. Neurosci. Lett. 2009;450:252–757. doi: 10.1016/j.neulet.2008.11.031. [DOI] [PubMed] [Google Scholar]
- 6.Sakakibara S, Imai T, Hamaguchi K, Okabe M, Aruga J, Nakajima K, Yasutomi D, Nagata T, Kurihara Y, Uesugi S, et al. Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev. Biol. 1996;176:230–242. doi: 10.1006/dbio.1996.0130. [DOI] [PubMed] [Google Scholar]
- 7.Sakakibara S, Okano H. Expression of neural RNA-binding proteins in the postnatal CNS: implications of their roles in neuronal and glial cell development. J. Neurosci. 1997;17:8300–8312. doi: 10.1523/JNEUROSCI.17-21-08300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Good P, Yoda A, Sakakibara S, Yamamoto A, Imai T, Sawa H, Ikeuchi T, Tsuji S, Satoh H, Okano H. The human Musashi homolog 1 (MSI1) gene encoding the homologue of Musashi/Nrp-1, a neural RNA-binding protein putatively expressed in CNS stem cells and neural progenitor cells. Genomics. 1998;52:382–384. doi: 10.1006/geno.1998.5456. [DOI] [PubMed] [Google Scholar]
- 9.Kawahara H, Imai T, Imataka H, Tsujimoto M, Matsumoto K, Okano H. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J. Cell Biol. 2008;181:639–653. doi: 10.1083/jcb.200708004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 11.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Nagata T, Kanno R, Kurihara Y, Uesugi S, Imai T, Sakakibara S, Okano H, Katahira M. Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1. J. Mol. Biol. 1999;287:315–330. doi: 10.1006/jmbi.1999.2596. [DOI] [PubMed] [Google Scholar]
- 13.Miyanoiri Y, Kobayashi H, Imai T, Watanabe M, Nagata T, Uesugi S, Okano H, Katahira M. Origin of higher affinity to RNA of the N-terminal RNA-binding domain than that of the C-terminal one of a mouse neural protein, musashi1, as revealed by comparison of their structures, modes of interaction, surface electrostatic potentials, and backbone dynamics. J. Biol. Chem. 2003;278:41309–41315. doi: 10.1074/jbc.M306210200. [DOI] [PubMed] [Google Scholar]
- 14.Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, Weinmaster G, Nakafuku M, Okano H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Battelli C, Nikopoulos GN, Mitchell JG, Verdi JM. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol. Cell Neurosci. 2006;31:85–96. doi: 10.1016/j.mcn.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Charlesworth A, Wilczynska A, Thampi P, Cox LL, MacNicol AM. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 2006;25:2792–2801. doi: 10.1038/sj.emboj.7601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horisawa K, Imai T, Okano H, Yanagawa H. 3′-Untranslated region of doublecortin mRNA is a binding target of the Musashi1 RNA-binding protein. FEBS Lett. 2009;583:2429–2434. doi: 10.1016/j.febslet.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 18.Spears E, Neufeld KL. Novel double-negative feedback loop between adenomatous polyposis coli and Musashi1 in colon epithelia. J. Biol. Chem. 2011;286:4946–4950. doi: 10.1074/jbc.C110.205922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohyama J, Tokunaga A, Fujita Y, Miyoshi H, Nagai T, Miyawaki A, Nakao K, Matsuzaki Y, Okano H. Visualization of spatiotemporal activation of Notch signaling: live monitoring and significance in neural development. Dev. Biol. 2005;286:311–325. doi: 10.1016/j.ydbio.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Kageyama R, Ohtsuka T, Shimojo H, Imayoshi I. Dynamic regulation of Notch signaling in neural progenitor cells. Curr. Opin. Cell Biol. 2009;21:733–740. doi: 10.1016/j.ceb.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 21.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 22.Miki T, Yasuda SY, Kahn M. Wnt/β-catenin signaling in embryonic stem cell self-renewal and somatic cell reprogramming. Stem Cell Rev. 2011 doi: 10.1007/s12015-011-9275-1. doi:10.1007/s12015-011-9275-1. [DOI] [PubMed] [Google Scholar]
