Abstract
Characterization of small non-coding ribonucleic acids (sRNA) among the large volume of data generated by high-throughput RNA-seq or tiling microarray analyses remains a challenge. Thus, there is still a need for accurate in silico prediction methods to identify sRNAs within a given bacterial species. After years of effort, dedicated software were developed based on comparative genomic analyses or mathematical/statistical models. Although these genomic analyses enabled sRNAs in intergenic regions to be efficiently identified, they all failed to predict antisense sRNA genes (asRNA), i.e. RNA genes located on the DNA strand complementary to that which encodes the protein. The statistical models enabled any genomic region to be analyzed theorically but not efficiently. We present a new model for in silico identification of sRNA and asRNA candidates within an entire bacterial genome. This model was successfully used to analyze the Gram-negative Escherichia coli and Gram-positive Streptococcus agalactiae. In both bacteria, numerous asRNAs are transcribed from the complementary strand of genes located in pathogenicity islands, strongly suggesting that these asRNAs are regulators of the virulence expression. In particular, we characterized an asRNA that acted as an enhancer-like regulator of the type 1 fimbriae production involved in the virulence of extra-intestinal pathogenic E. coli.
INTRODUCTION
The number of metabolic pathways in eubacteria known to be controlled by regulatory small RNAs (sRNAs) is growing. These pathways often regulate gene expression post-transcriptionally by modulating mRNA translation and/or mRNA stability through antisense mechanisms involving base pairing interactions with dedicated mRNA targets (1). Mechanistic studies revealed that sRNAs also modulate protein activity by sequestering them to modify their structures (2) or control the quality of the protein synthesis (3). Most of the characterized bacterial sRNA genes have been found in the intergenic regions (IGRs) of the core genome; in mobile genetic elements, such as insertion sequences, plasmids and phages (4); or in pathogenicity islands (PAI) (5,6). Previous studies have shown that sRNAs can regulate both bacterial metabolism as well as pathogenicity (7).
Recent data from high-throughput sequencing of the transcriptome (RNA-seq) and tiling microarray analyses have demonstrated the expression of many complementary sRNA/mRNA transcript pairs in Listeria monocytogenes (8), Helicobacter pylori (9) and Escherichia coli (10). These results highlight that the number of sRNA genes located at the same genomic locus as protein coding genes (CDS), but on the DNA opposite strand, was underestimated. The sRNA molecules encoded by these genes are referred to antisense RNAs (asRNA) or naturally occurring RNAs. It was deduced from these studies that the diversity of sRNAs is likely to be much greater than expected, most particularly for asRNA genes, which in turn raises a plethora of questions about their functions (11). Few recent studies have indicated that asRNA genes encoding molecules that are partially (12) or fully complementary to a CDS (13) have a physiological role but the contribution of asRNAs to regulation of metabolism and pathogenicity has not been studied extensively. RNA-seq and tiling microarrays represent significant technical advances for the identification of sRNAs because the whole transcriptome could be analyzed. However, both techniques have strong limitations, particularly in terms of experimental costs and the cumbersome nature of the data analysis and experimental procedure, which includes the crucial choice of relevant strains and growth conditions. Thus, in silico methods remain of great interest for screening of a large number of genomes without high cost and time consuming tasks.
Many methods for in silico identification of sRNAs exist, but only a few algorithms can efficiently predict sRNA gene loci in the full bacterial genome sequence (14). Different in silico methods based on comparative genomics (15–19), statistics/probability analyses (20–24), and RNA secondary structure analyses (16,25) have been developed but they vary considerably in efficacy. The most recent algorithms for identification of sRNA genes are combinations of several pre-existing independent methods, for increasing their sensitivity and predictive potentials. However, most of these sRNA gene finders were first designed for and mainly applied to Gram-negative bacteria and they require significant adjustments to analyze genomes of unrelated bacteria. Most of the methods based on comparative genomics to identify small (<500 nt) conserved gene structures, including promoter sequences, were highly bacterial order dependent (15). Indeed, transcription promoters are highly diversified and DNA recognition consensus sequences among bacterial species were often divergent or not known. Only Rho-independent terminators (RITs) identification seemed to be a valuable search for building an almost general sRNA gene finder and can constitute the basis of a gene signature research algorithm. Restriction of the computational searches for novel sRNA genes located in the IGRs constitutes another important limitation of the current algorithms. Studies using machine learning algorithms [i.e. stochastic context free grammar (16), neural networks (20), boosted genetic programming (22), gapped Markov model (23) and support vector machine (24) methods] enabled the detection of new sRNAs in protein-coding regions but the number of putative asRNAs identified are variable between studies and some of these studies lacked of in vivo validation. Comparison of the data obtained by the application of these mathematical models with those recently obtained by RNA-seq or tilling microarray analyses demonstrated that the efficiencies of these in silico analyses need improvements. The defect of these methods to identify most asRNAs partially or fully overlapping protein-coding genes, probably related to their low efficiency to discriminate sequence conservations due to the presence of a protein coding sequence from conservations due to the presence of an asRNA gene. While these strategies are interesting, their limitations are inherent to RNA secondary structure diversities that impaired the efficiency of the co-variance model, especially for unstructured sRNAs (16). Despite all efforts made, current methods could be perfected and a number of strategies remain to be tested.
We report here the development and validation of a new in silico strategy, that successfully identifies known and new sRNA genes based on the analysis of the complete genome sequence of Gram-negative and Gram-positive bacteria, including those located in intergenic and CDS regions. Improvement of current RIT searches and covariation identification by our new algorithms enhanced sRNAs discovery. For example, analysis of the genomes of extra-intestinal pathogenic E. coli (ExPEC) and Streptococcus agalactiae, two opportunistic pathogens in which gene regulation undoubtly plays an important role in pathogenesis, led to the identification of numerous new sRNAs, including asRNA genes specific for the ExPEC strains or the Group B Streptococci. Transcription analysis of sRNAs located close to pathogenicity-associated gene clusters and functional characterization of two asRNAs suggested that they might control the expression of pathogenicity-related genes in both bacteria which confirmed the efficiency of our new method.
MATERIALS AND METHODS
Genome and pathogenicity island sequences
All genome sequences of E. coli and S. agalactiae were obtained from the Genbank database (http://www.ncbi.nlm.nih.gov/genbank/). The PAI-IAL862 of E. coli AL862 strain was sequenced at the Pasteur Institute and was deposited to Genbank under accession number GQ497943.
Identification of RITs
For Gram-negative bacteria, RITs were predicted with the RNAMotif program (26) by a slightly modified version of the previously described method (27). We used the perfect stem loop structure template as described, except that we permitted no more than one mismatch within the stem structure. We also used the same scoring formula, excepted that the ΔG037 of the RNA:DNA hybrid duplex of the poly-uracil tail and its complementary genomic sequence were scored with Melting4 software, using nearest neighbor thermodynamic parameters (28). All candidates with a score greater than −4.0 kcal/mol were removed. For Gram-positive bacteria, Rho-independent terminators were predicted by TransTermHP (29).
Bacterial strains and growth conditions
All E. coli strains (Table 1) were cultured in Luria Bertani (LB) or M9 supplemented with 0.4% of sodium pyruvate media. S. agalactiae NEM316 was grown in Todd Hewitt (TH) or RPMI1640 medium supplemented with 0.4% glucose and 5% 1M HEPES buffer. Antibiotics for plasmid selection were used at the following concentrations: for E. coli, carbenicillin, 100 µg/ml, kanamycin, 50 µg/ml, and chloramphenicol, 12.5 µg/ml; for S. agalactiae, erythromycin, 5 µg/ml. The 536 Δhfq::KmFRT strain was constructed by the allelic exchange recombination protocol using the thermosensitive plasmid pKOBEG-Apra (36). The 500 nucleotides adjacent to the 5′ and 3′ regions of the hfq gene were amplify and assembled with the kanamycin FRT flanked cassette from the pKD4 plasmid by PCR prior to strain transformation (38).
Table 1.
Strains and plasmids used in this study
| Name | Description | Genotype/Resistancea | Reference |
|---|---|---|---|
| Strains | |||
| E. coli AL862 | Sepsis-associated ExPEC isolate | afa8+ | (30) |
| E. coli 536 | Pyelonephritis-associated ExPEC isolate (O6:K15:H31) | pap+, fim+ | (31) |
| E. coli 536 Δfim::cat | Deletion of the full fim gene cluster | CmR | (32) |
| E. coli 536 Δhfq::KmFRT | Allelic exchange of the hfq gene with kanamycin FRT cassette | KmR | This study |
| S. agalactiae NEM316 | Human septicaemia isolate | (33) | |
| E. coli TOP10 | Laboratory strain | fim- | (34) |
| E. coli TOP10Δhfq::KmFRT | Hfq-deficient strain JVS-2001 | KmR | (34) |
| E. coli TOP10Δhfq::FRT | Hfq-deficient strain JVS-2001 with the FRT flanked kanamycin resistance cassette removed by action of the FLP flipase from pCP20 plasmid | KmS | This study |
| Plasmids | |||
| pCP20 | Thermosensitive plasmid expressing the flp flippase gene | CbR, CmR | (35) |
| pKOBEG-Apra | Thermosensitive recombination plasmid used for allelic exchange | pSC101ts, ApraR | (36) |
| pZE21-gfp | gfp gene under the control of the PLtetO-1 promoter | ColE1, KmR | (37) |
| pZE2R-gfp | Replacement of the PLtetO-1 promoter from pZE21-gfp by the Pλ constitutive promoter | ColE1, KmR | (37) |
| pZE21-null | pZE1-gfp derivative expressing a non sense sRNA | ColE1, KmR | This study |
| pZE2R-null | pZE2R-gfp derivative expressing a non sense sRNA | ColE1, KmR | This study |
| pZE2R-fimR | Insertion of fimR gene into the EcoRI/XbaI sites of the pZE2R-gfp plasmid | ColE1, KmR | This study |
| pZE21-antifimR | Insertion of fimR antisense sequence into the EcoRI/XbaI sites of the pZE21-gfp plasmid | ColE1, KmR | This study |
| pZE2R-SQ18 | Insertion of SQ18 gene into the EcoRI/XbaI sites of the pZE2R-gfp plasmid | ColE1, KmR | This study |
| pXG-0 | Luciferase-expressing plasmid | pSC101*, CmR | (34) |
| pXG-10 | Translational fusion of lacZ and gfp genes | pSC101*, CmR | (34) |
| pXGfimD::gfp | pXG10 derivative with a fimD::gfp translational fusion | pSC101*, CmR | This study |
| pXGgbs0031::gfp | pXG10 derivative with a gbs0031::gfp translational fusion | pSC101*, CmR | This study |
| pTCV-erm-ΩPtet | Shuttle low-copy vector to analyze regulatory elements in Gram-positive bacteria under the control of the constitutive promoter Ptet | pAMβ1, ErmR | S. Dramsi |
| pTCV-SQ18 | Insertion of the SQ18 sRNA gene into the BamHI/PstI sites of pTCVerm-Ptet plasmid. | pAMβ1, ErmR | This study |
| pTCV-SQ485 | Insertion of the SQ485 sRNA gene into the BamHI/PstI sites of pTCVerm-Ptet plasmid. | pAMβ1, ErmR | This study |
| pTCV-SQ893 | Insertion of the SQ893 sRNA gene into the BamHI/PstI sites of pTCVerm-Ptet plasmid. | pAMβ1, ErmR | This study |
aApra, Cb, Cm, Erm, Km were resistance to apramycin, carbenicillin, chloramphenicol, erythromycin and kanamycin, respectively.
