Abstract
Eukaryotic telomeres are transcribed into telomeric repeat-containing RNA (TERRA). Telomeric transcription has been documented in mammals, birds, zebra fish, plants and budding yeast. Here we show that the chromosome ends of Schizosaccharomyces pombe produce distinct RNA species. As with budding yeast and mammals, S. pombe contains G-rich TERRA molecules and subtelomeric RNA species transcribed in the opposite direction of TERRA (ARRET). Moreover, fission yeast chromosome ends produce two novel RNA species: C-rich telomeric repeat-containing transcripts (ARIA) and subtelomeric transcripts complementary to ARRET (αARRET). RNA polymerase II (RNAPII) associates with pombe chromosome ends in vivo and the telomeric factor Rap1 negatively regulates this association, as well as the cellular accumulation of RNA emanating from chromosome ends. We also show that the RNAPII subunit Rpb7 and the non-canonical poly(A) polymerases Cid12 and Cid14 are involved in the regulation of TERRA, ARIA, ARRET and αARRET transcripts. We confirm the evolutionary conservation of telomere transcription, and reveal intriguing similarities and differences in the composition and regulation of telomeric transcripts among model organisms.
INTRODUCTION
Telomeres, the heterochromatic structures protecting the integrity of eukaryotic chromosome ends, are transcribed into telomeric repeat-containing RNA (TERRA) (1–3). TERRA has been identified in different mammals, birds, Saccharomyces cerevisiae, Danio rerio and Arabidopsis thaliana (4–8). Individual TERRA transcripts contain G-rich telomeric RNA repeats and RNA tracts corresponding to adjacent subtelomeric sequences. In mammalian cells with telomeres up to 20 kb, TERRA ranges in size between ∼100 and more than 9000 bases (5,7). In budding yeast strains with telomeres of ∼400 bp, most TERRA molecules are below 500 bases in length (6). A C-rich telomeric repeat-containing RNA complementary to TERRA has not been detected in mammalian and budding yeast cells (5–7). Budding yeast also contains RNA molecules dubbed ARRET, which are complementary to the subtelomeric tract of TERRA molecules but are devoid of detectable telomeric repeats (6). Whether ARRET also exists outside of budding yeast is unclear.
TERRA transcription is mainly sustained by the DNA-dependent RNA polymerase II (RNAPII) (5–7). In cultured mammalian cells, the RNAPII inhibitor alpha amanitin diminished TERRA cellular levels (7). Analogously, temperature sensitive (ts) alleles of the budding yeast RNAPII subunit Rpb3p failed to sustain TERRA expression at restrictive temperatures (6). In addition, active RNAPII was found to associate with mammalian and budding yeast telomeres in vivo (6,7,9). In particular in humans, active RNAPII binds to specific promoter regions dedicated to the transcription of TERRA (9,10). Human TERRA promoters lie within the subtelomere of various chromosome ends and are embedded within CpG dinucleotide-rich islands. These CpG islands are methylated by the concerted activity of DNA methyltransferase (DNMT) 1 and 3b to repress their transcriptional activity (9,10).
Human and yeast TERRA molecules contain 7-methylguanosine cap structures at their 5′-ends and are at least in part (∼11% in human and ∼100% in budding yeast cells) 3′ polyadenylated (6,7,11,12). Interestingly, human poly(A)+ TERRA molecules displayed a longer half-life than their poly(A)− counterparts (12), and inactivation of the Pap1p poly(A) polymerase in budding yeast led to TERRA disappearance (6).
While the cellular localization of TERRA has not been determined in yeast, RNA fluorescence in situ hybridization (FISH) studies demonstrated that a fraction of human and mouse TERRA stably associates with telomeric heterochromatin throughout the cell cycle, including transcriptionally inactive metaphase (2,5,7). Total TERRA cellular levels are nonetheless down-regulated during S-phase progression when chromosomal DNA is replicated, suggesting a coordination between TERRA regulation and telomere replication (12,13). Indeed, recent evidence indicates that during S-phase in human cancer cells, TERRA and heterogeneous nuclear protein A1 (hnRNPA1) regulate timing of an exchange at telomeres between replication protein A (RPA) and POT1, both ssDNA-binding proteins (13). TERRA might also play a role in telomeric heterochromatin establishment. Transfection of human cancer cells with short interference RNA molecules against TERRA UUAGGG repeats resulted in a ∼40% reduction of TERRA levels and in decreased density of the heterochromatin marks di- and tri-methylated histone H3K9 at telomeres (14). It has also been proposed that TERRA negatively regulates telomerase-mediated telomere elongation. RNA oligonucleotides comprising the TERRA-like sequence (UUAGGG)3 inhibited human telomerase activity in vitro (7,15). Moreover, forced transcription of a yeast chromosome end led to shortening of its telomeric tract, although it remains unclear whether this shortening resulted from compromised telomerase function (16).
Here, we present the first molecular characterization of telomere transcription in the fission yeast Schizosaccharomyces pombe. Fission yeast telomeric RNA species include: (i) telomeric G-rich TERRA; (ii) a novel complementary C-rich class that we name ARIA; (iii) ARRET; and (iv) a novel antisense ARRET that we call αARRET. We refer to the ensemble of these RNA classes as ‘the telomeric transcriptome’. Both TERRA and ARIA localize to the nucleus. RNAPII binds in vivo to TERRA transcription start sites, which are located subtelomerically, and ARRET and αARRET cellular levels decrease upon functional inactivation of the RNAPII subunit Rpb7. We also show that the telomeric factor Rap1 restricts RNAPII binding to TERRA TSS and the cellular accumulation of the telomeric transcriptome. Finally, at least a fraction of TERRA and the large majority of ARRET and αARRET molecules are polyadenylated and deletion of the non-canonical poly(A) polymerases cid12+ and cid14+ genes differentially impact on the steady-state levels of the different telomeric RNA species.
MATERIALS AND METHODS
Strains and media
Original Schizosaccharomyces pombe strains used in this study were kind gifts from Julie Promisel Cooper, Karl Ekwall and Marc Bühler or were purchased from Bioneer Corporation (Table 1). Strains were manipulated according to standard methods (17,18). Strains were grown in yeast extract medium with amino acid supplements (YES) and 150 µg ml−1 Geneticin was added to the medium when required. CAF55, CAF101 and CAF103 strains were generated by replacement of the complete SPBC1778.02 ORF with a ΔSPBC1778.02::kanMX4 resistance cassette PCR amplified from CAF39. All strains were validated for correct genomic locus arrangement by PCR, RT–PCR and appropriate phenotypic analyses.
