Abstract
While telomere repeat-containing non-coding RNA has been identified in a variety of eukaryotes, its biological role is not yet clear. We have identified telomeric transcripts in fission yeast, a model system that combines precise genetic manipulability with telomeres remarkably similar to those of human. Like human and budding yeast, fission yeast harbours a population of telomeric RNA molecules containing G-rich telomeric repeats transcribed from the subtelomere to the telomere. In addition, we detect substantial levels of C-rich telomeric RNA whose appearance is independent of the RNA-dependent RNA polymerase, suggesting that the telomere repeats themselves serve as promoter sites; multiple distinct subtelomeric RNAs are also present. The regulation of these transcripts depends on the telomere-associated proteins Taz1 and Rap1, as deletion of taz1+ or rap1+ leads to increased levels of both telomere repeat-containing and subtelomeric transcripts. In contrast, loss of the heterochromatin proteins Swi6 or Clr4 or the telomerase regulator Rif1 results in elevated subtelomeric RNA levels while telomere-repeat containing transcript levels remain repressed. Coupled with the large body of knowledge surrounding the functions of telomeric and heterochromatin factors in fission yeast, these in vivo analyses suggest testable models for the roles of TERRA in telomere function.
INTRODUCTION
The telomere field was rocked in 2007 by the revelation that rather than being transcriptionally silent, as had been implied by the long-recognized existence of the telomere position effect, human telomeres are transcribed to produce non-coding RNAs (1,2). These RNA species, dubbed TERRA, generally correspond to the telomeric G-rich strand, originate from promoters in the subtelomere, range in length from 0.1 to ≥9.0 kb, and associate at least to some extent with telomeres in the nucleus. TERRA appears widely conserved, having since been identified not only in mammals, but also in zebrafish, birds, plants and budding yeast (1–5).
Tremendous progress has been made in defining TERRA biogenesis and its regulation. In human cells, G-rich telomeric RNA is transcribed from the subtelomere towards the telomere by RNA Pol II, and a fraction of TERRA is polyadenylated. Cytosine methylation of CpG dinucleotide-rich DNA islands comprising at least a subset of TERRA promoters inhibits transcription; TERRA is also modulated by histone methylation (6–8). TERRA can be detected as telomeric foci by RNA fluorescence in situ hybridization (1,2) and these foci increase in intensity upon depletion of components of the nonsense-mediated RNA decay (NMD) pathway (1). The budding yeast telomere- and promoter-binding protein Rap1 also negatively regulates TERRA expression via its interaction with the telomeric factors Rif1/2 and the silencing factors Sir2/3/4 (9).
Despite our ever-expanding knowledge of how TERRA biogenesis is controlled, a clear biological role for TERRA remains largely elusive. A role in telomerase inhibition has been proposed based on a number of experimental manipulations that reveal a correlation between TERRA accrual and telomere shortening. In human cells, the TERRA accumulation that accompanies depletion of NMD components is associated with sporadic telomere loss, suggesting that TERRA removal is important for telomere length maintenance (1). In budding yeast, mutating the 5′-to-3′ RNA exonuclease Rat1 leads to TERRA accumulation and telomere shortening. Interestingly, this telomere shortening can be rescued by overexpression of RNaseH, suggesting that RNA/DNA hybrids contribute to the TERRA overabundance phenotype (3). While it is formally possible that these effects could be indirect, an earlier study demonstrated that transcription of a single budding yeast telomere leads to shortening of that telomere (10), supporting the notion that an accumulation of telomeric transcripts limits telomerase action in cis. The sequence complementarity between TERRA and the template region of the RNA subunit of telomerase suggests a possible mechanism of inhibition through base pairing. This idea is supported by in vitro studies in which TERRA inhibits telomerase activity (11,12). Furthermore, TERRA is found in association with telomerase in human cell extracts, suggesting that a direct interaction between TERRA and telomerase could mediate telomerase inhibition (11). However, whether the anti-correlation between TERRA levels and telomere length reflects in vivo interactions with telomerase or with other players in telomere replication remains an open question.