RNA sample preparation
All cultures were established with a 1/50 dilution of an overnight culture, incubated at 37°C under shaking at 140 rpm. Samples were prepared from cultures stopped during the exponential phase of growth OD600 of 0.6 for E. coli or OD600 of 0.4 for S. agalactiae, or stationary phase after 24 h for both bacteria. Total RNAs was isolated from E. coli strains with Trizol (Invitrogen), used according to the manufacturer's protocol except that the bacteria were harvested by centrifugation at 4000g for 5 min at room temperature, to prevent cold shock stress. Total RNAs was extracted from S. agalactiae with hot phenol as described (Pichon 2005 5). RNA samples were treated twice, with 30 units of DNase I (Amersham) for 90 min at 37°C and extracted by phenol/chloroform treatment and precipitated in ethanol. The RNA was re-suspended in DEPC-treated water and checked for putative degradations on 2% agarose gel. Genomic DNA contaminations were analyzed by PCR amplification of the 5S RNA using the 5S.Fw and 5S.RT primers.
RACE experiments
The determination of the 5′-end of sRNAs were done as previously described (39).
Nested and classic RT–PCR
Chimeric DNAs (cDNA) were synthesized from 5 µg of heat-denatured total RNAs with 200 units of Superscript III reverse transcriptase enzyme (Invitrogen). For analyses of sRNA expression, the reaction was performed at 55°C for 1 h with 2 pmol of gene specific primer (Sigma Proligo) (Supplementary Table S1) to maintain stringent conditions and synthesized strand specific products. For mRNA expression analysis, the reaction was performed at 42°C for 1 h with 200 ng of random hexamer according to supplier's protocol. Reactions were inactivated by heating at 70°C for 10 min. The cDNA was amplified by PCR done with 0.4 units of Taq polymerase (QBiogen), 100 nM of each primer pair (gene.RT and gene.Fw or gene.Nested and gene.Fw for nested PCR), 200 µM dNTP and 2 µl of the RT reaction. The thermal cycling were 94°C, 3 min, followed by 40 cycles of 94°C, 30 s; 55°C, 30 s; and 72°C for 30 s. and final extension of 72°C, 7 min. PCR products were analyzed by electrophoresis in 4% ethidium bromide-stained agarose gels.
Northern blot hybridization
Northern blot membranes were prepared and hybridization was carried out as described (5). Briefly, RNA samples were separated by urea denaturating polyacrylamide gel electrophoresis and transferred to Zeta probe GT membranes (Biorad). Membranes were hybridized with 32P 5′-end-labeled oligonucleotides in ExpressHyb (Clontech) and scanned with a PharosFX system (Biorad).
Analysis of small RNA and mRNA interaction
The pZE2R-null and pZE21-null plasmids were constructed by digesting the pZE2R-gfp and pZE21-gfp plasmids with EcoRI (Invitrogen) and XbaI (Roche). The DNA fragments containing the kanamycin resistance gene and the origin of replication were separated by gel electrophoresis and extracted from the agarose with the Qiagen gel extraction kit. We treated 200 ng of the two cleaved plasmid DNA fragments with Klenow enzyme (NEB) for 1 h at room temperature, followed by re-circularization with T4 DNA ligase (Fermentas) and transformed in the TOP10 strain.
For expression of the FimR and SQ18 sRNAs in E. coli, we amplified the fimR gene from E. coli 536 and the SQ18 gene from S. agalactiae NEM316 genomic DNAs by PCR using Taq DNA polymerase (MPbio) with cl.fimR.EcoRI and cl.fimR.XbaI or cl.SQ18.EcoRI and cl.SQ18.XbaI primers, respectively. The two PCR products were inserted to pCRII-TOPO plasmid (Invitrogen). The pCRII-fimR or pCRII-SQ18 plasmids were digested with EcoRI and XbaI. The DNA band containing the sRNA gene was purified from the gel and ligated with pZE2R DNA digested with EcoRI and XbaI, with T4 DNA ligase. The ligation products were transformed in the TOP10 strain, generating the pZE2R-fimR and pZE2R-SQ18 plasmids. The pZE21-antifimR plasmid was constructed in the same way as pZE2R-fimR, except that we used the cl.antifimR.EcoRI and cl.antifimR.XbaI primers for PCR.
The fimD::gfp and gbs0031::gfp fusion genes were expressed by inserted the fimD and gbs0031 CDSs depleted of stop codons into the pXG10 plasmid as described (34). The DNA fragments containing the fimD and gbs0031 CDSs were amplified with LA Taq (Takara) with fimD.NheI and fimD.Mph1103I or gbs0031.NheI and gbs0031.Mph1103I primers, respectively. The other steps and Western blotting were done as described (34).
For expression of the SQ18, SQ485, SQ893 sRNAs in S. agalactiae, we amplified the three sRNA genes from S. agalactiae NEM316 genomic DNAs by PCR using Taq DNA polymerase (MPbio) with cl.SQ18.BamHI and cl.SQ18.PstI or cl.SQ485.BamHI and cl.SQ485.PstI or cl.SQ893.BamHI and cl.SQ893.PstI couple of primers, respectively. The PCR products were first cloned into the pCRII-TOPO plasmid (Invitrogen) and recloned into the BamHI/PstI sites of the shuttle vector pTCV-erm-ΩPtet plasmid, giving the pTCV-SQ18, pTCV-SQ485, pTCV-SQ893 expression plasmids. These vectors were introduced by electroporation in S. agalactiae NEM316.
Analysis of expression by quantitative real-time PCR
Total RNAs were reverse-transcribed as described in the section on RT–PCR, except that 10 µg of total RNA were used. All primers were designed with Primer3 (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). We determined mRNA and 5 S RNA levels from cDNAs synthesized with random primers. The sRNA levels were analyzed with cDNAs synthesized with specific primers. All cDNA samples were analyzed using iQ SYBR green supermix (BioRad) according to manufacturer protocol and were run on a MyiQ thermal cycler (BioRad) with the following thermal cycling conditions, 95°C 5 min, 40 cycles of 95°C, 30 s; 60°C for 60 s. All experiments were carried out with at least two duplicate RNA samples. The 5S rRNA was used as reference and the gene and relative level of expression between samples were calculated by the ΔΔCt method (40).
Yeast agglutination, motility and biofilm assays
All assays were carried out with E. coli strains cultured in LB broth and incubated overnight at 37°C without shaking. The culture medium was eliminated by centrifugation and bacteria were washed once with 1X PBS. Yeast agglutination assays and motility tests were performed as described (41). Biofilm formation assays were conducted in polypropylene microtiter plates. Bacteria were grown statically in LB and M63 glucose media for 48 h, and biofilms were visualized by crystal violet staining as described (42).
RESULTS
Design and validation of an sRNA genefinder based on the identification of orphan RITs
We hypothesized that the core prediction system for a versatile sRNA genefinder algorithm that predicted preferentially non-coding sRNAs should combine several functionalities. First, it should predict the signatures composed of recognition sites for sRNA-binding proteins, for example RIT. Second, it should be able to inspect the flanking nucleic acid sequences using comparative genomic and RNA structure predictions plus a scoring method based on covariation analysis, to provide a strong phylogenetic evidence for the existence of RNA stems (2,14).
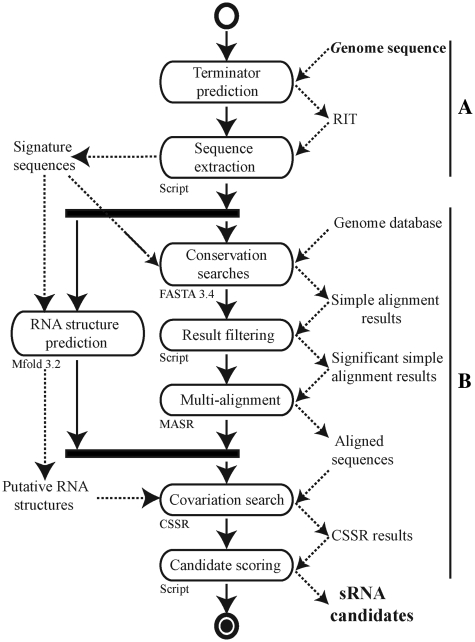
The RIT site, which is often involved in the termination processes of sRNA genes in E. coli (∼70%) and in other bacteria such as Staphylococcus aureus (5), was used as a starting point for our sRNA search model (Figure 1). By applying it to the genome of the extensively studied E. coli MG1655, we detected 16 959 putative terminators with a ΔG037 ≤ −4 kcal/mol score. The 1504 RIT located close to the stop codon (from −25 to +60 nt) on the same DNA strand as a CDS were automatically removed from the data set. The remaining putative terminators and the 200-nt upstream sequences were considered as sRNA candidate signatures. Their sequence conservation was analyzed using FASTA 3.4 software (43) against 44 complete genomes of Enterobacteria (Genbank database, 24/07/2007). Insignificant hits with an e-value >0.0001 were excluded. MASR software was used to transform FASTA pairwise alignments into multi-alignment. RNA structure predictions of sRNA signature candidates were done with the Mfold 3.2 program (44). The CSSR program, by combining MASR multiple alignments and Mfold predictions, detects the RNA structure conservations and presence of covariations (see supplementary data for a description of MASR and CSSRTo identify the most probable sRNA genes, candidates were ranked according to their RIT scores (Supplementary Table S2).
Figure 1.
UML activity diagram for our in silico sRNA prediction model. (A) The first part of this process involves the prediction of sRNA protein-binding sites (RIT prediction in this study) and extraction of the flanking sequences. (B) Core software for sRNA analysis and discovery based on a combination of comparative genomics, RNA prediction and covariation analysis.
Our model identified sRNA candidates associated with an RIT within CDSs. However, the large number of candidates identified in E. coli MG1655 (>2000 antisense and >3000 sense sRNA candidates) suggested that these included high number of false-positives. We therefore filtered-out sense and antisense candidates in which the ΔG°37 score of the RIT was less that −8 kcal/mol. Finally, we scored sRNA candidates from E. coli MG1655 on the basis of their RIT, which were weighted by the number of covariation pairs found by CSSR. Threshold values of −4 kcal/mol or −8 kcal/mol for the RIT score and a requirement for at least two covariations, including one in the RIT stem, led to the prediction of 1867 sRNA candidates that could be classified into eight different groups according to their position relative to adjacent CDSs (Table 2). In order to maximize the prediction of non-coding sRNAs, small CDSs were tentatively predicted using Glimmer2 software (45).