- 23.Devgan V, Mammucari C, Millar SE, Brisken C, Dotto GP. p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation. Genes Dev. 2005;19:1485–1495. doi: 10.1101/gad.341405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- 25.Bai J, Ramos RL, Ackman JB, Thomas AM, Lee RV, LoTurco JJ. RNAi reveals doublecortin is required for radial migration in rat neocortex. Nat. Neurosci. 2003;6:1277–1283. doi: 10.1038/nn1153. [DOI] [PubMed] [Google Scholar]
- 26.de Sousa Abreu R, Sanchez-Diaz PC, Vogel C, Burns SC, Ko D, Burton TL, Vo DT, Chennasamudaram S, Le SY, Shapiro BA. Genomic analyses of musashi1 downstream targets show a strong association with cancer-related processes. J. Biol. Chem. 2009;284:12125–12135. doi: 10.1074/jbc.M809605200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakano A, Kanemura Y, Mori K, Kodama E, Yamamoto A, Sakamoto H, Nakamura Y, Okano H, Yamasaki M, Arita N. Expression of the neural RNA-binding protein Musashi1 in pediatric brain tumors. Pediatr. Neurosurg. 2007;43:279–284. doi: 10.1159/000103307. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez-Diaz PC, Burton TL, Burns SC, Hung JY, Penalva LO. Musashi1 modulates cell proliferation genes in the medulloblastoma cell line Daoy. BMC Cancer. 2008;8:280. doi: 10.1186/1471-2407-8-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toda M, Iizuka Y, Yu W, Imai T, Ikeda E, Yoshida K, Kawase T, Kawakami Y, Okano H, Uyemura K. Expression of the neural RNA-binding protein Musashi1 in human gliomas. Glia. 2001;34:1–7. doi: 10.1002/glia.1034. [DOI] [PubMed] [Google Scholar]
- 30.Kanemura Y, Mori K, Sakakibara S, Fujikawa H, Hayashi H, Nakano A, Matsumoto T, Tamura K, Imai T, Ohnishi T, et al. Musashi1, an evolutionarily conserved neural RNA-binding protein, is a versatile marker of human glioma cells in determining their cellular origin, malignancy, and proliferative activity. Differentiation. 2001;68:141–152. doi: 10.1046/j.1432-0436.2001.680208.x. [DOI] [PubMed] [Google Scholar]
- 31.Ma YH, Mentlein R, Knerlich F, Kruse ML, Mehdorn HM, Held-Feindt J. Expression of stem cell markers in human astrocytomas of different WHO grades. J. Neurooncol. 2008;86:31–45. doi: 10.1007/s11060-007-9439-7. [DOI] [PubMed] [Google Scholar]
- 32.Seigel GM, Hackam AS, Ganguly A, Mandell LM, Gonzalez-Fernandez F. Human embryonic and neuronal stem cell markers in retinoblastoma. Mol. Vis. 2007;13:823–832. [PMC free article] [PubMed] [Google Scholar]
- 33.Sakatani T, Kaneda A, Iacobuzio-Donahue CA, Carter MG, de Boom Witzel S, Okano H, Ko MS, Ohlsson R, Longo DL, Feinberg AP. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307:1976–1978. doi: 10.1126/science.1108080. [DOI] [PubMed] [Google Scholar]
- 34.Ito T, Kwon HY, Zimdahl B, Congdon KL, Blum J, Lento WE, Zhao C, Lagoo A, Gerrard G, Foroni L, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kharas MG, Lengner CJ, Al-Shahrour F, Bullinger L, Ball B, Zaidi S, Morgan K, Tam W, Paktinat M, Okabe R, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 2010;16:903–908. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishimoto Y, Okano H. New insight into cancer therapeutics: induction of differentiation by regulating the Musashi/Numb/Notch pathway. Cell Res. 2010;20:1083–1085. doi: 10.1038/cr.2010.122. [DOI] [PubMed] [Google Scholar]
- 37.Kawahara H, Okada Y, Imai T, Iwanami A, Mischel PS, Okano H. Musashi1 cooperates in abnormal cell lineage protein 28 (Lin28)-mediated let-7 family microRNA biogenesis in early neural differentiation. J. Biol. Chem. 2011;286:16121–16130. doi: 10.1074/jbc.M110.199166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Werner MH, Gupta V, Lambert LJ, Nagata T. Uniform 13C/15N-labeling of DNA by tandem repeat amplification. Methods Enzymol. 2001;338:283–304. doi: 10.1016/s0076-6879(02)38225-9. [DOI] [PubMed] [Google Scholar]