A positive role for TERRA in promoting telomere integrity has also been suggested, as treatment of human cells with siRNA directed against TERRA leads to telomere dysfunction-induced foci, telomere loss and genomic instability. However, indirect effects of TERRA siRNAs on telomere integrity have thus far not been addressed (13). TERRA siRNA treatment also leads to a reduction of telomeric heterochromatin marks as assessed by chromatin immunoprecipitation (ChIP), highlighting a potential role for TERRA in heterochromatin regulation at the telomere. Finally, through its interaction with the heterogeneous nuclear ribonucleoprotein hnRNPA1, TERRA may regulate the fate of the 3′ single strand (ss) telomeric overhang by modulating the relative binding capacities of the major ss telomere binding proteins Pot1 and RPA, thus playing an important role in telomere replication and end protection (14). Verification of the biological validity of these ideas awaits the ability to manipulate TERRA specifically in vivo without simultaneously altering additional telomere components.
Studies of the fission yeast telomere have provided essential insights not only into the composition of mammalian telomeres, but also into mechanisms of telomerase recruitment, chromosome end-protection and semi-conservative telomeric DNA replication (15–18). Fission yeast telomeres and general heterochromatin recapitulate their human counterparts closely, for example harbouring the canonical heterochromatic histone mark H3K9Me3 as well as associated chromodomain proteins (Swi6/HP1), features that are absent from budding yeast. Here we investigate transcription of fission yeast telomeres and show that not only are they transcribed, but also multiple distinct RNA species exist and are regulated differentially by the telomere proteins Taz1, Rap1 and Rif1 as well as the heterochromatin assembly machinery. These observations add weight to the notion that telomeric transcripts are conserved and biologically important, and provide powerful tools for future studies into this mysterious non-coding RNA.
MATERIALS AND METHODS
Fission yeast strains
Standard media and growth conditions were used. Gene deletion and C-terminal tagging were achieved by one-step gene replacement (19,20). Strains used in this study are listed in Supplementary Table S1.
DNA isolation and Southern blotting
DNA isolation and Southern blot analysis were performed as described previously (17). The ds Telo probe was generated by amplifying telomere sequence in a PCR reaction with RedTaq polymerase, primers oC and oG and wt genomic DNA as a template. PCR conditions were 3′ 95°C, 35 cycles of 95°C for 30 s, 62°C for 20 s, 72°C for 20 s, followed by 72°C for 5′. The product (a smear) was then gel isolated. Loading control probe was generated by PCR for an internal genomic sequence in gene SPAC4A8.02c using primers 5′-CTCGTGACAACAACTTAGTC and 5′-ACTTTTGGGCGTTCAGAAGC.
RNA isolation and northern blotting
Cells of the indicated strains were grown in 10–50 ml rich media to OD600 of 0.6–0.8, and RNA was isolated using hot-phenol extraction (21). RNA (10–40 µg) was electrophoresed through 1.2% formaldehyde agarose gels and transferred to Hybond N+. membranes (Amersham Pharmacia). A total of 25 ng double-stranded telomere and subtelomeric probes were 32P labelled by random priming using the Prime-it Kit (Agilent Technologies) and 50 µM oligonucleotide probes were 5′-end labelled using T4 polynucleotide kinase (NEB). Probe Telo1 is derived from a genomic telomere that was digested with ApaI and cloned into pIRT2 and contains ∼270 bp telomere sequence and 32 bp of STE1. Tel80 comprises synthetic telomere repeats that were ligated and cloned into pBluescript, and thus contains no STE sequence (17). Probe Telo (Figure 1C) corresponds to the PCR product generated from the C-oligo and G-oligo primers using wt genomic DNA as a template. The STE1 and STE2 probes were made by digesting pNSU70 with ApaI + EcoRI and NsiI, respectively, and gel isolating the ∼750 bp and ∼3.5 kb bands, respectively. Oligonucleotide probe sequences are as follows: C-rich probe: 5′-GTACCCCTGTAACCCCTGTAACCTGTAACCGC and G-rich probe: 5′-GGTTACAGGTTACAGGGGTTACAGGGGTAC. Following pre-hybridization in Church Gilbert buffer, probes were added and hybridized for 16 h at 65°C for double-stranded probes, or 45°C for oligonucleotide probes. Blots were washed in 1× SSC, 0.1% SDS at the hybridization temperature.
Figure 1.