Table 2.
Summary of sRNA candidates identified in silico
| Strain | Disease | IGR | asRNA | 5′ asRNA | 3′ asRNA | 5′ & 3′ asRNA | 5′ UTR | 3′ UTR | sense RNA |
|---|---|---|---|---|---|---|---|---|---|
| Escherichia coli | |||||||||
| MG1655 | L. S. | 195 | 452 | 74 | 142 | 73 | 89 | 199 | 643 |
| UTI89 | Cys. | 199 | 398 | 66 | 95 | 77 | 96 | 170 | 527 |
| 536 | Pyl. | 191 | 388 | 66 | 107 | 54 | 73 | 140 | 496 |
| AL862 | Sep. | 9 | 6 | 2 | 2 | 0 | 3 | 3 | 4 |
| S88 | Men. | 212 | 430 | 63 | 103 | 85 | 90 | 154 | 532 |
| Streptococcus agalactiae | |||||||||
| NEM316 | Sep. | 41 | 63 | 12 | 24 | 6 | 5 | 21 | 25 |
IGR, intergenic region; asRNA, sRNA antisense to a CDS; 5′ asRNA, antisense to the 5′-end of a CDS ; 3′ asRNA, antisense to the 3′-end of a CDS; 5′ UTR, 5′ untranslated region of a CDS; 3′ UTR, 3′ untranslated region of a CDS. For classification of the sRNA candidates into one of these categories, the first nucleotide of the RIT was used as the position reference of the candidate. This nucleotide had to be on the opposite DNA strand, between nucleotides −50 nt to +15 nt around the ATG codon (5′ asRNA), from position +15 nt with respect to the ATG codon to position –50 nt near the stop codon (asRNA) or from –50 nt to +15 nt around the stop codon (3′ asRNA). When candidates were on the same DNA strand as the CDS, the window around the first RIT nucleotide was < –100 nt before the ATG codon (5′ UTR), < +200 nt after the stop codon (3′ UTR) and from +50 nt after the ATG to –50 nt before the stop codon (seRNA). All candidates outside a CDS not included in a previous category are referred to IGR candidates. All candidates had to have a RIT with a score of ΔG°37 < -4 kcal/mol and at least two covariations had to be present in the RNA structure including the stem of the RIT. For asRNA and seRNA candidates, ΔG°37 had to be below -8 kcal/mol. L. S., laboratory strain; Cys., cystitis; Pyl., pyelonephritis; Sep., sepsis; Men., meningitis. Only the PAI-IAL862 sequence of the AL862 strain was analyzed.
Efficiency of the in silico model
We first tested whether the use of covariations efficiently selected true positive sRNAs and rejected true negative candidates by using our in silico model to analyze the 101 known sRNAs from the E. coli MG1655 strain (Supplementary Table S4), which included 18 asRNAs. All the sRNA sequences were submitted directly to the core software by bypassing the RIT predictions (Figure 1B). The core software identified 77 (92.7%) of the sRNAs located in the IGR and 16 (88.9%) of the asRNAs as putative candidates. The statistical significance of the covariation identified by the Covation Search in Small RNAs software (CSSR) was evaluated by shuffling the 101 sRNA multi-alignments using the Altschul and Erikson shuffle algorithm (25). In these conditions, the total number of covariations found by CSSR in sRNAs was 73.7% lower than for the unshuffled data set, suggesting that most of the predicted covariations were statistically significant.
We assessed the efficiency of our in silico model as an sRNA genefinder by its ability to re-predict known bona fide sRNAs with RIT in six complete genome sequences (Table 3). Globally, our in silico model detected known sRNAs with efficiencies of 70.1% and 71.3% for IGR-located sRNAs and asRNAs, respectively. In the case of E. coli MG1655, among the sRNAs with a RIT that were not identified, rybB and rydC genes have a RIT with a loop size that exceeds the maximum length tolerated by our method. Other candidates among those not identified the rdlA, rdlC, sokA, sokC, sokE and sokX were all cis-regulatory sRNAs. We suggested that putative structural constraints were applied to these sRNAs leading to the use of atypical RIT. The E. coli MG1655 strain transcriptome was recently analyzed in an RNA-seq experiment and 5 out of the 10 newly confirmed sRNAs were re-predicted by our in silico analysis (47). Confirmed sRNAs from published RNA-seq analysis of S. aureus N315 were compared to our data and 62.5% of the transcribed sRNAs (with and without RIT) were re-predicted (48). Given that our in silico model was able to predict candidates irrespective of their expression, we were able to re-identify four known sRNAs (RNAIII, Sau-02, Sau-30, RsaE) that were absent from RNA-seq data (48).
Table 3.
Efficiency of the in silico process for predicting previously known sRNAs in six bacterial species
| Gram | Strains | Total known sRNAs | sRNA genes in IGR |
asRNA genes in CDS |
||
|---|---|---|---|---|---|---|
| Known sRNA with RIT | Success (%) | Known asRNA with RITa | Success (%)b | |||
| − | E. coli MG1655 | 101 | 60 | 86.7 | 5 | 60 |
| − | S. typhimurium LT2 | 79 | 51 | 70.6 | 0 | NA |
| − | V. cholerae O1 | 40 | 31 | 90.4 | 9 | 55.5 |
| − | P. aeruginosa PAO1 | 24 | 24 | 66.7 | 0 | NA |
| + | S. aureus N315 | 55 | 38 | 76.3 | 1 | 100 |
| + | L. monocytogenes EGD-e | 50 | 27 | 29.6 | 10 | 70 |
aThe RITs of the published asRNA genes were not characterized by authors.
The efficiency of sRNAs prediction was calculated from data for bona fide sRNA genes. Only sRNAs that had been experimentally validated by Northern blots, 5′ RACE and RT–PCR were taken into account. We excluded unconfirmed sRNAs from RNA-seq or tiling microarray data and 5′ or 3′ UTRs from mRNAs.
bNA, Not Applicable.
Screening for new sRNAs from ExPEC Escherichia coli isolates
Escherichia coli is a species encompassing a broad variety of commensal and pathogenic strains that have diverged due to a high rate of genetic exchange (49). Using an exhaustive and hand-curated database of sRNA genes found in the genera Escherichia, we recently updated the annotation of known sRNAs in the genome of the MG1655 strain (Supplementary Table S4). We also reported that these genes were structurally well conserved in the genome of 6 pathogenic and commensal strains recently sequenced, although their copy number may vary (49). These data suggested that unidentified sRNAs that are absent from the MG1655 strain might be involved in regulatory pathways specific to pathogenic isolates.
We thus focused our searches for sRNAs on ExPEC strains, a group of major human pathogens responsible for urinary tract infections, meningitis, sepsis, etc. (50). Despite extensive studies, no gene or pool of genes specifically linked to extra-intestinal virulence has been identified in these strains. This strongly suggests that virulence results from multi-factorial processes depending on the expression of both core-genome and strain-specific genes (49). We thus investigated the possible role of ExPEC specific sRNAs in virulence control by applying our in silico model to the entire genomes of three clinical isolates (UTI89, 536 and S88) which are associated with cystitis, pyelonephritis and newborn meningitis, respectively (49,51,52). We also analyzed the sequence of the tRNAPhe inserted PAI from AL862 strain (PAI-IAL862), a sepsis isolate (30).
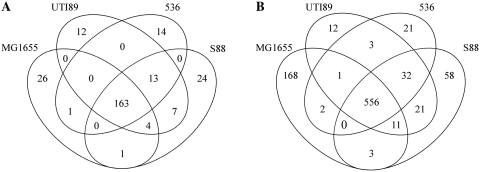
The RIT-associated sRNA candidates from the whole genomes or PAI-IAL862 sequences were collected with our model and classified according to their genomic coordinates (Supplementary Table S3), as summarized in Table 2. In each genome, we identified more than 1500 sRNA candidate genes. The number of putative sRNA genes located in the IGRs did not exceed 200 (∼10% of all candidates), a finding consistent with other in silico searches (19). Most of these candidate genes were located in the core genome (∼81.8% on average) rather than in PAIs (data not shown) suggesting that they may regulate the general cell metabolism (Figure 2A). We detected numerous asRNA among sRNA candidates (∼40% of all candidates), partially or fully antisense to a CDS, that were dispersed throughout the genome sequences, including their PAIs (data not shown). The partially asRNA candidates (∼15% of all candidates) overlaps either the upstream or downstream regions of a CDS, suggesting that they control the translation and/or stability of the complementary mRNA. In the case of the 59 000 bp PAI-IAL862 sequence, 29 sRNA gene candidates were predicted, 10 (34.5%) being asRNAs, a percentage similar to that found in other ExPEC genomes. As shown for MG1655 analysis, many candidates were found in sense orientation within CDSs (∼34% of all candidates).
Figure 2.
Comparative analysis of sRNAs identified by our in silico model based on sequence conservation among ExPEC strains. Venn diagram representations of the number of sRNA predicted in IGR (A) and of asRNAs predicted in CDSs (B).