Table 1.
Yeast strains used in this study
| Strains | Genotype | Source |
|---|---|---|
| CAF1 | h+ ade6-M210 ura4-D18 leu1-32 (wt) | Bioneer Corporation |
| CAF2 | h+ ade6-M216 ura4-D18 leu1-32 (wt) | Bioneer Corporation |
| CAF4 | h+ ade6-M210 (or ade6-M216) ura4-D18 leu1-32 ΔSPBC543.03c::kanMX4 | Bioneer Corporation |
| CAF13 | h− (strain 972) | J. P. Cooper |
| CAF39 | h+ ade6-M216 ura4-D18 leu1-32 ΔSPBC1778.02::kanMX4 | Bioneer Corporation |
| CAF55 | h− ΔSPBC1778.02::kanMX4 | This study |
| CAF97 | h+ leu1-32 ura4-ΔS/E ade6-ΔNcoI cc2SphII::ura4+ | K. Ekwall |
| CAF98 | h− rpb7-G150D (csp3ts) | K. Ekwall |
| CAF101 | h+ leu1-32 ura4-ΔS/E ade6-ΔNcoI cc2SphII::ura4+ ΔSPBC1778.02::kanMX4 | This study |
| CAF103 | h− rpb7-G150D (csp3ts) ΔSPBC1778.02::kanMX4 | This study |
| CAF109 | h90 mat3::ura4+ ura4-DS/E leu1-32 ade6-M210 ΔSPAC12G12.13c::kan | M. Bühler |
| CAF117 | h+ ade6-M210 ura4-D18 leu1-32 ΔSPAC19D5.03::kanMX4 | Bioneer Corporation |
| CAF118 | h+ ade6-M216 ura4-D18 leu1-32 ΔSPBC1685.06::kanMX4 | Bioneer Corporation |
| CAF28 | h+ ade6-M216 ura4-D18 leu1-32 ΔSPCC663.12::kanMX4 | Bioneer Corporation |
| CAF119 | h+ ade6-M216 ura4-D18 leu1-32 ΔSPAC17H9.01::kanMX4 | Bioneer Corporation |
RNA isolation and analysis
Total RNA was isolated as described previously (19) from exponentially growing yeasts (OD595: 0.5–1.0) and treated twice with RNase-free DNase (Qiagen) to eliminate DNA contaminations. Poly (A)+ RNA was isolated from total RNA using the GenElute mRNA miniprep kit (Sigma) according to the manufacturer instructions. For northern blot analysis, RNA was electrophoresed in 1.2% formaldehyde agarose gels, transferred to positively charged nylon membranes (GE Osmonics) and hybridized to radioactively labeled probes at 45–60°C for 18 h. Oligonucleotide probes were 5′-end labeled with T4 polynucleotide kinase (New England Biolabs) in the presence of [γ-32P]ATP. Strand-specific ARRET and αARRET probes were generated using genomic PCR products as template DNA for primer extension reactions carried out with each oligonucleotide used for PCR in presence of Klenow Fragment (New England Biolabs) and [α-32P]dCTP. The DNA probe detecting simultaneously S. pombe TERRA and ARIA was a DNA fragment excised from the plasmid pIRT2-Telo (kind gift of Julie P. Cooper) and labeled by random priming in presence of [α-32P]dCTP. After hybridization, membranes were washed twice in 2× SSC, 0.2% SDS and once in 1× SSC, 0.2% SDS at the same temperature as for hybridizations. Radioactive signals were detected using a Typhoon FLA 9000 Biomolecular Imager (GE Healthcare) and analyzed using ImageJ (National Institutes of Health, MD, USA) and Adobe Photoshop software. For RT–PCR experiments, total RNA was treated three times with RNase-free DNase (Qiagen) and reverse transcribed using the SuperScript III First-Strand Synthesis System (Invitrogen) according to manufacturer instructions. cDNA was PCR amplified using Taq DNA Polymerase (New England Biolabs) with an activation step at 98°C (30 s) followed by 28 cycles at 98°C (10 s), 58°C (15 s) and 72°C (20 s) and a final extension step at 72°C (5 min). PCR products were electrophoresed in 1.6% agarose gels. Oligonucleotide sequences are listed in Supplementary Table S1.
5′- and 3′-RACE
The FirstChoice RLM-RACE kit (Ambion) was used to determine S. pombe TERRA transcription start sites and ARRET 3′-ends. Nearly, 1–10 µg of total RNA was used as starting material. All steps were carried out according to the manufacturer instructions using the oligonucleotides listed in Supplementary Table S1. For TERRA TSS determination, RNA was reverse transcribed using oligonucleotides oC and cDNA was amplified by nested PCR using o5 and o3 oligonucleotides in combination with inner and outer RACE primers supplied with the kit. For ARRET 3′-end determination, RNA was reverse transcribed using a modified oligo d(T) supplied with the kit and cDNA was amplified by nested PCR using o3 and o3i oligonucleotides in combination with inner and outer RACE primers supplied with the kit. PCR products were electrophoresed in agarose gels, gel purified and cloned into pDRIVE cloning vector (Qiagen). Eight to 18 independent plasmids were sequenced for each RACE reaction.
RNA FISH
Schizosaccharomyces pombe cells were grown to mid-log phase and fixed in 4% paraformaldehyde for 1 h at RT. Cells were washed three times in 0.1 M sodium phosphate buffer (pH 6.0) and treated with 1 mg/ml Novozym (Sigma Aldrich) and 5 mg/ml Zymolyase 20 T (Seikagaku Biobusiness) dissolved in PEMS buffer (100 mM PIPES pH 6.9, 0.1 mM MgCl2, 1 mM EGTA, 1.2 M sorbitol) for 20 min at 37°C. Spheroplasts were washed three times in 300 µl of PEMS buffer without enzymes and 50 µl of cell suspension were spotted onto poly-l-lysine coated multiwell slides. Cells were incubated successively in 70, 85 and 100% ethanol and air-dried. Cells were blocked in prehybridization solution [4× SSC, 5× Denhardt solution, 1 mg/ml E. coli tRNA (Sigma Aldrich)] for 30 min at RT and successively hybridized using 2.5 ng/ml cy3-conjugated oligonucleotides oC or oG (Supplementary Table S1) at 42°C for 16 h in a humid chamber. After hybridization, slides were washed four times in 4× SSC at 42°C for 10 min each, stained with 100 ng/ml DAPI dissolved in 1× PBS and mounted in Vectashield medium (Vector Laboratories). Images were acquired using the Axiovert 200 M epifluorescence microscope (Zeiss) equipped with an ORCA-ER CCD camera (Hamamatsu) and analyzed using the Adobe Photoshop software.