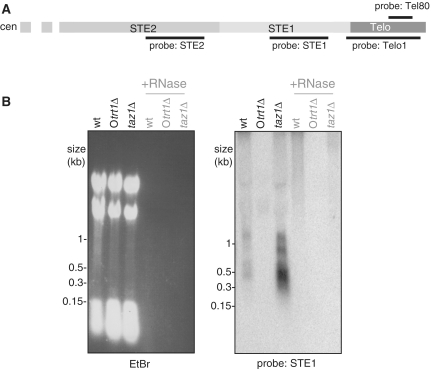
Telomeric RNA is detectable in wt and taz1Δ cells. (A) Schematic of the three fission yeast chromosomes with telomeres indicated by black rectangles versus the circular chromosomes of Otrt1Δ strains that lack telomeres entirely. (B) Northern blot analysis was performed on 40 µg total cellular RNA from each strain, with or without RNase A. The ethidium bromide (EtBr)-stained gel prior to transfer is shown on the left. The blot was hybridized with Telo1, which contains 250 bp of telomere repeats plus 32 bp of STE sequence (see text); hybridization to ribosomal RNA is also observed. The smear that appears above the telomeric RNA in wt and taz1Δ lanes is not RNase-sensitive, and therefore likely to represent residual DNA in the sample. (C) Southern blot of ApaI-digested genomic DNA hybridized with Telo. The blot was stripped and re-probed with an internal region to control for loading (below). (D) Telomeric RNA in taz1Δ cells is predominantly C-rich. Northern blot analysis of 20 µg total cellular RNA hybridized with strand-specific telomere probes. dsTelo: a dsDNA telomere-repeat containing fragment that allows comparison of hybridization intensity between G- and C-rich probes. Samples were run on the same gel and transferred, then the membrane was cut and probed with either a C-rich probe (left panel) to detect G-rich RNA, or a G-rich (right panel) probe to detect C-rich RNA. The G-rich probe detects telomeric RNA in taz1Δ samples, whereas the C-rich probe only detects bands that are present in Otrt1Δ samples as well and are therefore not telomere-specific; background hybridization to rRNA provides a loading control.
Reverse transcription PCR
RNA was treated with RNase-free DNase (Invitrogen) and 2.5 µg RNA was used as template in first strand synthesis by Superscript III (Invitrogen). The denaturation step was performed as follows: for act1R and o3, RNA + primer were incubated at 65°C for 5 min and placed on ice for >1 min; for oC and oG, 90°C for 1′ followed by cooling to 55°C over 1′. Reverse transcription (RT) reactions were treated with RNase A and 1 µl of the RT reaction was used in a PCR reaction with the primers indicated in Figure 2A. PCR with subtelomeric primers was carried out using RedTaq polymerase (Thermo Scientific), for a 3′ 95°C step, then 35 cycles of 95°C for 30 s, 62°C for 20 s, 72°C for 20 s, followed by 72°C for 5′. PCR for act1 was performed the same way, except an annealing temperature of 58°C was used for 20–25 cycles.
Figure 2.
Rdp1 is required neither to generate TERRA nor C-rich telomeric RNA. (A) RT-PCR reveals telomeric and subtelomeric RNA transcribed from the subtelomere outwards. Top: Schematic of telomeric region. Oligonucleotides used are designated as arrows pointing in the 5′-to-3′ direction. First strand synthesis with or without reverse transcriptase (RT+/−) using the primers indicated (first strand) was performed on RNA from the indicated strains. cDNA was then amplified with the oligonucleotides indicated (PCR). Genomic DNA (DNA ctrl) used as template was included as a control. o3 primes cDNA from RNA transcribed in a telomere-to-subtelomere direction, whereas oC primes cDNA from RNA of the opposite polarity. oG likely primes C-rich telomeric cDNA, but subtelomeric primers would not amplify this cDNA. This serves as an important negative control, as under standard denaturation conditions, endogenous priming of telomeric RNA can occur; such endogenous priming was removed under the conditions used here (see ‘Materials and Methods’ section). The act1 control demonstrates the presence of RNA in all samples. (B) Transcription of the telomeric C-strand is independent of Rdp1. Strand-specific northern blot analysis of total cellular RNA as in Figure 1D. Telomere-specific signals are absent from the blot probed with C-rich RNA, as all signals are present in the Otrt1Δ sample. In contrast, the G-rich probe detects telomere-specific C-strand signal in taz1Δ and taz1Δrdp1Δ samples. C-rich and G-rich probes hybridize approximately equally to ds telomeric DNA (data not shown).