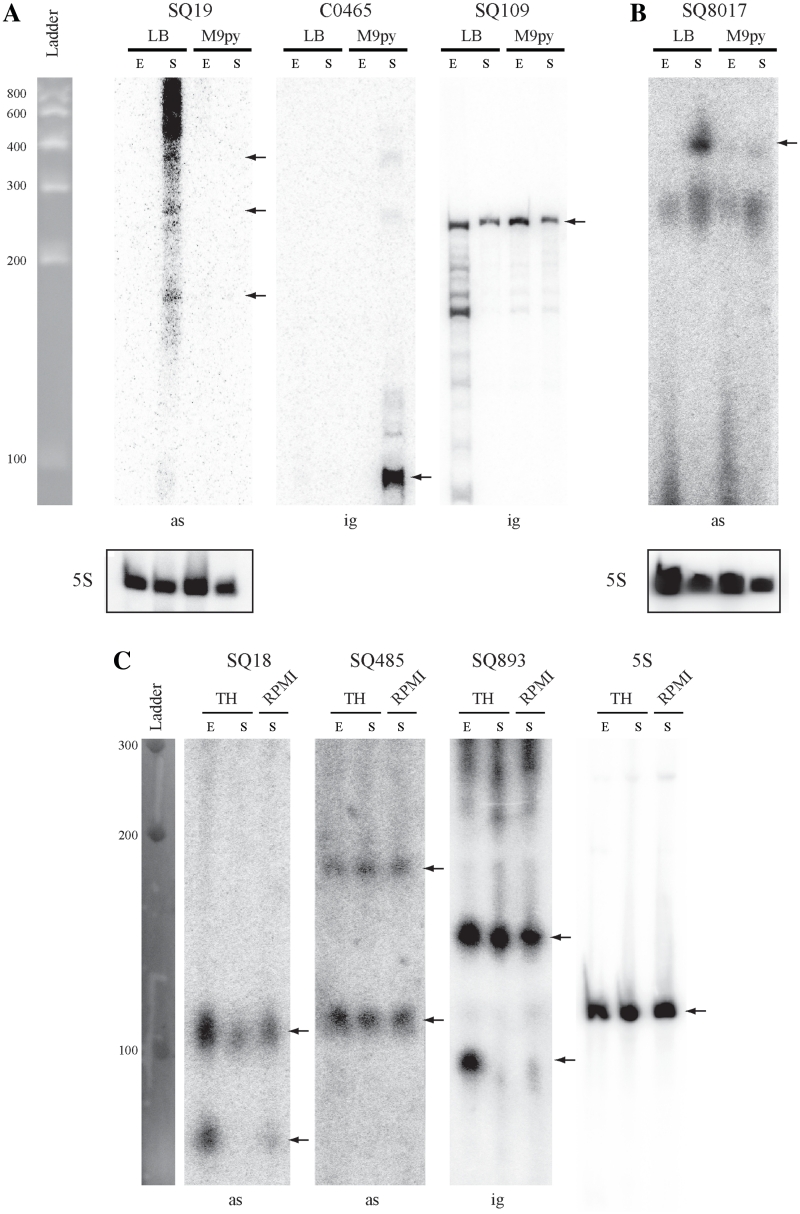
Given the large number of sRNA candidates, we focused on those genetically associated with clusters of genes known to be involved in extra-intestinal virulence, in particular the ExPEC-specific PAI-IAL862 (E. coli AL862), PAI-II536 (E. coli 536) and the fim gene cluster encoding type 1 fimbriae (E. coli 536). Screening by RT–PCR analysis revealed that six out of the seven sRNA candidates from PAI-IAL862 were transcribed: one candidate was located in an IGR and five were asRNAs (Supplementary Figure S1A). We evaluated the sensitivity of our RT–PCR method by carrying out hemi-nested RT–PCR experiments (53) (Supplementary Figure S2A). This analysis did not confirm expression of the SQ24 and SQ27 asRNAs, both targeting a putative transposase CDSs (Supplementary Figure S2B). Expression and size of the two of four remaining sRNAs was analyzed by Northern blot due to their co-localization with pathogenic factor genes (Figure 3A). The same transcription analysis was carried out for 10 sRNA candidates from the genome of the E. coli 536 strain, including nine candidates located in PAI-II536 sequence and 1 in the fim gene cluster: two candidates were located in IGRs and 8 were asRNAs. All candidates were expressed in our growth conditions as shown by our expression screening by RT–PCR (Supplementary Figure S1B) associated with hemi-nested RT–PCR performed to confirm the specificity of RT–PCR reactions for all sRNAs (data not shown). Northern blot analyses of several candidates were done to confirm size and expression of selected relevant sRNA (Figure 3B).
Figure 3.
Northern blot analysis of some sRNAs from E. coli AL862, 536 and S. agalactiae NEM316 strains. Expression analysis of 7 sRNA candidates co-localized with virulence factors (see Table 4) and identified in (A) E. coli AL862, (B) E. coli 536 and (C) S. agalactiae NEM316 strains. Expression was analyzed in two phases of growth (E, exponential; S, late stationary) in LB and M9 + 0.4% pyruvate (M9py) media for E. coli or TH and RPMI1640 + 0.4% glucose media for S. agalactiae. Expression of the constitutively transcribed 5S ribosomal gene was used as loading control. The C0465 sRNA which is expressed only in early stationary phase in E. coli MG1655 strain was used as a negative expression control (46). Notes, ig, sRNA gene located in the IGR; as, sRNA gene located a position antisense to a CDS (asRNA). Black arrows indicated hybridized sRNA molecules.
Comparative sequence analysis by our in silico model showed that all but one of the 14 validated sRNAs of E. coli AL862 and 536 were frequently found in the genome of sequenced ExPEC isolates but not in other E. coli pathotype strains. The remaining SQ8017 sRNA was located in the fim gene cluster encoding the virulence-associate type 1 fimbriae present in almost all commensal and pathogenic strains. Most of the new sRNA genes identified in this study are asRNAs genetically associated with a cluster of genes involved in ExPEC pathogenicity which suggests that they may be involved in virulence control (Table 4). Data for other expressed or not tested candidates are shown in Supplementary Data.
Table 4.
List of validated sRNA genes located close to virulence-related genes
| Candidate | sRNA | Origin | Loc.a | 5′-endb | 3′-endc | Typed | Target genese | Target function | O. g.f | ExPEC specific?g | Scoreh N/ kcal/mol |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Escherichia coli | |||||||||||
| SQ8164 | IntP4R | E. c. 536 | PAI-II | 4 735 462 | 4 735 232 | asRNA | intP4 | PAI DNA mobility | < > | No | 10 / −26.28 |
| SQ7560 | PrfR | E. c. 536 | PAI-II | 4 747 389 | 4 747 630 | asRNA | prfF | Adhesion | > < | Yes | 3 / −12.64 |
| SQ7575 | HlyR | E. c. 536 | PAI-II | 4 763 726 | 4 763 963 | asRNA | hlyA | Hemolysis | > < | Yes | 2 / −5.76 |
| SQ7606 | HaeR | E. c. 536 | PAI-II | 4 783 731 | 4 783 731 | asRNA | ECP_4580 | Filamentous haemagglutinin | > < | Yes | 9 / −6.52 |
| SQ8017 | FimR | E. c. 536 | Core | 4 852 969* | 4 852 518 | asRNA | fimD | Adhesion | < > | No | 15 / −8.49 |
| SQ109 | AfaR | E. c. AL862 | PAI-I | 56 564* | 56 332 | IGR | afa8 | Adhesion | > < | Yes | 2 / −5.2 |
| SQ19 | IntR | E. c. AL862 | PAI-I | 58 845 | 59 076 | asRNA | Int | PAI DNA mobility | < > | No | 12 / −14.94 |
| Streptococcus agalactiae | |||||||||||
| SQ18 | SQ18 | S. a. NEM316 | Core | 47 857* | 47 734 | asRNA | gbs0031 | Surface exposed protein | > < | N.A. | 3 / −10 |
| SQ340 | SQ340 | S. a. NEM316 | PAI-X | 1 163 702* | 1 163 779 | IGR | gbs1118 | Transposase of TnGBS2 | > < | N.A. | 3 / −10.5 |
| SQ893 | SQ893 | S. a. NEM316 | Core | 13 00 661 | 1 300 360 | IGR | gbs1263 | Fibronectin binding protein | < > | N.A. | 3 / −4 |
| SQ407 | SQ407 | S. a. NEM316 | PAI-XII | 1 350 419 | 1 350 658 | asRNA | Lmb | Laminin binding protein | > < | N.A. | 11 / −11.5 |
| SQ485 | SQ485 | S. a. NEM316 | Core | 1 655 610 | 1 655 852 | asRNA | gbs1588/ gbs1589 | Putative ABC transporter | > < | N.A. | 9 / −10.3 |
| SQ1004 | SQ1004 | S. a. NEM316 | PAI-XIII | 2 052 153 | 2 052 383 | IGR | gbs1987 | Streptomycin resistance | > < | N.A. | 3 / −7.6 |
aLocalization of the sRNA gene. Core, core genome; PAI, pathogenicity islands.
bThe 5′-end of the sRNA candidate is arbitrarily located 200 bp upstream from the first nucleotide of the predicted RIT. An asterisk indicates the 5′ triphosphates RNA end determined by 5′ RACE. The 5′ ends of SQ109 (E. coli AL862) and SQ340 (S. agalactiae NEM316) sRNAs were determined in another study (C.P., personal communication).
cThe 3′-end of the sRNA candidate is defined as the last nucleotide of the RIT poly-uracil tail.
dType of sRNA candidate gene locus. IGR, intergenic region; asRNA, sRNA antisense to a CDS.
eAntisense sRNA predicted target mRNA. The sRNA genes located in an IGR may regulate adjacent genes by an antisense mechanism.
fO. g., Orientation of genes (order sRNA/mRNA).
gSpecificity was determined by FASTA analysis against the Genbank database.
hN, number of covariations identified/RIT score in kcal/mol.
E.c., Escherichia coli; S.a., Streptococcus agalactiae
The FimR asRNA from E. coli 536 up-regulates the expression of type 1 fimbriae
In E. coli, type 1 fimbriae play a role in the development of urinary tract infections by mediating adhesion to specific receptors on the uroepithelium. During the pathogenesis of cystitis, type 1 fimbriae promote the invasion of bladder cells and the formation of intra cellular communities (54) but they are also involved in biofilm formation (55). The fim gene cluster is composed of nine genes (Supplementary Figure S4) whose expression is controlled by phase variation and various regulators. As SQ8017 asRNA and fimD CDS are located in the same genomic locus, we hypothesized that this asRNA controlled the expression of the fim gene cluster and we therefore renamed it FimR. Mapping of the transcription start site of fimR by 5′ RACE was determined at position T4852969 in the sequence of the E. coli 536 strain (Table 4). Analysis of fimR promoter region revealed the presence of a putative σE promoter. The ‘AA’ tract from the -35 box, the invariable C-residue from the -10 box, the 17 bp spacer, the 6 bp discriminator sequence and the -1 T-residue were observed, indicating such prediction may be reliable. Thus, it suggested that FimR expression is controlled by environmental stimuli (56). Given the position of fimR promoter and RIT, the calculated RNA size was ∼440 nt compatible with the ∼410 nt long RNA observed by Northern blot (Figure 3).
Type 1 fimbriae mediate adhesion to mannose-containing receptors, a biological trait quantified in vitro with the yeast agglutination assay (57,41). The specificity of the assay for evaluating the expression of the fim gene cluster of E. coli 536 was confirmed with a 536 Δfim mutant that does not agglutinate. We tested our hypothesis by constructing derivatives of strain 536 over-expressing FimR or a FimR antisense sRNA (antiFimR) and assessing the yeast agglutination titer. The expression of antiFimR should inactivate the FimR regulation pathway by competing with FimR mRNA substrate. The primary transcript of the fimR gene including its RIT was cloned under the control of the Pλ promoter of pZE2R-gfp to give the pZE2R-fimR plasmid. We also constructed pZE21-antifimR by cloning in antisense the same primary transcript under the control of the PLtetO-1 promoter. These fimR, antifimR and mock plasmids were introduced into the 536 and 536 Δfim strains. As expected, FimR and antiFimR over-expression in E. coli 536 significantly modified the agglutination titer (4-fold increase and 4-fold decrease, respectively; Table 5). These findings indicate that FimR upregulates the production of type 1 fimbriae.
Table 5.