Chromatin immuno-precipitation
For each sample, 150 ml of yeast culture (OD595: ∼1) was cross-linked in 1% formaldehyde for 30 min and successively quenched in 125 mM Glycine for 5 min. Cross-linked material was resuspended in 400 µl of lysis buffer [50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, protease inhibitor cocktail (Roche)] and subjected to mechanical lysis with glass beads using the FastPrep FP220 apparatus (three times 4.5 m/s for 45 s). Lysates were centrifuged for 30 min at 16 000g and pellets were resuspended in 500 µl of lysis buffer and sonicated in a Bioruptor UCD-200. Sonicated material was centrifuged for 5 min at 10 000g and supernatant containing fragmented chromatin was recovered. For each IP, an equivalent of 25 ODs was immunoprecipiated using 2.4 µg of anti-Phospho RNA Polymerase II (S2) antibody (A300-654A, Bethyl Laboratories), or 2.4 µg of anti-Phospho RNA Polymerase II (S5) antibody (A300-655 A, Bethyl Laboratories), or 3 µl of anti-RNA Polymerase II antibody (17-672, Millipore). Immunoprecipitations were performed on a rotating wheel at 4°C in presence of 25 µl Protein A/G sepharose beads (GE-Healthcare). Beads were washed three times in lysis buffer, once in lysis buffer containing 500 mM NaCl, once in wash buffer (10 mM Tris–HCL, 0.25 M LiCl, 0.5% Nonidet P40, 05% sodium deoxycholate) and once in lysis buffer. Immunoprecipitated chromatin was eluted in elution buffer (1% SDS, 100 mM Sodium Bicarbonate, 40 µg/ml RNAse A) and incubated at 37°C for 1 h. Cross-links were reversed at 65°C for 16 h. DNA was purified with the Wizard SV Gel and PCR Clean-Up System (Promega). Quantitative real-time PCR was performed using the Rotor-Gene Q light cycler (Qiagen) and the LightCycler 480 SYBR Green I master mix (Roche). Primers for TERRA TSS were TSS-Fw and TSS-Rv; primers for act1 were act1RT-Fw act1RT-Rv (Supplementary Table S1). The levels of cellular RNAPII molecules were monitored by standard western blot procedures using the same antibodies used for chromatin immuno-precipitation (ChIP).
RESULTS
Independent transcripts originate from fission yeast chromosome ends
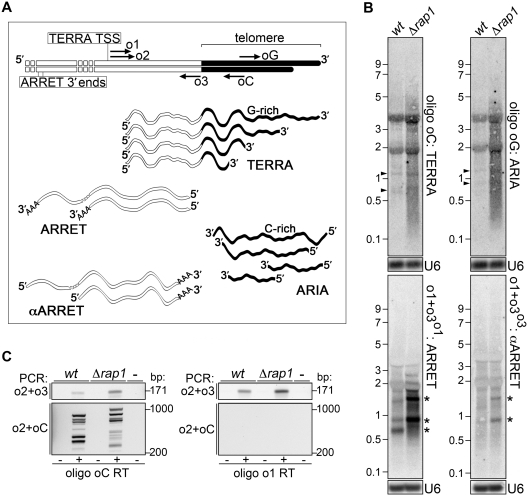
To test whether fission yeast chromosome ends are transcribed, we extracted total RNA from wild-type (wt) and from mutant yeasts deleted for the telomeric factor Rap1. This deletion leads to telomere elongation and derepression of telomere position effect (TPE) (20,21), conditions expected to facilitate detection of TERRA. RNA hybridization using random primer-labeled probes corresponding to the S. pombe telomeric sequence produced a prominent, disperse RNase-sensitive hybridization smear primarily between 2 and 0.1 kb but extending up to ∼5 kb in Δrap1 samples. A hybridization signal around 0.5 kb was barely detectable in wt samples (Supplementary Figure S1A). We then hybridized identical RNA membranes with oligonucleotides complementary to the G-rich or the C-rich strand of S. pombe telomeres (oligonucleotides oC and oG, respectively; see Figure 1A). The two oligonucleotide probes hybridized to digested genomic DNA with similar efficiencies (Supplementary Figure S1B). Unexpectedly, hybridization smears similar in size distribution and intensity were observed with both oligonucleotide probes in Δrap1 RNA samples (Figure 1B). Two discrete faint bands of ∼0.7 and 1.1 kb were also observed in wt and Δrap1 samples with both oligonucleotides (see arrowheads in Figure 1B). Thus, unlike budding yeast and mammals (5–7), fission yeast contains not only TERRA but also C-rich telomeric RNA repeats and Rap1 restricts the cellular levels of both telomeric repeat-containing RNA species. We named this new C-rich telomeric RNA ARIA (the Italian noun for air, the complementary element of earth, in Italian TERRA). Given the repetitive nature of TERRA and ARIA sequences, the telomeric oligonucleotides are expected to base pair more often with longer molecules, thereby producing a stronger hybridization signal. Considering that the intensity of TERRA and ARIA hybridization smears are similar throughout the covered range of molecular weights (Figure 1B), we conclude that the median length of TERRA and ARIA molecules is shorter than the one of the entire telomeric tract. Interestingly, similar experiments performed in Δrap1 strains generated in an independent genetic background revealed that ARIA steady-state levels are higher than the ones of TERRA (data not shown; see also Greenwood and Cooper, this issue), suggesting strain-specific mechanisms of regulation.
Figure 1.