Primer sequences: oC: 5′-GTAACCCCTGTAACCGTAACCC oG: 5′-GGGTTACGGTTACAGGGGTTAC, o1: 5′-TAGGAAGTGCGGTAAGTTG, o2: 5′-GTGTAATACAGTAGTGCAGTG, o3: 5′-GTGTGGAATTGAGTATGGTGAA, o4: 5′-CGGCTGACGGGTGGGGCCCAATA, act1F: 5′-ATGGAAGAAGAAATCGCAG act1R: 5′-CAAAACAGCTTGAATAGC.
RESULTS
Telomeric RNA in Schizosaccharomyces pombe
To detect telomere repeat-containing RNA, we performed northern blot analysis (Figure 1B) on total cellular RNA from wild-type (wt) cells using a probe comprising 250 bp of telomeric DNA along with 32 bp of the subtelomere. As a negative control, we included total cellular RNA from a strain lacking telomeric DNA sequences, Otrt1Δ. This strain was derived as a ‘survivor’ of the loss of telomeres following deletion of trt1+, the gene encoding the catalytic subunit of telomerase; in these strains, death due to telomere loss was bypassed via fusion between either end of each chromosome. The resulting circular chromosomes lack telomere repeats as well as distal subtelomeric sequences (Figure 1A) (22,23). We also included total RNA from a strain that harbours extremely long telomeres (∼2 kb to ∼5 kb versus ∼300 bp in wt cells) due to deletion of the gene encoding the double-stranded telomere repeat-binding protein Taz1 (28). An RNase-sensitive diffuse band centred around 0.4 kb is detectable in wt cells but, importantly, this band is absent in Otrt1Δ cells (Figure 1B). Telomeric RNA is not only detectable but also highly elevated (by >10-fold) in strains lacking Taz1. Intriguingly, however, we do not observe a large increase in the average size of the telomeric RNA, the majority of which remains in the 0.2–0.7 kb range in taz1Δ cells despite their elongated telomeres (Figure 1B and C). Hence, loss of Taz1 confers a change in the quantity, rather than the identity, of the telomeric RNA species.
Telomeric RNA corresponds primarily to the C-rich telomeric strand
To determine whether fission yeast telomeric RNA is G-rich, like human TERRA, or C-rich, we conducted northern blot analysis using ss oligonucleotide probes specific for the G-rich (C-oligo probe) or C-rich (G-oligo probe) strands of the telomere (Figure 1D). Using the C-rich probe, we do not detect any unique RNA species in wt or taz1Δ cells—the background hybridization that we detect using the C-oligo probe is also present in Otrt1Δ cells, indicating that these bands are not telomeric. Surprisingly, however, C-rich telomeric RNA is readily detectable in taz1Δ cells. The lower specific activities of end-labelled oligonucleotide probes versus the double-stranded random prime-labelled probe used in Figure 1B accounts for the absence of a signal in wt samples.
Our observation that the majority of telomeric RNA is C-rich contrasts with the predominance of G-rich transcripts in other species studied thus far. We therefore turned to a more sensitive method, RT-PCR, to determine whether G-rich telomeric RNA also exists in fission yeast (Figure 2A). A C-rich telomeric oligonucleotide, oC, was used for first strand cDNA synthesis. This primes reverse transcription of G-rich telomeric RNA species. The repetitive nature of the telomere repeat region prohibits the use of a telomeric primer for amplification of a unique fragment, so we used a primer that anneals 124 bp upstream of the telomere, o1, along with a primer that anneals near the junction of the subtelomere and the telomere, o4; these subtelomeric primers will detect G-rich transcripts originating in the subtelomere. Indeed, products of the expected size were detected with this primer combination in both wt and taz1Δ cells (Figure 2A), indicating that canonical TERRA exists in fission yeast. As predicted by northern blot analysis (Figure 1B), TERRA is more abundant in taz1Δ cells than in wt cells.
The RT-PCR approach was also used to further investigate transcripts comprising the C-strand and therefore originating within the telomere repeats themselves. Such transcripts can be detected by using subtelomeric primers in the first-strand cDNA synthesis step as well as in the amplification step. Indeed, using o3 to prime first-strand synthesis, we can detect subtelomeric RNA at the subtelomere/telomere boundary (o2 + o4); while the level of this species increases in the absence of Taz1, it is clearly present in wt strains. Hence, transcription in the telomere-to-subtelomere direction occurs not only in a taz1Δ background (Figure 2A) but also in wt cells.