Yeast agglutination assays for E. coli 536 derivatives
| Strain | Yeast agglutination titer |
|---|---|
| 536 + pZE2R-null | 1/16 |
| 536 + pZE2R-fimR | 1/64 |
| 536 Δfim::cat + pZE2R-null | NO |
| 536 Δfim::cat + pZE2R-fimR | NO |
| 536 + pZE21-null | 1/16 |
| 536 + pZE21-antifimR | 1/4 |
| 536 Δfim::cat + pZE21-null | NO |
| 536 Δfim::cat + pZE21-antifimR | NO |
| 536 | 1/16 |
| 536Δhfq::KmFRT | NO |
The level of expression of type 1 fimbriae was assessed in E. coli 536 wild type and mutant strains expressing the FimR sRNA, the antiFimR sRNA or mock plasmids. No 536 Δfim strains agglutinated yeasts indicating that the agglutination phenotypes resulted from the expression of type 1 fimbriae. NO: not observable.
FimR asRNA binds the fimD mRNA and positively regulates type 1 fimbriae expression
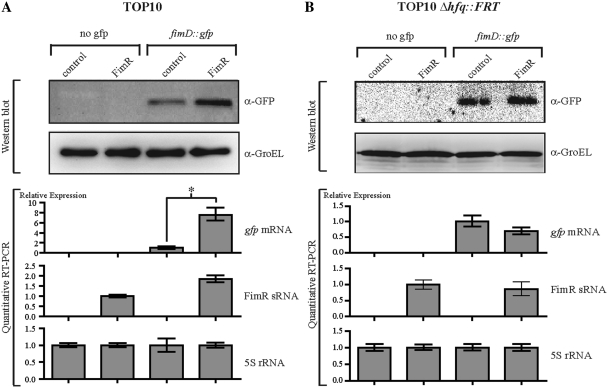
We assessed the putative base-pairing interaction of FimR and fimD mRNA using a translational control and target recognition system (34). A translational fusion of fimD and gfp genes was constructed by fusing the full stop-codon-less fimD CDS to the ATG-less gfp gene from pXG10 plasmid. Expression of the fimD::gfp fusion was monitored by quantitative RT–PCR and Western blot in E. coli TOP10 (a Δfim strain) harboring pXGfimD::gfp target plasmid or pXG-0 (no target control) and either pZE2R-fimR or pZE2R-null plasmids (Figure 4). Comparison of the relative levels of expression of fimD::gfp mRNA in pZE2R-fimR and pZE2R-null bearing strains showed that FimR over-expression was associated with a 8-fold increase of the amount of fusion mRNA (Figure 4A). Western blot experiments with antibodies directed against the GFP protein revealed a 2-fold increase in FimD::Gfp protein expression, consistent with the transcriptome analysis (Figure 4A). Accumulation of the fimD::gfp and FimR transcripts strongly suggested that these RNA molecules may be stabilized when co-expressed (Figure 4A). A post-transcriptional regulation of fimD mRNA by FimR likely occurs through a putative antisense base-pairing between the two RNA molecules.
Figure 4.
Over-expression of FimR and SQ18 antisense sRNAs regulates the fimD and gbs0031 target genes, respectively. (A) Analysis by Western blot and quantitative RT–PCR of gfp and FimR gene expression in E. coli strain TOP10 harboring pZE2R-fimR or pZE2R-null plasmids combined with pXG-0 (no gfp target control) or pXGfimD::gfp target expression plasmids. The four isolates were cultured in LB medium at 37°C until they reached an OD600 of 0.9. Quantitative expression of the gfp fusion gene was normalized to 1.0 for the TOP10 + pZE2R-null + pXGfimD::gfp strain. FimR expression was normalized to 1.0 for the TOP10 + pZE2R-fimR + pXG-0 strain. (B) Western blot and quantitative RT–PCR analysis were performed as described in (A) but in a Δhfq context. Asterisks indicate a significant difference between mean values in unpaired t-tests (P < 0.01).
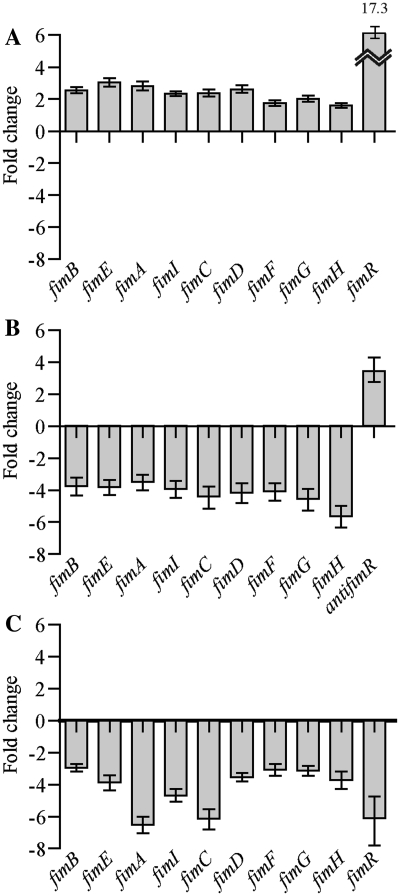
We investigated the role of FimR in vivo by carrying out a more detailed analysis of expression of the fimBE and fimAICDFGH operons and of FimR asRNA of E. coli 536 carrying pZE2R-fimR, pZE21-antifimR, or mock plasmids by quantitative RT–PCR. Over-expression of FimR from a multicopy plasmid (∼17 copies per chromosome equivalent) increased 2.34-fold the expression of fimB to H (Figure 5A). This result suggests that FimR positively regulates not only fimD, but also of the entire fim gene cluster. This hypothesis was confirmed by analyzing the relative expression level of fim genes in strain 536 which carries pZE21-antifimR. The antiFimR over-expression decreased 4.18-fold fim expression to reach a value lower than that obtained with mock plasmid (Figure 5B) indicating that FimR inhibition down-regulated fim gene expression. Furthermore, yeast agglutination assays with E. coli 536 + pZE2R-fimR cultured in human urine for 24 h showed that FimR increased the agglutination titer to the levels found with bacteria grown in LB medium (data not shown). It is thus likely that FimR controls type 1 mediated adhesion in vivo during host colonization.
Figure 5.
FimR sRNA up regulates type 1 fimbriae gene expression in vivo. Quantitative real-time RT–PCR analysis of expression of the fimBEAICDFGH gene cluster was performed in (A) E. coli 536 + pZE2R-fimR relatively to E. coli 536 + pZE2R-null, (B) E. coli 536 + pZE21-antifimR relatively to E. coli 536 + pZE21-null and (C) 536 Δhfq::KmFRT relatively to 536 strains, cultured in LB medium statically at 37°C for 24 h (stationary phase).
Hfq is required for fimD/FimR base pairing
About 40% of the known sRNAs from E. coli require the Hfq protein to interact with their targets. Since Hfq contributes to the virulence of the ExPEC E. coli UTI89 strain (57), we investigated the requirement of this protein for FimR regulation in E. coli 536. We investigated the requirement of Hfq protein for FimR/fimD interaction by introducing the pXGfimD::gfp or pXG-0 plasmids into the TOP10 Δhfq::FRT strain harboring either pZE2R-fimR or pZE2R-null plasmids. In contrast to the variations in gene expression observed in TOP10 cells, quantitative expression analysis of fimR and gfp genes in TOP10 Δhfq::FRT revealed no significant differences in either the RNA or protein levels in the presence or absence of FimR (Figure 4B). The loss of FimR-dependent regulation indicated that the Hfq protein was required for the binding of FimR to fimD::gfp mRNA.
We investigated the role of Hfq in vivo by constructing the E. coli 536 Δhfq::KmFRT strain and assessed adhesion mediated by type 1 fimbriae with a yeast agglutination assay. As expected, loss of hfq expression induced the loss of visible agglutination, suggesting that fewer type 1 fimbriae were produced in the hfq- mutant (Table 5). Next, we assessed the relative expression levels of the fimBE and fimAICDFGH operons and of FimR asRNA of E. coli 536 Δhfq::KmFRT by quantitative RT–PCR. As expected, loss of hfq expression decreased of fimBE and fimAICDFGH mRNA production by an average ∼4-fold and that of FimR asRNA by ∼6-fold. The fimA gene encoding the major structural subunit of type 1 fimbriae (∼1000 to 10 000 monomers per fimbriae) was impacted more severely and decreased ∼7-fold. Taken together, these results suggest that Hfq regulated type 1 fimbriae synthesis by mediating base pairing of FimR with fimD mRNA.
The FimR regulon controls biofilm development and bacterial motility
We checked whether the expression of fim genes was linked to FimR regulation and controlled virulence by investigating various fimbriae-associated phenotypes in E. coli 536 expressing the fimR and antifimR genes.
The adhesion mediated by type 1 fimbriae is an important factor in biofilm formation (55). As FimR enhanced type 1 fimbriae production, we investigated the effect of FimR on biofilm formation for E. coli 536 derivatives carrying pZE2R-fimR, pZE21-antifimR, or mock plasmids. In our conditions, the strains that expressed the pZE2R-fimR or the mock plasmids displayed similar levels of biofilm formation whereas the E. coli 536 + pZE21-antifimR isolate formed no detectable biofilm (data not shown). These observations suggest that FimR is required for biofilm development.
The productions of type 1 fimbriae and flagella have been shown to be co-regulated in various pathogenic E. coli isolates (55). We therefore analyzed the relation between FimR and motility by performing motility tests on various E. coli 536-derived strains. Compared to a null plasmid-bearing strain, motility was unaffected by the over-expression of FimR but significantly decreased by over-expression of antiFimR, resulting in virtually non-motile bacteria (data not shown). Thus, under laboratory growth conditions, fimR expression is linked to type 1 fimbriae-mediated biofilm formation, and bacterial motility; two phenotypes known to be important in the urovirulence of ExPEC strains.
Identification of sRNAs from S. agalactiae
The Gram-positive bacterium S. agalactiae (also referred to as Group B Streptocccus, GBS) is a major cause of bacterial sepsis, pneumonia and meningitis in newborns and is also responsible for pregnancy-related morbidity (58). As our in silico model is based on the recognition of RIT-associated signatures found in both Gram-negative and Gram-positive bacteria, we assessed whether our program was efficient for predicting asRNAs also in Gram-positive bacteria. We assessed its efficiency by searching sRNAs in S. agalactiae strain NEM316. All steps of the process were identical to those used for E. coli except the following modification: TransTerm HP was used to predict RITs and comparative genomics analyses were carried out with a database of Lactobacillale genome sequences (Genbank release of 07/06/2008). The data collected from our in silico search revealed the existence of 197 sRNA candidates with genes located in the IGRs while others were partially or fully antisense to CDSs (Table 2). In addition, some candidates were located upstream or downstream from a CDS and were putative mRNA encoded regulatory elements (e.g. Riboswitch). Interestingly, as in the E. coli analysis, sense RNA candidates were also predicted.
The genes of sRNA candidates were distributed throughout the genome and we analyzed by RT–PCR the expression of 30 out of 197 sRNA candidates located both in the core genome and PAIs. The expression of the TmRNA and 5S sRNA genes was used as positive controls. The analysis revealed that 26 out of the 30 predicted sRNA candidates were expressed thus demonstrating the versatility and efficiency of our in silico model (Supplementary Figure S1C).