(A) Schematic representation of S. pombe chromosome ends and telomeric transcriptome. Telomeric repeats are in black and subtelomeric sequences in white. Oligonucleotides used for RT–PCR and/or northern blotting are indicated by arrows. The sketch is not in scale. (B) RNA isolated from wt and Δrap1 cells was hybridized with the indicated probes. Exposures and signal detection were performed in parallel for TERRA and ARIA as well as for ARRET and αARRET. The black arrowheads indicate two discrete bands of ∼0.7 and 1.1 kb detected by oC and oG oligonucleotides and of so far unknown origin. The asterisks indicate major hybridization bands corresponding to ARRET and αARRET RNA species. U6 snRNA was used as loading control. Molecular weights are on the left in kb. (C) Total RNA from wt and Δrap1 strains was reverse transcribed (RT) using oC (left panels) or o1 (right panels) oligonucleotides and cDNA was PCR amplified using o2+o3 or o2+oC oligonucleotides. The most right samples correspond to control reactions where template was omitted. Molecular weights are on the right in bp.
The 5′-rapid amplification of cDNA ends (RACE) experiments performed using oC oligonucleotides for reverse transcription identified a unique transcription start site (TSS) for TERRA both in wt and in Δrap1 cells. TERRA TSS lies within the TAS1 telomere-associated sequences present on chromosomes I and II ends and it coincides with a subtelomeric Adenine positioned 211 nt upstream of the first telomeric repeat (Figure 1A and Supplementary Figure S2). Therefore, TERRA also exists in wt yeast and—like in other organisms—TERRA transcription starts within subtelomeric regions and proceeds toward chromosome ends (5,6). Given the high sequence conservation among TAS1 sequences from different chromosome ends, TERRA molecules originating from independent chromosomes are expected to be identical. In addition, because our RACE protocol only allows amplification of RNA molecules with a methylated 5′ cap, at least a fraction of S. pombe TERRA is 5′ methylated. Finally, while TERRA TSS is only a few hundreds base pair away from the telomeric tract, TERRA molecules are up to ∼5 kb long in Δrap1 cells (Figure 1B and Supplementary Figure S1A). We conclude that single TERRA molecules in the mutant strain are mostly composed of telomeric RNA repeats and their length heterogeneity stems mainly from 3′-ends.
We then hybridized wt and Δrap1 RNA using strand-specific probes corresponding to a 171 bp subtelomeric genomic sequence comprised between TERRA TSS and the telomeric tract. The 171 bp fragment was PCR amplified from genomic DNA with o1+o3 oligonucleotides and radioactively labeled by extension of either o1 or o3 primer (Figure 1A and Supplementary Figure S2). Hybridizations with probes labeled with o1 oligonucleotides (probe o1+o3o1) produced discrete RNase-sensitive hybridization fragments of ∼1.6 and 0.8 kb both in wt and Δrap1 samples. An additional band of ∼0.6 kb was detected in wt samples (Figure 1B and Supplementary Figure S1A). These subtelomeric RNA species transcribed in the opposite direction of TERRA likely correspond to ARRET, originally described in budding yeast (6). The 3′-RACE experiments identified two genomic Guanines placed 1904 and 1897 nt upstream of the first telomeric repeat both in wt and Δrap1 strains and few base pairs away from two putative polyadenylation signal sequences (Figure 1A and Supplementary Figure S2). These 3′-ends can account for the ∼1.6 kb ARRET molecules detected by northern blot (Figure 1B). Because shorter ARRET species were also detected, we infer that alternative 3′-ends should be located downstream to the ones identified in our RACE experiments. Indeed, while the identified 3′-ends are embedded within TAS2 elements, highly homologous (∼90%) sequences exist within TAS1 elements and extend for ∼400 bp upstream of the first telomeric repeat.
Hybridizations with complementary probes (o1+o3o3) revealed faint discrete hybridization products of ∼1.6 and 0.8 kb mostly evident in Δrap1 samples (Figure 1B and Supplementary Figure S1A). No obvious hybridization smear was generated by these probes, which are expected to detect TERRA molecules through base pairing with their subtelomeric tract. Our interpretation is that o1+o3o3 probes primarily detect subtelomeric RNA molecules that are more abundant than TERRA but transcribed in the same orientation. The lengths of these molecules are strikingly similar to the ones of ARRET molecules (Figure 1B) and we dubbed these RNA anti-ARRET (αARRET). While ARRET-associated hybridization signal is obviously more intense than the one associated to αARRET, o1+o3o1 probes generated less intense signals than o1+o3o3 probes when hybridized to genomic DNA (Supplementary Figure S1B). Therefore, ARRET is considerably more abundant than αARRET. As with TERRA and ARIA, Rap1 negatively regulate the steady-state levels of ARRET (Figure 1B), and to much lower extents of αARRET.
We then performed RT-PCR experiments using different combinations of oligonucleotides for RT and successive PCR reactions (Figure 1A and Supplementary Figure S2). First, we reverse transcribed total RNA using oC oligonucleotides and performed PCR amplifications with o2+o3 oligonucleotides. We obtained amplicons of the expected 171 bp in both wt and Δrap1 RNA samples (Figure 1C), confirming the existence in both strains of canonical TERRA molecules composed of subtelomeric and telomeric sequences. We then reverse transcribed RNA with o1 oligonucleotides and performed o2+o3 PCR amplifications. Again, we obtained the expected 171 bp amplicons both in wt and Δrap1 strains (Figure 1C). These amplification products could derive from either ARRET or ARIA molecules, if ARIA contained subtelomeric sequences. To distinguish, we PCR amplified the same cDNA samples with o2+oC oligonucleotides. No amplification product was produced with cDNA prepared with o1 oligonucleotides, while several discrete amplicons were produced with cDNA prepared with oC oligonucleotides (Figure 1C). The sequence of these amplicons corresponded to DNA fragments initiating with the o2 oligonucleotide and ending at different positions within the telomeric tract (data not shown). This confirmed that the 171 bp amplification products obtained using o2+o3 oligonucleotides for PCR of cDNA prepared using oC oligonucleotides are derived from RNA molecules containing both telomeric and subtelomeric sequences (i.e. TERRA). In contrast, the 171 bp amplification products obtained using o2+o3 oligonucleotides for PCR of cDNA prepared using o1 oligonucleotides likely originate from RNA molecules with subtelomeric sequences devoid of telomeric repeats (i.e. ARRET). Moreover, ARIA molecules appear to be largely or completely devoid of subtelomeric sequences. In summary, four independent RNA species emanate from fission yeast chromosome ends: TERRA, ARIA, ARRET and αARRET (see sketch in Figure 1A). We refer to the ensemble of these transcripts as ‘the telomeric transcriptome’.