We were intrigued by the predominance of C-rich telomeric RNA, and considered the possibility that it could be generated by an RNA-dependent RNA polymerase that uses G-rich telomeric RNA as a template. We therefore deleted the gene encoding Rdp1, the only known RNA-dependent RNA polymerase in fission yeast, in wt and taz1Δ backgrounds. We found that C-rich telomeric RNA levels are unchanged in the absence of Rdp1 (Figure 2A and B). Therefore, the template for C-rich telomeric RNA is not likely to be G-rich RNA, but rather the chromosomal telomeric DNA, and due to RNA polymerase directionality, transcription must initiate within the telomere repeat region.
Multiple subtelomeric RNAs in S. pombe
The detection of telomeric transcripts by RT-PCR with subtelomeric primers indicates that subtelomeric sequences are also transcribed in fission yeast. The region designated as STE1 encompasses ∼1.8 kb of ∼85 bp AT-rich semi-repetitive elements immediately centromere-proximal to the telomere repeats on chromosomes I, II and sometimes III. We investigated whether STE1 RNA could be detected by northern blot analysis (Figure 3B). Indeed, STE1-containing RNA species of distinct sizes, approximately 400, 600, 800 and 1200 bases, can be detected in wt and taz1Δ cells. These RNAs are not detected in Otrt1Δ cells, as would be expected based on the absence of STE1 sequences in these circular chromosome-containing survivors. Like transcripts comprising telomeric repeats, subtelomeric RNA also appears to be highly upregulated in cells lacking Taz1.
Figure 3.
Detection of subtelomeric RNA. (A) Schematic representation of the ends of Chromosomes I and II. Telomeres (Telo) are ∼270 bp in length and the subtelomeric elements are ∼1.8 kb (for STE1) and ∼5 kb (STE2) in length. Probes are indicated by black solid lines. (B) Northern blot analysis was performed as in Figure 1B, except that a subtelomeric sequence (STE1) probe was used. RNase-sensitive bands of ∼0.4, 0.6, 0.8 and 1.2 kb are detected in wt and taz1Δ but not Otrt1Δ samples.
Differential elevation of telomeric and subtelomeric RNAs in telomere and heterochromatin mutants
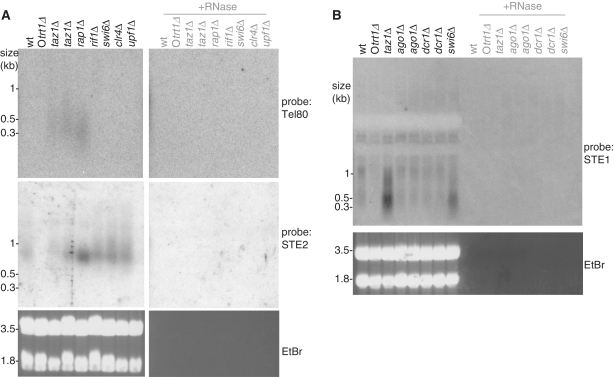
taz1Δ telomeres sustain numerous defects, including not only dramatic elongation but also stalled semi-conservative replication, loss of protection from end-joining and recombination and loss of the telomere position effect. We investigated whether other mutants with elongated telomeres and various subsets of the additional taz1Δ phenotypes also exhibit upregulation of telomeric and subtelomeric RNAs. Deletion of rap1+ or rif1+ results in long telomeres of ≥5 and ∼0.5–1 kb, respectively. Northern analysis with a telomeric probe that lacks any subtelomeric sequence, Tel80, reveals telomeric RNA in taz1Δ and rap1Δ cells (Figure 4). Wt telomeric RNA is not detectable as the Tel80 probe is weaker than that used in Figure 1B. As we observed for taz1Δ cells, average telomeric transcript lengths do not increase in rap1Δ cells despite extremely long telomeres. In contrast, cells lacking Rif1 harbour moderately elongated telomeres without a detectable upregulation of telomeric RNA, thus ruling out the possibility that such RNAs are upregulated by increased telomere length per se.
Figure 4.