To confirm the RT–PCR results, we further characterized by Northern blot analysis with 32P labeled oligonucleotides the 26 RT–PCR positive sRNA candidates. Ten candidates gave a strong hybridization signal. The absence or weak signal obtained for the other candidates may be due to lower sensitivity of the Northern blot technique compared to RT–PCR (data not shown). The SQ18, SQ485, SQ655 and SQ893 sRNAs gave multiple bands suggesting a cleavage by ribonucleases or a transcription initiated from multiple promoters (Figure 3, Supplementary Figure S5). Four validated sRNAs were found to be located close or antisense to CDS involved in the pathogenicity of S. agalactiae (Table 4). Comparative genomic analysis using FASTA3 indicated that none of the sRNAs described here were present in sequenced strains of the phylogenetically related pathogen Streptococcus pyogenes and that none of the sRNAs previously described in S. pyogenes were present in S. agalactiae, suggesting that these molecules display a high degree of species specificity in the genus Streptococcus. However, as recently reported, one of our sRNA candidates (SQ517) has an ortholog (csRNA12) in Streptococcus pneumoniae (59).
The SQ18, SQ485 and SQ893 sRNAs from S. agalactiae NEM316 modulate expression of adjacent genes
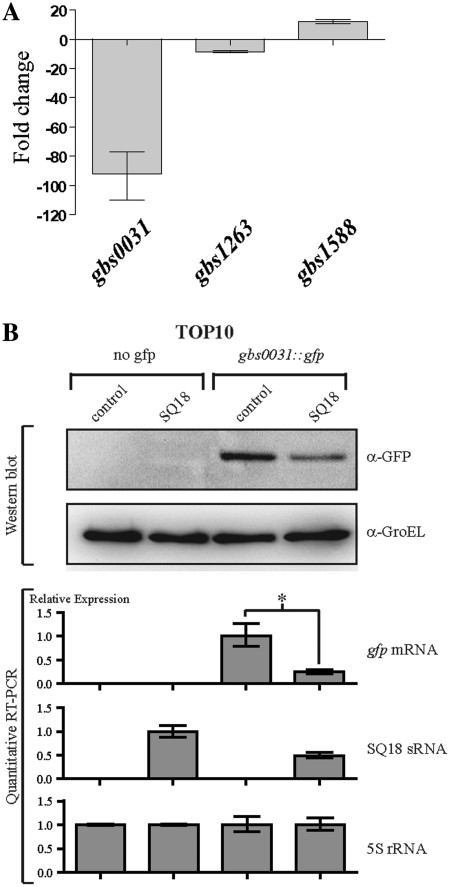
As shown for the ExPEC strains, some sRNAs were found to be near virulence-related gene clusters. So we investigated whether the SQ18 and SQ485 asRNAs and the SQ893 sRNA over-expression regulated the expression of other genes in the S. agalactiae NEM316 strain. The primary RNA transcripts of adjacent antisense genes to SQ18, SQ485 and SQ893 sRNAs were determined by searching in silico for putative promoters and terminators. This analysis revealed that the adjacent mRNA transcripts of gbs0031, gbs1588 and gbs1263 were putative antisense targets of SQ18, SQ485 and SQ893 sRNAs, respectively. To test these hypotheses, we cloned each of the three sRNA genes downstream the strong promoter Ptet in the shuttle vector pTCV-erm-ΩPtet, giving pTCV-SQ18, pTCV-SQ485 and pTCV-SQ893 plasmids. These plasmids were introduced into the S. agalactiae NEM316 strain and the expression of the putative target genes was analyzed by qRT–PCR (Figure 6A). Over-expression of the SQ18 asRNA and the SQ893 sRNAs significantly decreased the levels of their respective target mRNAs gbs0031 and gbs1263, suggesting that both sRNAs act as negative regulators. In contrast, over-expression of the SQ485 asRNA led to an increase in the amount of gbs1588 mRNA, suggesting that this asRNA acts as a positive regulator (Figure 6A).
Figure 6.
SQ18, SQ893 and SQ485 sRNAs controlled the gbs0031, gbs1263 and gbs1588 target genes expression, respectively. (A) Quantitative real-time RT–PCR analysis of expression of gbs0031, gbs1263 and gbs1588 gene. The relative expression of the three mRNA genes were determined by comparing over-expressing strains S. agalactiae NEM316 + pTCV-SQ18 or pTCV-SQ485 or pTCV-SQ893 against the wild-type S. agalactiae NEM316 isolate. (B) Analysis by Western blot and quantitative RT–PCR of the expression of the gfp and SQ18 gene expression in E. coli TOP10 strain harboring pZE2R-SQ18 or mock plasmids combined with pXG-0 (no gfp target control) or pXGgbs0031::gfp expression plasmids. SQ18 expression was normalized to 1.0 for the TOP10 + pZE2R-SQ18 + pXG-0 strain. Asterisks indicate a significant difference between mean values in unpaired t tests (P < 0.01).
The SQ18 asRNA from S. agalactiae NEM316 down-regulates expression of the Sip gene by an antisense mechanism
A translational control and target recognition system (34) was used for investigating the putative base pairing between SQ18 asRNA and gbs0031 mRNA which encodes a surface immunogenic protein (Sip) that elicits protective immunity against group B streptococci (60). We first characterized the 5′-end of the primary transcript of SQ18 by 5′ RACE. The 5′ triphosphate end was determined at G47857 and was associated with a putative σA promoter (Table 4). The SQ18 gene was inserted into pZE2R-gfp to give pZE2R-SQ18 and the stop-codon-less gbs0031 CDS was fused to the ATG-less gfp gene from pXG10, giving the pXGgbs0031::gfp plasmid. Four TOP10 strains harboring pZE2R-SQ18 or pZE2R-null plasmids combined with pXGgbs0031::gfp or pXG-0 plasmids were constructed. The expressions of the sRNA and the fusion mRNA were analyzed by quantitative RT–PCR and Western blot. Comparison of the relative levels of expression of gbs0031::gfp mRNA in pZE2R-SQ18 and pZE2R-null bearing strains showed that SQ18 over-expression was associated with a 4-fold decrease in the amount of the fusion mRNA (Figure 6B). Consistently, Western blot experiments carried out with antibodies directed against GFP (Gbs0031::Gfp) indicated a 2.6-fold decrease of the amount of Gfp fusion in the strain over-expressing SQ18 (Figure 6B). Thus, SQ18 is a negative post-transcriptional antisense regulator of gbs0031::gfp gene activity when expressed in E. coli.
DISCUSSION
High-throughput sequencing of bacterial transcripts (RNA-seq) or tilling microarray experiments showed that sRNA gene diversity is far greater than expected (8,9,61,62). In particular, these data revealed the existence of mRNA and asRNA pairs transcribed from genes present at the same locus, but on opposite DNA strands. There is a growing interest in the analysis of bacterial sRNAs in particular their contribution to gene regulation including the expression of virulence factors, but the identification of the full set of sRNA genes as performed by RNA-seq or tiling microarray remains a difficult task and the experimental costs remain high. We have thus designed and validated a new in silico model that efficiently identifies sRNA genes, including asRNAs, in any bacterial genomes, including both IGR and CDS regions. Our analysis of genome sequences from ExPEC and S. agalactiae, two major human pathogens, predicted the existence of numerous sRNAs, including asRNAs co-localized with virulence-associated genes.
Previous in silico methods for identifying de novo sRNAs in bacterial genomes increased in efficiency over time, but they are still limited for the analysis of IGR and do not predict asRNAs that partially or totally overlap neighboring CDS. Several sRNAs have been described in E. coli and other species (1), but few data are available for asRNAs (4,12,13). Our combination of RIT prediction, comparative genomics, RNA structure prediction with an implemented scoring system based on a RIT score and the analysis of covariations, identified ∼1800 and ∼200 sRNA candidates for E. coli and S. agalactiae genomes, respectively. The mean efficiency of our in silico model, based on the analysis of six genomes and expressed as the percentage of predicted versus known sRNAs, was estimated to be 70.1% and 71.5% for sRNAs located in the IGR and asRNAs, respectively (Table 3) which suggests that it is an efficient tool for analyzing any bacterial genomes. Up to now, few innovative in silico models were able to identify asRNA genes. The corresponding algorithms, based on comparative genomic approaches or mathematical/statistical analyses of the RNA secondary structures, were validated only with E. coli genomes (20,23,24,25) and only a few asRNA candidates were identified. In addition, these tools were either unable to predict sRNA genes de novo (25) or lacked validation data supporting their use as reliable asRNAs finders (20,23,24). Our study suggests that our in silico model can predict asRNA genes fully transcribed from CDS regions in antisense and possibly in sense orientation. Recent RNA-seq data suggested the existence of sense sRNAs but no biological functions were identified to date (9). Globally, we identified here sRNA and asRNA candidates evenly distributed throughout the genome. Based on the recognition efficiency of known E. coli sRNAs (Table 3), our approach appears as reliable as all currently available algorithms.
The main limitation of our approach is that it requires RIT prediction to detect sRNAs. We initially used RIT prediction to demonstrate that our in silico model efficiently identified known sRNAs in E. coli because 72.3% of known sRNA genes located in IGRs have an RIT. As a consequence, sRNA genes that utilize atypical RITs or a different termination process were not predicted with our model. We had hypothesized that any protein binding sites in sRNA could be the starting point of our predictive model. Thus, identification of the Rho protein or the Hfq binding sites may be good alternatives to enhance our sRNAs prediction model especially as RNA-seq data for E. coli (10) and Salmonella species (62) showed that RIT seemed to be less frequent in asRNA genes (<∼50%). On the other hand, we used two distinct RIT prediction models, which might exhibit variable predictive efficiencies for different bacteria. This approach is also limited by the number of fully sequenced genomes available and the requirement that the genetic divergence among these sequences be minimal to allow covariation identification. During our study, 15 E. coli and 3 S. agalactiae sequences were available and the mutation frequency among the genomes within these two species was not the same. The sequence conservation among S. agalactiae strains was higher than it was for the E. coli strains. Thus, the different RIT prediction efficiencies obtained for these two bacteria may explain why we identified ten times more candidates in E. coli than S. agalactiae.