RNAPII is involved in the biogenesis of the S. pombe telomeric transcritptome
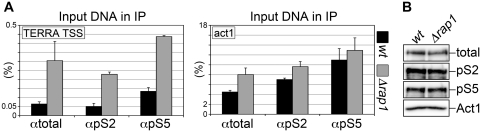
To test whether RNAPII sustains transcription of S. pombe chromosome ends, we first performed ChIPs using antibodies directed against total RNAPII or against RNAPII C-terminal domain (CTD) repeats phosphorylated at Serine 2 (pS2) or Serine 5 (pS5). Co-immunoprecipitated DNA was subjected to real-time quantitative PCR using oligonucleotides spanning TERRA TSS. Although at low levels (0.05–0.15% of input DNA), TERRA TSS DNA was reproducibly recovered in wt ChIP samples with all antibodies above control ChIPs performed using only beads. A 6- to 9-fold increase over wt was measured in Δrap1 samples, while the cellular levels of total or phosphorylated RNAPII were not altered by rap1+ deletion (Figure 2A and B). PCR amplifications using primers specific for the highly transcribed act1+ gene validated our ChIP protocols (Figure 2A). Hence, RNAPII is bound to TERRA TSS in vivo and Rap1 negatively regulates such binding. We conclude that the increased levels of TERRA—and perhaps of the entire telomeric transcriptome—observed in Δrap1 strains (Figure 1B) are likely to derive at least in part from increased RNAPII-mediated transcription of chromosome ends.
Figure 2.
(A) Chromatin isolated from wt and Δrap1 cells was immuno-precipitated using antibodies against RNAPII C terminal domain repeats either unmodified (αtotal) or phosphorylated at Serine 2 (αpS2) or Serine 5 (αpS5). Quantitative real-time PCR was performed using primers flanking TERRA TSS (left graph) or amplifying a fragment from the highly transcribed RNAPII substrate gene act1 (right graph; positive control). Graphs show the fraction of input DNA retrieved in the different samples, after subtraction of the background signal measured for control reactions performed using only beads. Bars and error bars are averages and standard deviations from three independent experiments. (B) Total proteins were extracted from wt and Δrap1 strains and analyzed by western blot with the same antibodies used for ChIP. Act1 was used as loading control.
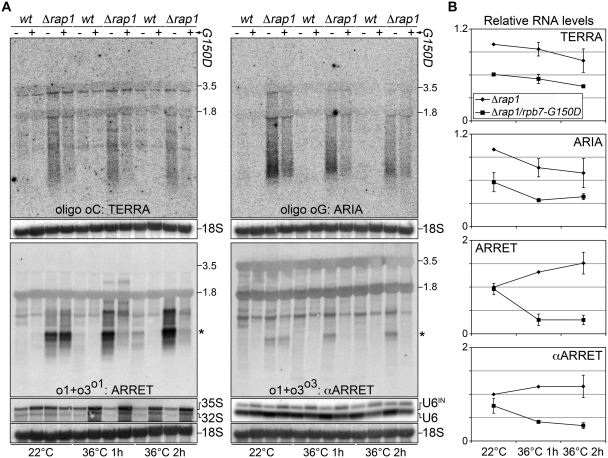
We next exploited a ts allele of the RNAPII subunit Rpb7 (rpb7-G150D), which becomes fully dysfunctional when cells are grown at the restrictive temperature 36°C (22). We generated rap1+ deletion strains carrying the rpb7-G150D mutation and prepared total RNA from cells grown at the permissive temperature 22°C, or incubated at 36°C for 1 or 2 h. TERRA and ARIA steady-state levels were already reduced at permissive temperatures in Δrap1/rpb7-G150D cells as compared to single rap1+ deletion counterparts. In addition, the temperature shift induced a gradual loss of TERRA and ARIA levels, independent of the Rpb7 status (Figure 3A and B). In contrast, ARRET and αARRET transcripts were unaffected in rpb7-G150D mutants at permissive temperatures, while a sudden decline of their levels was observed upon shift to restrictive temperatures only in strains carrying the mutation (Figure 3A and B). The steady-state levels of the unstable 35S and 32S precursor ribosomal RNA and unspliced U6 snRNA, which are transcribed by RNAPI and RNAPIII, respectively, were not diminished in rpb7-G150D strains at restrictive temperatures (Figure 3A) indicating that RNAPI and RNAPII were still functional. We conclude that Rpb7 most probably sustains ARRET and αARRET transcription, while it remains to be carefully determined whether and to what extent it participates in TERRA and ARIA biogenesis.
Figure 3.
(A) Wt and Δrap1 strains carrying intact Rpb7 or the G150D mutation were grown at permissive temperature (22°C) for several generations and then shifted to restrictive temperature (36°C) for 1 or 2 h. Total RNA was isolated and subjected to northern blot analyses as in Figure 1B. After signal detection, membranes were stripped and hybridized using probes detecting 35S and 32S precursor rRNAs (unstable RNAPI transcripts), intron-containing U6 snRNA (U6IN; unstable RNAPIII transcript) and 18S rRNA (loading control). Molecular weights are on the left in kb. The asterisks indicate the ARRET and αARRET hybridization signals used for quantifications. (B) Quantifications of TERRA, ARIA, ARRET and αARRET levels at permissive and restrictive temperatures. Values were normalized through the relative 18S values and expressed as fold increase over Δrap1 at 22°C. Values and error bars are averages and standard deviations from two independent experiments.
Polyadenylation of the S. pombe telomeric transcriptome
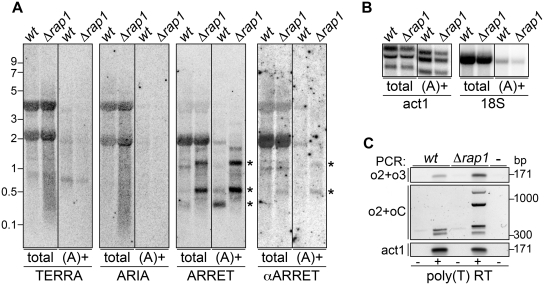
To analyze the polyadenylation state of the S. pombe telomeric transcriptome, we prepared poly(A)+ RNA from wt and Δrap1 cells, and subjected it to northern blot hybridization (Figure 4A and B). Confirming the efficiency of our fractionation protocol, poly(A)+ act1+ mRNA was readily detected in all poly(A)+ fractions while only traces of poly(A)− 18S ribosomal RNA (rRNA) were detected in the same fractions (Figure 4B). Hybridizations with oC and oG oligonucleotides produced only very faint signals in the poly(A)+ fractions of wt and Δrap1 samples. In contrast, ARRET and αARRET species were present in the poly(A)+ fractions at ratios comparable to that measured for act1+ mRNA (Figure 4A and B).