Telomeric and heterochromatin proteins regulate telomeric and subtelomeric RNA. (A) Telomeric and subtelomeric RNAs are differentially regulated by telomeric and heterochromatin proteins. Total cellular RNA of 20 µg was loaded in each lane, blotted and hybridized with Tel80, which contains only telomere sequence. The blot was then stripped and hybridized with the STE2 probe. (B) Northern analysis was carried out as in A on total cellular RNA from the indicated strains.
To determine whether subtelomeric transcripts are subject to regulation by telomere proteins in a manner analogous to telomeric transcripts, we re-probed the northern blot in Figure 4 with a double-stranded STE2 probe, which hybridizes to subtelomeric RNAs just upstream of STE1 (Figures 3A and 4A). We detect an ∼800 base STE2-containing RNA in wt cells that is absent in Otrt1Δ cells. This STE2 RNA is only slightly upregulated by taz1+ or rap1+ deletion (Figure 4A), and unlike the telomere repeat-containing transcripts, it is also upregulated in a rif1Δ background (Figure 4A). These data indicate the existence of multiple telomeric and subtelomeric transcripts whose regulation is genetically distinct, with Taz1 and Rap1 regulating both transcript types and Rif1 regulating the subtelomeric species uniquely.
While fission yeast subtelomeres are incorporated into canonical heterochromatin containing methylated H3-lys9 and Swi6/HP-1 (24), the role of subtelomeric heterochromatin remains unclear, as telomere length and protection are intact in its absence. To determine whether this heterochromatin regulates telomeric and subtelomeric transcript accumulation, we deleted the genes encoding Clr4, the single fission yeast histone H3-lys9 methyltransferase, and Swi6. We also included Upf1 in our analysis, as the orthologue of this NMD pathway component has been shown to regulate human TERRA. We observe upregulation of STE2- and STE1-containing RNA in swi6Δ and clr4Δ strains (Figure 4A, Supplementary Figure S1); however, these mutants do not exhibit an increase in telomere repeat-containing RNA levels. Deletion of the genes encoding the RNAi pathway components Ago1 or Dcr1 compromises heterochromatin formation at pericentromeres and proximal subtelomeric regions (≥10 kb from the chromosome end). However, STE1 transcript levels are not grossly affected by ago1+ or dcr1+ deletion (Figure 4B). Finally, deletion of upf1+ has no effect on the levels of telomeric or subtelomeric RNAs as detected by northern blot analysis (Figure 4A). Thus, the presence of functional heterochromatin restrains subtelomeric RNA expression or stability. However, a basal level of subtelomeric transcription must be permitted in this heterochromatic region, as STE RNA is detectable in wt cells.
DISCUSSION
Based on the close compositional and mechanistic similarities between fission yeast and human telomeres, it is perhaps not surprising that we detect several fission yeast telomeric RNA species, paving the way towards detailed functional analysis using this system's battery of tools for genetic manipulation. The fission yeast telomeric transcriptome comprises C-rich telomere repeat-containing RNA molecules, as well as conserved G-rich TERRA molecules transcribed from the subtelomere into the telomere repeats. Our observations are consistent with those in an accompanying study (Bah et al., this issue) in which RNA species that originate in the subtelomere and contain subtelomeric and G-rich telomeric sequences are termed TERRA, C-rich telomeric RNA is termed ARIA, centromere-proximal subtelomeric RNA complementary to the subtelomeric region of TERRA and lacking telomere repeats is termed ARRET, and the complement of ARRET is termed α-ARRET. The correspondence between the transcripts we observe and those described by Bah et al. is summarized in Supplementary Figure S2.
Cells lacking the heterochromatin assembly machinery exhibit enhanced levels of subtelomeric transcripts while leaving telomere repeat-containing transcript levels untouched. In contrast, cells lacking Taz1 or Rap1 upregulate both subtelomeric and telomeric RNAs. Remarkably, transcripts comprising the telomeric C-strand accumulate in taz1Δ cells, suggesting that the telomere itself acts as a promoter and posing questions as to how local transcription may contribute to the telomeric defects seen in taz1Δ cells as well as to normal telomere maintenance, considered below.