The Hfq protein is the chaperone for sRNAs found in numerous bacterial species that is involved in the regulation of general cell metabolism and virulence (1,2,7). It has recently been shown that Hfq contributes to the virulence of E. coli strains causing urinary tract infection, a subgroup of the ExPEC pathotype suggesting that sRNAs have an important regulatory role on the expression of ExPEC virulence (57). We analyzed multiple genome sequences of ExPEC strains which revealed that there is a set of sRNA genes specific to this pathotype. Species-specific sRNAs have been identified in other bacteria, such as S. aureus (5) or S. typhimurium (6), but they are mostly located in IGR and their distribution could not be often easily associated with a function and a degree of virulence. In particular, this is the case for the virulence associated sRNA genes like RNAIII (63) and SprD from S. aureus (64) and FasX from S. pyogenes (65). In contrast, the identification of FimR, HlyR, and PrfR asRNAs in clusters of genes required for the pathogenesis of cystitis and pyelonephritis (50) suggested the possible association of these asRNAs with these pathologies as observed for the AmgR asRNA from S. enterica (66). In contrast, the Hfq-dependent FimR regulation constitutes a rare case of an asRNA acting as a positive regulator of gene expression, thus revealing the importance of this new asRNA function. However, the molecular mechanisms by which FimR regulates type 1 fimbriae production is still a matter of debate despite the fact that it was extensively studied (11). Recent models of the post-transcriptional activation of collagenase mRNAs by VR-RNA in clostridia or of the streptokinase mRNA by FasX in Group A Streptococci (67) provides insight into some of the possible mechanism of regulation by FimR asRNA.
The control of expression of virulence genes during pathogenesis is critical for the opportunistic pathogen S. agalactiae. As only three complete genome sequences are currently available for the group B streptococci, the distribution of sRNA genes in this species remains largely unknown. We analyzed the genome sequence of the virulent strain NEM316 and identified 197 sRNA/asRNA genes and validated the expression of 26 of them. One putative sRNAs previously reported to interact with the CiaRH regulatory system from S. agalactiae NEM316 has been also identified in our analyses (59). Distribution of sRNA genes was uniform along the S. agalactiae NEM316 genome including the core genome and PAIs. Moreover, the location of sRNA genes in the PAI of S. agalactiae suggest that this may be a common feature in pathogenic bacteria as reported for S. aureus (5) and S. typhimurium (6). These observations indicated that pathogenesis of Group B Streptococci may be controlled by sRNAs, as demonstrated in Group A Streptococci (65,68,69). The regulatory roles of the SQ18, SQ485 and SQ893 sRNAs on adjacent mRNAs expression involved in virulence, as demonstrated in this study, provide additional support to this hypothesis. However, the role of sRNAs/asRNAs in the control of the virulence of Group B Streptococci remains to be characterized and our list of candidates may facilitate these studies.
This report demonstrated that an sRNA gene finder approach can efficiently identify sRNAs located within IGRs, asRNAs and putative sense RNAs transcribed within CDSs. The main advantage of in silico approaches over in vivo techniques (tiling microarrays and RNA-seq) is the capability to search for sRNAs in an unlimited number of strains irrespective of their growing conditions. This catalog may then be used to select the most valuable strains for in vivo studies and should facilitate the post-screening identification of expressed sRNAs and asRNAs in large collections of data. Accordingly, the results of our analysis of the genomes of two major human pathogens, E. coli and S. agalactiae, suggest that sRNAs as well as asRNAs are key elements in the control of their virulence.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1–S5, Supplementary Figures S1–S5, Supplementary Materials and Supplementary References [5,10,15,23,43,46,63,70–97].
FUNDING
This work was supported by Institut Pasteur (PTR165 to C.P.); Agence National de la Recherche for the ERA-NET Pathogenomics project (ANR-06-PATHO-002-03); Postdoctoral fellowship by the French Region Ile-de-France (DIM Malinf to C.P.). Funding for open access charge: Institut Pasteur.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Shaynoor Dramsi for pTCV-erm-ΩPtet plasmid gift, Ulrich Dobrindt for E. coli strains gift, Christophe Beloin for pZE21-gfp and biofilm analysis, Christiane Bouchier for PAI-IAL862 sequencing and Jörg Vogel for the complete pXG plasmids system. We also thank Carmen Buchreiser for critical reading of the article.
REFERENCES
- 1.Waters LS, Storz G. Regulatory RNAs in Bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pichon C, Felden B. Proteins that interact with bacterial small RNA regulators. FEMS Microbiol. Rev. 2007;31:614–625. doi: 10.1111/j.1574-6976.2007.00079.x. [DOI] [PubMed] [Google Scholar]
- 3.Gillet R, Felden B. Emerging views on tmRNA-mediated protein tagging and ribosome rescue. Mol. Microbiol. 2001;42:879–885. doi: 10.1046/j.1365-2958.2001.02701.x. [DOI] [PubMed] [Google Scholar]
- 4.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Pichon C, Felden B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl Acad. Sci. USA. 2005;102:14249–14254. doi: 10.1073/pnas.0503838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfeiffer V, Sittka A, Tomer R, Tedin K, Brinkmann V, Vogel J. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol. Microbiol. 2007;66:1174–1191. doi: 10.1111/j.1365-2958.2007.05991.x. [DOI] [PubMed] [Google Scholar]
- 7.Romby P, Vandenesch F, Wagner EGH. The role of RNAs in the regulation of virulence-gene expression. Curr. Opin. Microbiol. 2006;9:229–236. doi: 10.1016/j.mib.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;18:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 9.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermüller J, Reinhardt R, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 10.Lorentz C, Gesell T, Zimmermann B, Schoeberl U, Bilusic I, Rajkowitsch L, Waldsich C, von Haeseler A, Schroeder R. Genomix SELEX for Hfq binding RNAs identifies genomic aptamers predominantly in antisense transcripts. Nucleic Acids Res. 2010;38:3794–3808. doi: 10.1093/nar/gkq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomason MK, Storz G. Bacterial antisense RNAs: how many are there, and what are they doing? Annu. Rev. Genet. 2010;44:167–188. doi: 10.1146/annurev-genet-102209-163523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawano M, Aravind L, Storz G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007;64:738–754. doi: 10.1111/j.1365-2958.2007.05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dühring U, Axmann IM, Hess W, Wilde A. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc. Natl. Acad. Sci. USA. 2008;103:7054–7058. doi: 10.1073/pnas.0600927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pichon C, Felden B. Small RNA genes identifications and mRNA targets predictions in Bacteria. Bioinformatics. 2008;24:2807–2813. doi: 10.1093/bioinformatics/btn560. [DOI] [PubMed] [Google Scholar]
- 15.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EGH, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 16.Rivas E, Klein RJ, Jones TA, Eddy SR. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr. Biol. 2001;11:1369–1373. doi: 10.1016/s0960-9822(01)00401-8. [DOI] [PubMed] [Google Scholar]
- 17.Pichon C, Felden B. Intergenic Sequence Inspector: searching and identifying bacterial RNAs. Bioinformatics. 2003;19:1707–1709. doi: 10.1093/bioinformatics/btg235. [DOI] [PubMed] [Google Scholar]
- 18.Axmann I, Kensche P, Vogel J, Kohl S, Herzel H, Hess W. Identification of cyanobacterial non-coding RNAs by comparative genome analysis. Genome Biol. 2005;6:R73. doi: 10.1186/gb-2005-6-9-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livny J, Fogel MA, Davis BM, Waldor MK. sRNAPredict: an integrative computational approach to identify sRNAs in bacterial genomes. Nucleic Acids Res. 2005;33:4096–4105. doi: 10.1093/nar/gki715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter R, Dubchak I, Holbrook S. A computational approach to identify genes for functional RNAs in genomic sequences. Nucleic Acids Res. 2001;29:3928–3938. doi: 10.1093/nar/29.19.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schattner P. Searching for RNA genes using base composition statistics. Nucleic Acids Res. 2002;30:2076–2082. doi: 10.1093/nar/30.9.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saetrom P, Sneve R, Kristiansen KI, Snove O, Grünfeld T, Rognes T, Seeberg E. Predicting non-coding RNA genes in Escherichia coli with boosted genetic programming. Nucleic Acids Res. 2005;33:3263–3270. doi: 10.1093/nar/gki644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yachie N, Numata K, Saito R, Kanai A, Tomita M. Prediction of non-coding and antisense RNA genes in Escherichia coli with Gapped Markov Model. Gene. 2006;372:171–181. doi: 10.1016/j.gene.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 24.Wang C, Ding C, Meraz R, Holbrook S. PSoL: a positive sample only learning algorithm for finding non-coding RNA genes. Bioinformatics. 2006;22:2590–2596. doi: 10.1093/bioinformatics/btl441. [DOI] [PubMed] [Google Scholar]
- 25.Uzilov A, Keegan J, Mathews D. Detection of non-coding RNAs on the basis of predicted secondary structure formation free energy change. BMC Bioinformatics. 2006;7:173–203. doi: 10.1186/1471-2105-7-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macke TJ, Ecker DJ, Gutell RR, Gautheret D, Case DA, Sampath R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lesnik EA, Sampath R, Levene HB, Henderson TJ, McNeil JA, Ecker DJ. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 2001;29:3583–3594. doi: 10.1093/nar/29.17.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Novère N. MELTING, computing the melting temperature of nucleic acid duplex. Bioinformatics. 2001;17:1226–1227. doi: 10.1093/bioinformatics/17.12.1226. [DOI] [PubMed] [Google Scholar]
- 29.Kingsford C, Ayandale K, Salsberg SL. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 2007;8:R22. doi: 10.1186/gb-2007-8-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lalioui L, Le Bouguénec C. The afa-8 gene cluster is carried by a pathogenicity island inserted into the tRNAPhe of human and bovine pathogenic Escherichia coli isolates. Infect. Immun. 2001;69:937–948. doi: 10.1128/IAI.69.2.937-948.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berger H, Hacker J, Juarez A, Hughes C, Goebel W. Cloning of the chromosomal determinants encoding haemolysin production and mannose resistant haemagglutination in Escherichia coli. J. Bacteriol. 1982;152:1241–1247. doi: 10.1128/jb.152.3.1241-1247.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holden NJ, Totsika M, Mahler E, Roe AJ, Catherwood K, Lindner K, Dobrindt U, Gally DL. Demonstration of regulatory cross-talk between P fimbriae and type 1 fimbriae in uropathogenic Escherichia coli. Microbiology. 2006;152:1143–1153. doi: 10.1099/mic.0.28677-0. [DOI] [PubMed] [Google Scholar]
- 33.Glaser P, Rusniok C, Chevalier F, Buchrieser C, Frangeul L, Zouine M, Couve E, Lalioui L, Msadek T, Poyart C, et al. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 2002;45:1499–1513. doi: 10.1046/j.1365-2958.2002.03126.x. [DOI] [PubMed] [Google Scholar]