- 39.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi N, Iwahara J, Koshiba S, Tomizawa T, Tochio N, Güntert P, Kigawa T, Yokoyama S. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J. Biomol. NMR. 2007;39:31–52. doi: 10.1007/s10858-007-9175-5. [DOI] [PubMed] [Google Scholar]
- 41.Clore GM, Gronenborn AM. Determining the structures of large proteins and protein complexes by NMR. Trends Biotechnol. 1998;16:22–34. doi: 10.1016/S0167-7799(97)01135-9. [DOI] [PubMed] [Google Scholar]
- 42.Bax A. Multidimensional nuclear magnetic resonance methods for protein studies. Curr. Opin. Struct. Biol. 1994;4:738–744. [Google Scholar]
- 43.Cavanagh J, Fairbrother WJ, Palmer AG, III, Skelton NJ. Protein NMR Spectroscopy, Principles and Practice. San Diego, CA: Academic Press; 1996. [Google Scholar]
- 44.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 45.Powers R, Garrett DS, March CJ, Frieden EA, Gronenborn AM, Clore GM. The high-resolution, three-dimensional solution structure of human interleukin-4 determined by multidimensional heteronuclear magnetic resonance spectroscopy. Biochemistry. 1993;32:6744–6762. doi: 10.1021/bi00077a030. [DOI] [PubMed] [Google Scholar]
- 46.Hennig M, Williamson JR, Brodsky AS, Battiste JL. Recent advances in RNA structure determination by NMR. Curr. Protoc. Nucleic Acid Chem. 2001;Chapter 7:Unit 7.7. doi: 10.1002/0471142700.nc0707s02. [DOI] [PubMed] [Google Scholar]
- 47.Tsuda K, Someya T, Kuwasako K, Takahashi M, He F, Unzai S, Inoue M, Harada T, Watanabe S, Terada T, et al. Structural basis for the dual RNA-recognition modes of human Tra2-beta RRM. Nucleic Acids Res. 2011;39:1538–1553. doi: 10.1093/nar/gkq854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 49.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 50.Clore GM, Tang C, Iwahara J. Elucidating transient macromolecular interactions using paramagnetic relaxation enhancement. Curr. Opin. Struct. Biol. 2007;17:603–616. doi: 10.1016/j.sbi.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 52.Case DA, Cheatham TE, III, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S, Allain FH. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perez-Canadillas JM. Grabbing the message: structural basis of mRNA 3′UTR recognition by Hrp1. EMBO J. 2006;25:3167–3178. doi: 10.1038/sj.emboj.7601190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mazza C, Segref A, Mattaj IW, Cusack S. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 2002;21:5548–5557. doi: 10.1093/emboj/cdf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myere JC, Shamoo Y. Human UP1 as a Model for Understanding Purine Recognition in the Family of Proteins Containing the RNA Recognition Motif (RRM) J. Mol. Biol. 2004;342:743–756. doi: 10.1016/j.jmb.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 57.Enokizono Y, Konishi Y, Nagata K, Ouhashi K, Uesugi S, Ishikawa F, Katahira M. Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J. Biol. Chem. 2005;280:18862–18870. doi: 10.1074/jbc.M411822200. [DOI] [PubMed] [Google Scholar]
- 58.Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol. Cell. 2006;23:49–59. doi: 10.1016/j.molcel.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu RM. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sakakibara S, Nakamura Y, Yoshida T, Shibata S, Koike M, Takano H, Ueda S, Uchiyama Y, Noda T, Okano H. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc. Natl Acad. Sci. USA. 2002;99:15194–15199. doi: 10.1073/pnas.232087499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 62.Iwahara J, Tang C, Clore GM. Practical aspects of (1)H transverse paramagnetic relaxation enhancement measurements on macromolecules. J. Magn. Reson. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang C, Schwieters CD, Clore GM. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature. 2007;449:1078–1082. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.