When first reported in 2007 by Azzalin and Lingner, human TERRA was described as G-rich, originating in the subtelomere and spanning a wide range of lengths up to ≥9 kb. More recent work from the Lingner lab (25) has shown that surprisingly, the length heterogeneity of TERRA stems from varying lengths of the subtelomeric portion while the majority of TERRA molecules contain less than 0.4 kb of telomere sequence, despite the considerable length heterogeneity of the human telomere repeat tract. This is particularly interesting given our finding that the majority of telomeric RNA in cells lacking either Taz1 or Rap1 does not appear to change in size despite extremely long telomeres. Conceivably, either the repetitiveness of the telomere sequence or the specialized chromatin structure into which it is assembled limits the processivity of RNA polymerase II as it moves through the telomere repeat region.
We detect subtelomeric STE1 sequence-containing RNAs whose variable lengths center around 400, 600, 800 and 1200 nucleotides (Figure 3B). The distinct sizes suggest either that transcription initiates at a single site in the subtelomere and the transcript is processed into multiple products, or that there are multiple initiation sites in the subtelomere. Further upstream in the STE2 region, only one distinct RNA product is detectable at ∼800 bases (Figure 4A), although a faint smear above this band suggests there are other less abundant STE2 RNAs. While the STE2 region is known to harbour heterochromatin whose generation depends on the RNAi pathway, small RNAs corresponding to the more distal subtelomere and the telomere itself have not been described, and mutations in RNAi pathway components such as Dcr1, Rdp1 and Ago1 fail to alter telomeric transcript levels (Figure 4B), telomere length or the telomere position effect (26). One of the proposed functions of TERRA in other systems is a role in heterochromatin formation akin to human X chromosome silencing, in which the Xist RNA is transcribed only from the future inactive X chromosome (Xi); transcripts coat the Xi and help recruit heterochromatin machinery to silence the majority of the chromosome (27). As RNAi does not appear to regulate heterochromatin formation in the fission yeast distal subtelomere, perhaps the subtelomeric RNAs described here contribute to heterochromatin formation in this region. In this scenario, the upregulation of subtelomeric RNAs that we observe in the absence of Swi6 and Clr4 could reflect the abrogation of a negative feedback mechanism that would normally restrict local transcription once heterochromatin was established.
Remarkably, while we detect TERRA molecules whose transcription originates in the subtelomere by RT-PCR (Figure 2A), these are the less abundant RNA species compared to C-rich telomeric RNA, at least in cells lacking Taz1 (Figure 1D). As deletion of the gene encoding Rdp1, the only known RNA-dependent RNA polymerase in fission yeast, does not affect the levels of C- or G-rich telomeric RNA (Figure 2A and B), the telomeric DNA itself appears to serve as template for C-rich RNA transcription despite the absence of canonical promoter sequences in this region. Several unusual features of the telomeric environment could confer its susceptibility to transcriptional initiation, particularly in a taz1Δ setting. These features include a DNA end, a 3′ overhang, the presence of stalled replication forks and a unique chromatin architecture (J. Greenwood and J. P. Cooper, manuscript in preparation); all of these features are controlled by Taz1. Understanding the molecular events controlling generation of C-rich telomeric RNA is an important area for future studies.
The discovery that telomeric RNAs are more abundant in the absence of Taz1 also prompts us to wonder whether a subset of the known taz1Δ phenotypes are caused by increased local transcription or RNA accumulation. For example, one could imagine that either ongoing transcription or the physical association of transcripts with the telomere could hamper replication fork progression and could constitute the basis for the telomeric replication fork stalling seen in taz1Δ cells. Our observation that telomeric and subtelomeric RNAs are elevated in rap1Δ cells, in which replication proceeds relatively smoothly through the telomere, at first glance argues against this possibility. However, further studies will be necessary to determine whether overabundant RNAs associate with taz1Δ telomeres but are excluded from such interactions by the continued presence of Taz1 at rap1Δ telomeres.
Recent work in mammalian systems suggests that TERRA plays a role in telomerase regulation by restricting telomerase activity in part through hybridizing to the RNA template. As telomeric RNA is elevated and telomerase is more active in taz1Δ cells, our findings seem to contradict this. However, the elevated telomeric RNA that we observe is primarily C-rich, and would therefore not be complementary to the telomerase RNA template. On the other hand, the relative enhancement of C-rich versus G-rich telomeric RNA does not necessarily imply that the C-rich is the functional strand. Hence, while we have uncovered some of the components necessary for telomeric RNA regulation, the functions of telomere-repeat containing RNAs and telomeric transcription remain a mystery. To unassailably assess the functions of telomeric or subtelomeric RNAs, new experimental tools will have to be developed to alter transcript levels without disrupting other telomere components. Identification of telomeric RNA species in fission yeast provides further evidence that transcription of the telomere serves an important function and should accelerate progress in defining this function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2 and Supplementary Table 1.