Figure 4.
(A) Total RNA from wt and Δrap1 strains was subjected to poly(A)+ fractionation and corresponding volumes of total and poly(A)+ RNA were hybridized to detect TERRA, ARIA, ARRET and αARRET. The asterisks indicate the major hybridization bands corresponding to ARRET and αARRET RNA species. Molecular weights are on the left in kb. (B) The same RNA as in A was hybridized using probes detecting the polyadenylated act1+ mRNA and the non-polyadenylated 18S rRNA. (C) Total and poly(A)+ RNA was RT using poly(T) oligonucleotides and cDNA was PCR amplified using o2+o3 or o2+oC oligonucleotides. The most right sample corresponds to a control reaction where template was omitted. Molecular weights are on the right in bp.
We then reverse transcribed wt and Δrap1 RNA with poly(T) oligonucleotides and amplified cDNA with o2+o3 oligonucleotides (Figure 1A and Supplementary Figure S2). For both strains we obtained the expected 171 bp amplicons. Amplification of the same cDNA using o2+oC oligonucleotides produced several amplification products in both cDNA samples (Figure 4C), confirming the presence of telomeric repeats at least in some cDNA molecules. These results imply that the majority of ARRET and αARRET molecules and a minor fraction of TERRA molecules are polyadenylated. Our experimental set up does not allow us to distinguish between amplification products originating from different RNA species, nor to estimate whether a minor fraction of ARIA is polyadenylated.
The non-canonical poly(A) polymerases Cid12 and Cid14 differentially regulate the cellular levels of the telomeric transcriptome
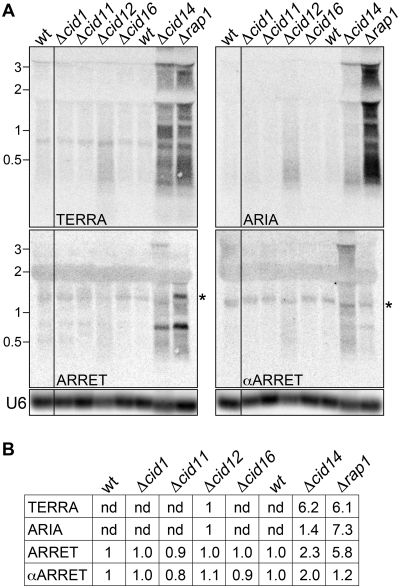
As already mentioned, inactivation of canonical poly(A) polymerase activity led to TERRA disappearance in budding yeast (6). In fission yeast, inactivation of the canonical polyA polymerase Pla1 is lethal (23) and conditional pla1+ mutants are not available. Therefore, we were unable to test to what extent this enzyme regulates the telomeric transcriptome. On the other side, several non-essential, non-canonical poly(A) polymerases, belonging to the Cid1 family, exist in fission yeast (24). We prepared total RNA from exponentially growing cells deleted for the non-canonical poly(A) polymerases Cid1, Cid11, Cid12, Cid16 and Cid14. Northern blot analysis revealed that the telomeric transcriptome was differentially affected by cid12+ and cid14+ deletions. In Δcid12 mutants, both TERRA and ARIA species were elevated when compared to wt cells, but they did not reach the levels measured in Δrap1 strains. On the contrary, the steady-state levels of ARRET and αARRET were not altered in a noticeable manner (Figure 5A and B). In Δcid14 cells, TERRA molecules were readily detected and were similar both in size and abundance to the ones detected in Δrap1 cells. ARIA and ARRET steady-state levels were also elevated as compared to wt, although to lower extents than in Δrap1 cells. Finally, αARRET RNA was elevated by 2-fold in the cid14+ deleted strain (Figure 5A and B).
Figure 5.
(A) RNA isolated from the indicated strains was hybridized as in Figure 1B. Two membranes obtained from blotting of gels run in parallel were first hybridized to detect TERRA and ARIA. After parallel exposures and signal detection, the same membranes were stripped and hybridized to detect ARRET and αARRET and successively to detect U6 snRNA (loading control). The asterisks indicate the hybridization bands used for ARRET and αARRET quantifications. Molecular weights are on the left in kb. (B) Quantifications of northern blots shown in (A). TERRA, ARIA, ARRET and αARRET hybridization signals were normalized through the relative U6 signals and expressed as fold increase over Δcid12 samples for TERRA and ARIA and wt samples for ARRET and αARRET. nd: not determined.
Schizosaccharomyces pombe TERRA and ARIA localize to the nucleus
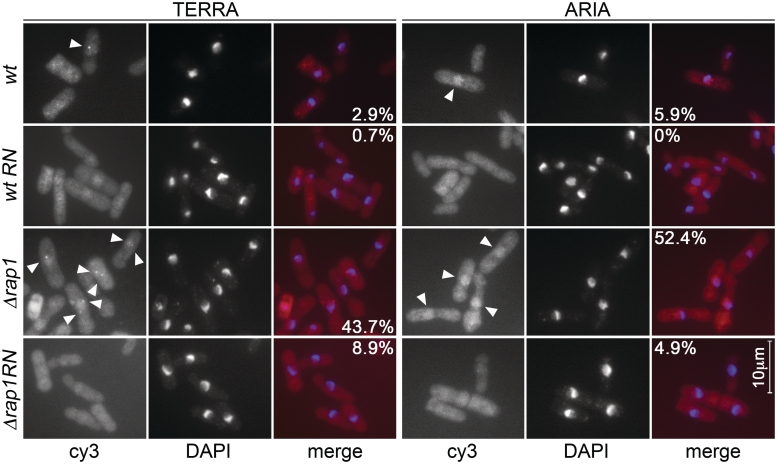
To determine the cellular localization of TERRA and ARIA, we performed RNA-FISH on fixed cells using cyanine 3 (cy3)-conjugated oC and oG oligonucleotides. Consistent with the northern blot results, rare (∼3–6%) stained cells were observed in wt samples. In approximately half of Δrap1 cells both probes generated positive hybridization signals confined to DAPI stained nuclear territories (Figure 6). RNase A treatment of fixed cells prior to hybridization substantially reduced the number of positive cells in both backgrounds, confirming that the probes hybridized to RNA molecules (Figure 6). While the signal detected with oG probes was diffused within the nucleus, the signal obtained with oC probes formed 1–3 discrete foci in all positive cells (Figure 6). We conclude that ARIA, or most of it, is nucleoplasmic while TERRA is recruited to discrete chromatin domains at least in part independently of Rap1. Attempts to combine RNA-FISH with immunostaining of telomeric proteins were unsuccessful. Nonetheless, bearing in mind that mammalian TERRA remains associated to telomeres throughout the cell cycle (2,5), we suspect that the TERRA foci visualized in our experiments localize to chromosome ends.