- 34.Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassette with the option of Flp-catalyzed excision of the antibiotic resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 36.Chaveroche MK, Ghigo JM, d'Enfert C. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 2000;28:e97. doi: 10.1093/nar/28.22.e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, The TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antal M, Bordeau V, Douchin V, Felden B. A small bacterial RNA regulates a putative ABC transporter. J. Biol. Chem. 2005;280:7901–7908. doi: 10.1074/jbc.M413071200. [DOI] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Pichon C, Héchard C, du Merle L, Chaudray C, Bonne I, Guadagnini S, Vandewalle A, Le Bouguénec C. Uropathogenic Escherichia coli AL511 requires flagellum to enter renal collecting duct cells. Cell. Microbiol. 2009;11:616–628. doi: 10.1111/j.1462-5822.2008.01278.x. [DOI] [PubMed] [Google Scholar]
- 42.Schrembri MA, Klemm P. Biofilm formation in a hydrodynamic environment by novel fimH variants and ramifications for virulence. Infect. Immun. 2001;69:1322–1328. doi: 10.1128/IAI.69.3.1322-1328.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearson WR. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol. 2000;132:185–219. doi: 10.1385/1-59259-192-2:185. [DOI] [PubMed] [Google Scholar]
- 44.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 45.Delcher A, Harmon D, Kasif S, White O, Salzberg S. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tjaden B, Saxena RM, Stolyar S, Haynor D, Kolker E, Rosenow C. Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res. 2002;30:3732–3738. doi: 10.1093/nar/gkf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raghavan R, Groisman EA, Ochman H. Genome-wide detection of novel regulatory RNAs in E. coli. Genome Res. 2011;21:1487–1497. doi: 10.1101/gr.119370.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beaume M, Hernandez D, Farinelli L, Deluen C, Linder P, Gaspin C, Romby P, Screnzel J, Francois P. Cartography of methicillin-resistant S. aureus transcripts: detection, orientation and temporal expression during growth phase and stress conditions. PLoS One. 2010;5:e10725. doi: 10.1371/journal.pone.0010725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, et al. Organised genome dynamics in the Escherichia coli species: the path to adaptation. PLoS Genetics. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 51.Chen SL, Hung CS, Xu J, Reigstad CS, Magrini V, Sabo A, Blasiar D, Bieri T, Meyer RR, Ozersky P, et al. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc. Natl. Acad. Sci. USA. 2006;103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hochhut B, Wilde C, Balling G, Middendorf B, Dobrindt U, Brzuszkiewicz E, Gottschalk G, Carniel E, Hacker J. Role of pathogenicity island-associated integrases in the genome plasticity of uropathogenic Escherichia coli strain 536. Mol. Microbiol. 2006;61:584–595. doi: 10.1111/j.1365-2958.2006.05255.x. [DOI] [PubMed] [Google Scholar]
- 53.Goode T, Ho WZ, O'Connor T, Busteed S, Douglas SD, Shanahan F, O'Connell J. Nested RT-PCR. In: O'Connell J, editor. Methods in Molecular Biology, RT-PCR Protocols. Humana Press; 2002. [DOI] [PubMed] [Google Scholar]
- 54.Wright KJ, Seed PC, Hultgren SJ. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell. Microbiol. 2007;9:2230–2241. doi: 10.1111/j.1462-5822.2007.00952.x. [DOI] [PubMed] [Google Scholar]
- 55.Pratt L, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type 1 pili. Mol. Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 56.Raivio T. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol. Microbiol. 2005;56:1119–1128. doi: 10.1111/j.1365-2958.2005.04625.x. [DOI] [PubMed] [Google Scholar]
- 57.Kulesus R, Diaz-Perez K, Slechta S, Eto DS, Mulvey MA. Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect. Immun. 2008;76:3019–3026. doi: 10.1128/IAI.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poyart C, Réglier-Poupet H, Tazi A, Billoët A, Dmytruk N, Bidet P, Bingen E, Raymond J, Trieu-Cuot P. Invasive group B streptococcal infections in infants, France. Emerg. Infect. Dis. 2008;14:1647–1649. doi: 10.3201/eid1410.080185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marx P, Nuhn M, Kovacs M, Hakenbeck R, Brückner R. Identification of genes for small non-coding RNAs that belongs to the regulon of the two component regulatory system CiaRH in Streptococcus. BMC Genomics. 2010;11:e661. doi: 10.1186/1471-2164-11-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brodeur B, Boyer M, Charlebois I, Hamel J, Couture F, Rioux C, Martin D. Identification of group B streptococcal Sip protein, which elicits cross protective immunity. Infect. Immun. 2000;68:5610–5618. doi: 10.1128/iai.68.10.5610-5618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mendoza-Vargas A, Olvera L, Olvera M, Grande R, Vega-Alvarado L, Taboada B, Jimenez-Jacinto V, Salgado H, Juarez K, Contreras-Moreira B, et al. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS ONE. 2009;4:e7526. doi: 10.1371/journal.pone.0007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chinni SV, Raabe CA, Zakaria R, Randau G, Hock Hoe C, Zemann A, Brosius J, Tang TH, Rozhdestvensky TS. Experimental identification and characterization of 97 novel npcRNA candidates in Salmonella enterica serovar Typhi. Nucleic Acids Res. 2010;38:5893–5908. doi: 10.1093/nar/gkq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chabelskaya S, Gaillot O, Felden B. A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule. PLoS Pathog. 2010;6:e1000927. doi: 10.1371/journal.ppat.1000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klenk M, Koczan D, Guthke R, Nakata M, Thiesen HJ, Podbielski A, Kreikenmeyer B. Global epithelial cell transcriptional responses reveal Streptococcus pyogenes Fas regulator activity association with bacterial aggressiveness. Cell. Microbiol. 2005;7:1237–1250. doi: 10.1111/j.1462-5822.2005.00548.x. [DOI] [PubMed] [Google Scholar]
- 66.Lee EJ, Groisman EA. An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol. Microbiol. 2010;76:1020–1033. doi: 10.1111/j.1365-2958.2010.07161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Podkaminski D, Vogel J. Small RNAs promote mRNA stability to activate the synthesis of virulence factors. Mol. Microbiol. 2010;78:1327–1331. doi: 10.1111/j.1365-2958.2010.07428.x. [DOI] [PubMed] [Google Scholar]
- 68.Halfmann A, Kovacs M, Hakenbeck R, Brückner R. Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol. Microbiol. 2007;66:110–126. doi: 10.1111/j.1365-2958.2007.05900.x. [DOI] [PubMed] [Google Scholar]
- 69.Perez N, Trevino J, Liu Z, Ho SCM, Babitzke P, Sumby P. A genome-wide analysis of small regulatory RNAs in the human pathogen group A Streptococcus. PLOS One. 2009;4:e7668. doi: 10.1371/journal.pone.0007668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brown S, Fournier MJ. The 4.5 S RNA gene of Escherichia coli is essential for cell growth. J. Mol. Biol. 1984;178:533–550. doi: 10.1016/0022-2836(84)90237-7. [DOI] [PubMed] [Google Scholar]
- 71.Brownlee GG. Sequence of 6S RNA of E. coli. Nature New Biol. 1971;229:147–149. doi: 10.1038/newbio229147a0. [DOI] [PubMed] [Google Scholar]
- 72.Okamoto K, Freundlich M. Mechanism for the autogenous control of the crp operon: Transcriptional inhibition by a divergent RNA transcript. Proc. Natl. Acad. Sci. USA. 1986;83:5000–5004. doi: 10.1073/pnas.83.14.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu MY, Gui G, Wei B, Preston JF, Oakford L, Yuksel U, Giedroc DP, Romeo T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 74.Wassarman K, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes & Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tetart F, Bouche JP. Regulation of the expression of the cell cycle gene ftsZ by DicF antisense RNA. Division does not require a fixed number of FtsZ molecules. Mol. Microbiol. 1992;6:615–620. doi: 10.1111/j.1365-2958.1992.tb01508.x. [DOI] [PubMed] [Google Scholar]
- 76.Sledjeski D, Gottesman S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1995;92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen S, Lesnik EA, Hall TA, Sampath R, Griffrey RH, Ecker DJ, Blyn LB. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. BioSystems. 2002;65:157–177. doi: 10.1016/s0303-2647(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 78.Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- 79.Rivas E, Eddy SR. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:1–19. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cole ST, Honore N. Transcription of the sulA-ompA region of Escherichia coli during the SOS response and the role of an antisense RNA molecule. Mol. Microbiol. 1989;3:715–722. doi: 10.1111/j.1365-2958.1989.tb00220.x. [DOI] [PubMed] [Google Scholar]
- 81.Vogel J, Argaman L, Wagner EGH, Altuvia S. The Small RNA IstR Inhibits Synthesis of an SOS-Induced Toxic Peptide. Curr. Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 82.Jain SK, Gurevitz M, Apirion D. A small RNA that complements mutants in the RNA processing enzyme ribonuclease P. J. Mol. Biol. 1982;162:515–533. doi: 10.1016/0022-2836(82)90386-2. [DOI] [PubMed] [Google Scholar]
- 83.Mizuno T, Chou MY, Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA) Proc. Natl. Acad. Sci. 1984;81:1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Argaman L, Altuvia S. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 2000;300:1101–1112. doi: 10.1006/jmbi.2000.3942. [DOI] [PubMed] [Google Scholar]
- 85.Kawano M, Oshima T, Kasai H, Mori H. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol. Microbiol. 2002;45:333–349. doi: 10.1046/j.1365-2958.2002.03042.x. [DOI] [PubMed] [Google Scholar]
- 86.Majdalani N, Chen S, Murrow J, St John K, Gottesman S. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- 87.Douchin V, Bohn C, Bouloc P. Down-regulation of porins by a small RNA bypasses the essentiality of the RIP protease RseP in Escherichia coli. J. Biol. Chem. 2006;281:12253–12256. doi: 10.1074/jbc.M600819200. [DOI] [PubMed] [Google Scholar]
- 88.Bosl M, Kersten H. A novel RNA product of the tyrT operon of Escherichia coli. Nucleic Acids Res. 1991;19:5863–5870. doi: 10.1093/nar/19.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 90.Kawano M, Reynolds A, Miranda-Rios J, Storz G. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 2005;33:1040–1050. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Polayes DA, Rice PW, Dahlberg JE. DNA polymerase I activity in Escherichia coli is influenced by spot 42 RNA. J. Bacteriol. 1988;170:2083–2088. doi: 10.1128/jb.170.5.2083-2088.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager J, Huttenhofer A, Wagner EGH. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 94.Geissman T, Chevalier C, Cros MJ, Boisset S, Fechter P, Noirot C, Schrenzel J, François P, Vandenesch F, Gaspin C, et al. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 2009;37:7239–7257. doi: 10.1093/nar/gkp668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marchais A, Naville M, Bohn C, Bouloc P, Gautheret D. Single-pass classification of all non-coding sequences in a bacterial genome using phylogenetic profiles. Genome Res. 2009;19:1084–1092. doi: 10.1101/gr.089714.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bohn C, Rigoulay C, Chabelskaya S, Sharma CM, Marchais A, Skorski P, Borezée-Durant E, Barbet R, Jacquet E, Jacq A, et al. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 2010;38:6620–6636. doi: 10.1093/nar/gkq462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Abu-Qatouseh LF, Chinni SV, Seggewiss J, Proctor RA, Brosius J, Roshdestvensky TS, Peters G, von Eiff C, Becker K. Identification of differentially expressed small non-protein-coding RNAs in Staphylococcus aureus displaying both the normal and the small-colony variant phenotype. J. Mol. Med. 2010;88:565–575. doi: 10.1007/s00109-010-0597-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.