FUNDING
Funding for open access charge: Cancer Research UK and the European Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank their lab members, Claus Azzalin and Joachim Lingner for discussions, and Claus Azzalin for sharing unpublished data.
REFERENCES
- 1.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 2.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 3.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 4.Solovei I, Gaginskaya ER, Macgregor HC. The arrangement and transcription of telomere DNA sequences at the ends of lampbrush chromosomes of birds. Chromosome Res. 1994;2:460–470. doi: 10.1007/BF01552869. [DOI] [PubMed] [Google Scholar]
- 5.Vrbsky J, Akimcheva S, Watson JM, Turner TL, Daxinger L, Vyskot B, Aufsatz W, Riha K. siRNA-mediated methylation of Arabidopsis telomeres. PLoS Genet. 2010;6:e1000986. doi: 10.1371/journal.pgen.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caslini C, Connelly JA, Serna A, Broccoli D, Hess JL. MLL associates with telomeres and regulates telomeric repeat-containing RNA transcription. Mol. Cell Biol. 2009;29:4519–4526. doi: 10.1128/MCB.00195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng Z, Campbell AE, Lieberman PM. TERRA, CpG methylation and telomere heterochromatin: lessons from ICF syndrome cells. Cell Cycle. 9:69–74. doi: 10.4161/cc.9.1.10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nergadze SG, Farnung BO, Wischnewski H, Khoriauli L, Vitelli V, Chawla R, Giulotto E, Azzalin CM. CpG-island promoters drive transcription of human telomeres. RNA. 2009;15:2186–2194. doi: 10.1261/rna.1748309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iglesias N, Redon S, Pfeiffer V, Dees M, Lingner J, Luke B. Subtelomeric repetitive elements determine TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO Rep. 12:587–593. doi: 10.1038/embor.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandell LL, Gottschling DE, Zakian VA. Transcription of a yeast telomere alleviates telomere position effect without affecting chromosome stability. Proc. Natl Acad. Sci. USA. 1994;91:12061–12065. doi: 10.1073/pnas.91.25.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 38:5797–5806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoeftner S, Blasco MA. Chromatin regulation and non-coding RNAs at mammalian telomeres. Semin. Cell Dev. Biol. 21:186–193. doi: 10.1016/j.semcdb.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flynn RL, Centore RC, O'Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 471:532–536. doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dehe PM, Cooper JP. Fission yeast telomeres forecast the end of the crisis. FEBS Lett. 584:3725–3733. doi: 10.1016/j.febslet.2010.07.045. [DOI] [PubMed] [Google Scholar]
- 16.Jain D, Cooper JP. Telomeric strategies: means to an end. Annu. Rev. Genet. 44:243–269. doi: 10.1146/annurev-genet-102108-134841. [DOI] [PubMed] [Google Scholar]
- 17.Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- 18.Rog O, Cooper JP. Telomeres in drag: Dressing as DNA damage to engage telomerase. Curr. Opin. Genet. Dev. 2008;18:212–220. doi: 10.1016/j.gde.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 20.Sato M, Dhut S, Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- 21.Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura TM, Cooper JP, Cech TR. Two modes of survival of fission yeast without telomerase. Science. 1998;282:493–496. doi: 10.1126/science.282.5388.493. [DOI] [PubMed] [Google Scholar]
- 23.Sadaie M, Naito T, Ishikawa F. Stable inheritance of telomere chromatin structure and function in the absence of telomeric repeats. Genes Dev. 2003;17:2271–2282. doi: 10.1101/gad.1112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanoh J, Sadaie M, Urano T, Ishikawa F. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr. Biol. 2005;15:1808–1819. doi: 10.1016/j.cub.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 25.Porro A, Feuerhahn S, Reichenbach P, Lingner J. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol. Cell Biol. 30:4808–4817. doi: 10.1128/MCB.00460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall IM, Noma K, Grewal SI. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl Acad. Sci. USA. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JT. The X as model for RNA's niche in epigenomic regulation. Cold Spring Harb. Perspect Biol. 2010;2:a003749. doi: 10.1101/cshperspect.a003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.