Figure 6.
Exponentially growing wt and Δrap1 cells were hybridized in situ using cy3-conjugated oC or oG oligonucleotides to detect TERRA and ARIA, respectively. Control cells were treated with RNase A (RN) prior to hybridization. In the merge panels TERRA and ARIA are in red, DAPI stained DNA in blue. Numbers indicate the fraction of cells displaying positive hybridization signals in the different samples. White arrowheads point to example of positively stained nuclei.
DISCUSSION
The telomeric transcriptome of the model organism S. pombe comprises a complex array of independent transcripts deriving from chromosome ends. As for budding yeast (6), fission yeast cells contain TERRA and ARRET molecules. In addition, we report here for the first time the existence of two novel classes of chromosome end-derived transcripts, ARIA and αARRET. The novel αARRET species should be investigated in other model systems, since we are not aware of published strategies that would have detected them. On the contrary, attempts to detect ARIA failed in mammalian and budding yeast cells (4–8). Nevertheless, it remains possible that rapidly degraded, unstable ARIA transcripts are produced also in mammals and budding yeast. A recent study from the Riha laboratory showed that the ends of several Arabidopsis chromosomes are transcribed into TERRA molecules. Furthermore, northern blot hybridization of Arabidopsis RNA revealed C-rich telomeric repeats forming a diffuse hybridization signal (8). Although the majority of this RNA appears to stem from transcription of centromeric loci containing relics of telomeric DNA, the authors of this study concluded that the telomeric fraction of this C-rich telomeric RNA corresponded to ARRET transcripts (8). Our results challenge this conclusion and suggest that ARIA molecules could be present at readily detectable levels also in plants. A detailed characterization of the telomeric transcriptome of Arabidopsis will clarify to what extent it resembles the one of S. pombe.
As with other organisms, RNAPII plays a major role in the biogenesis of the telomeric transcriptome of fission yeast. The physical association of active RNAPII to S. pombe TERRA TSS suggests that TERRA is indeed the outcome of RNAPII-mediated transcription of telomeric tracts. This hypothesis is further substantiated by the fact that the telomeric factor Rap1 negatively regulates the association of RNAPII to TERRA TSS and increased TERRA cellular levels are measured in Δrap1 strains as compared to wt. Because ARRET and αARRET molecules contain TERRA TSS sequences, it is likely that the detected binding of RNAPII to this region derives not only from RNAPII molecules actively transcribing TERRA, but also ARRET and/or αARRET. We have also shown that the essential RNAPII subunit Rpb7 sustains ARRET and αARRET cellular levels. The rpb7-G150D mutation used in this study was previously shown to compromise transcription initiation of reverse centromeric dg-dh repeats through binding to their promoters (22). Although the existence of ARRET and αARRET promoters have not been demonstrated yet, it is plausible that Rpb7 could mediate transcription initiation of ARRET, αARRET or both.
Complementary centromeric dg-dh repeat transcripts are precursors of centromeric siRNAs, which, in turn, mediate establishment of centromeric heterochromatin (25–27). The complementary nature of ARRET and αARRET raises the possibility that they might form double-stranded RNA intermediates, successively processed into siRNA molecules in vivo. Indeed, subtelomeric siRNAs have been identified in S. pombe, although genomic mapping assigned them to loci laying several kilo bases upstream of ARRET 3′-ends (28). It will be important to directly search for ARRET-like siRNA sequences and, if they exist, test their putative functions in establishing heterochromatin at chromosome ends.
TERRA and ARIA molecules could also base pair in vivo as to give rise to telomeric siRNA molecules similar to the ones described in mammalian and plant cells (8,29). Further supporting this hypothesis, both TERRA and ARIA are upregulated in Δcid12 strains. Cid12 associates with the RNA-directed RNA polymerase Rdp1 and with the RNA helicase Hrr1 to form a catalytically active RNA-directed RNA polymerase complex (RDRC) (30). Inactivation of Rdp1 or Cid12 leads to derepression of centromeric gene silencing and impaired production of centromeric siRNA molecules (30). It is possible that the accumulation of TERRA and ARIA in Δcid12 cells is the outcome of compromised processing of these molecules into siRNA. Alternatively, the RDRC complex could contribute to the establishment of telomeric heterochromatin and the elevated levels of TERRA and ARIA in Δcid12 mutants might underscore a lack of telomeric silencing. These results also suggest that canonical RDRC activity does not contribute to the transcription of the telomeric transcriptome, a notion further substantiated by the fact that the steady-state levels of TERRA, ARIA, ARRET and αARRET were not diminished in Δrdp1 cells (data not shown; see also Greenwood and Cooper, this issue).
ARRET and αARRET species are fully polyadenylated, while TERRA, and perhaps ARIA, only marginally. Polyadenylation of non-coding RNAs can lead to their destabilization through nuclear RNA degradation pathways in several eukaryotes. For example, S. cerevisiae strains deficient for the nuclear exosome component Rrp6p or for members of the TRAMP polyadenylation complex such as Trf4p accumulated polyadenylated transcripts originating from RNAPII-mediated transcription of intragenic regions (31–34). Similarly, budding yeast TERRA and ARRET transcripts accumulated in Δtrf4 strains (6). Because, Cid14 is the pombe functional homolog of Trf4p (24), our results suggest an evolutionary conservation of the TRAMP complex-associated roles in muting the telomeric transcriptome. Intriguingly, budding yeast TERRA was rapidly destabilized upon functional inactivation of the poly(A) polymerase Pap1p (6) and the poly(A)+ fraction of human TERRA had a longer half life than its poly(A)− counterpart (12). Altogether, these observations portray a scenario where the polyadenylation state of the different RNA species of the telomeric transcritpome is differentially dictated by alternative poly(A) polymerases as to direct them toward or protect them from degradation pathways.
In conclusion, our extensive analysis of the telomeric transcriptome of S. pombe has revealed that transcription of chromosome ends gives rise to an intricate array of RNA species more complex than previously anticipated. We have also started to unravel the molecular networks sustaining the biogenesis and regulation of the S. pombe telomeric transcriptome. We anticipate that our results will pave the way for a large body of research aiming at understanding the molecular roles associated to the telomeric transcriptome in a genetically tractable model organism bearing telomeres closely resembling those of humans.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures S1 and S2, Supplementary Table S1.
FUNDING
European Research Council (grant BFTERRA); the Swiss National Science Foundation (grant numbers 3100A0-120090 and PP00P3-123356); Fondazione Cariplo (grant number 2008–2507). Funding for open access charge: European Research Council (grant BFTERRA).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Julie P. Cooper and Jessica Greenwood (Cancer Research UK, London) for reagents and for sharing unpublished results. They thank Karl Ekwall (Karolinska Institutet, Stockholm) for the Rpb7-G150D strains, Marc Bühler (FMI, Basel) for the cid14+ deleted strain and the ETHZ Light Microscopy Center for microscope services. They are also grateful to the members of the Azzalin laboratory for helpful discussions and to Alicia Smith for critical reading of the manuscript.
REFERENCES
- 1.Arora R, Brun CM, Azzalin CM. TERRA: long noncoding RNA at eukaryotic telomeres. Prog. Mol. Subcell. Biol. 2010;51:65–94. doi: 10.1007/978-3-642-16502-3_4. [DOI] [PubMed] [Google Scholar]
- 2.Chawla R, Azzalin CM. The telomeric transcriptome and SMG proteins at the crossroads. Cytogenet. Genome Res. 2008;122:194–201. doi: 10.1159/000167804. [DOI] [PubMed] [Google Scholar]
- 3.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solovei I, Gaginskaya ER, Macgregor HC. The arrangement and transcription of telomere DNA sequences at the ends of lampbrush chromosomes of birds. Chromosome Res. 1994;2:460–470. doi: 10.1007/BF01552869. [DOI] [PubMed] [Google Scholar]
- 5.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 6.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 7.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell. Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 8.Vrbsky J, Akimcheva S, Watson JM, Turner TL, Daxinger L, Vyskot B, Aufsatz W, Riha K. siRNA-mediated methylation of Arabidopsis telomeres. PLoS Genet. 2010;6:e1000986. doi: 10.1371/journal.pgen.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nergadze SG, Farnung BO, Wischnewski H, Khoriauli L, Vitelli V, Chawla R, Giulotto E, Azzalin CM. CpG-island promoters drive transcription of human telomeres. RNA. 2009;15:2186–2194. doi: 10.1261/rna.1748309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farnung BO, Giulotto E, Azzalin CM. Promoting transcription of chromosome ends. Transcription. 2010;1:140–143. doi: 10.4161/trns.1.3.13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azzalin CM, Lingner J. Telomeres: the silence is broken. Cell Cycle. 2008;7:1161–1165. doi: 10.4161/cc.7.9.5836. [DOI] [PubMed] [Google Scholar]
- 12.Porro A, Feuerhahn S, Reichenbach P, Lingner J. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol. Cell. Biol. 2010;30:4808–4817. doi: 10.1128/MCB.00460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flynn RL, Centore RC, O'Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 2011;471:532–536. doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38:5797–5806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandell LL, Gottschling DE, Zakian VA. Transcription of a yeast telomere alleviates telomere position effect without affecting chromosome stability. Proc. Natl Acad. Sci. USA. 1994;91:12061–12065. doi: 10.1073/pnas.91.25.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabatinos SA, Forsburg SL. Molecular genetics of Schizosaccharomyces pombe. Methods Enzymol. 2010;470:759–795. doi: 10.1016/S0076-6879(10)70032-X. [DOI] [PubMed] [Google Scholar]
- 18.Bähler J, Wu J-Q, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 19.Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol. 2001;11:1624–1630. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- 21.Miller KM, Ferreira MG, Cooper JP. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 2005;24:3128–3135. doi: 10.1038/sj.emboj.7600779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19:2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tallada VA, Bridge AJ, Emery PA, Hagan IM. Suppression of the Schizosaccharomyces pombe cut12.1 cell-cycle defect by mutations in cdc25 and genes involved in transcriptional and translational control. Genetics. 2007;176: 73–83. doi: 10.1534/genetics.107.072090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevenson AL, Norbury CJ. The Cid1 family of non-canonical poly(A) polymerases. Yeast. 2006;23:991–1000. doi: 10.1002/yea.1408. [DOI] [PubMed] [Google Scholar]
- 25.Provost P, Silverstein RA, Dishart D, Walfridsson J, Djupedal I, Kniola B, Wright A, Samuelsson B, Radmark O, Ekwall K. Dicer is required for chromosome segregation and gene silencing in fission yeast cells. Proc. Natl Acad. Sci. USA. 2002;99:16648–16653. doi: 10.1073/pnas.212633199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volpe T, Schramke V, Hamilton GL, White SA, Teng G, Martienssen RA, Allshire RC. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11:137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- 27.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 28.Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 29.Cao F, Li X, Hiew S, Brady H, Liu Y, Dou Y. Dicer independent small RNAs associate with telomeric heterochromatin. RNA. 2009;15:1274–1281. doi: 10.1261/rna.1423309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to non-coding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 31.Carneiro T, Carvalho C, Braga J, Rino J, Milligan L, Tollervey D, Carmo-Fonseca M. Depletion of the yeast nuclear exosome subunit Rrp6 results in accumulation of polyadenylated RNAs in a discrete domain within the nucleolus. Mol. Cell. Biol. 2007;27:4157–4165. doi: 10.1128/MCB.00120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houseley J, Kotovic K, El Hage A, Tollervey D. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 2007;26:4996–5006. doi: 10.1038/sj.emboj.7601921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 34.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.