Abstract
Helicase–nuclease systems dedicated to DNA end resection in preparation for homologous recombination (HR) are present in all kingdoms of life. In thermophilic archaea, the HerA helicase and NurA nuclease cooperate with the highly conserved Mre11 and Rad50 proteins during HR-dependent DNA repair. Here we show that HerA and NurA must interact in a complex with specific subunit stoichiometry to process DNA ends efficiently. We determine crystallographically that NurA folds in a toroidal dimer of intertwined RNaseH-like domains. The central channel of the NurA dimer is too narrow for double-stranded DNA but appears well suited to accommodate one or two strands of an unwound duplex. We map a critical interface of the complex to an exposed hydrophobic epitope of NurA abutting the active site. Based upon the presented evidence, we propose alternative mechanisms of DNA end processing by the HerA-NurA complex.
INTRODUCTION
In all organisms, genetic material is continuously subjected to a wide variety of harmful lesions. Rapid and accurate repair of this damage is essential for maintaining chromosomal integrity, and for preserving cellular viability. Possibly the most detrimental form of DNA damage is the double-strand break (DSB), when both strands of the DNA helix are severed. DSBs are produced by genotoxic agents such as oxygen radicals or ionizing radiation; alternatively they can be generated during programmed cellular processes such as meiosis and assembly of immunoglobulin genes. In eukaryotic cells, DSBs are repaired using two distinct processes, namely non-homologous end-joining (NHEJ) and homologous recombination (HR). In the less accurate, homology-independent NHEJ pathway, the two ends of the broken chromosome are ligated back together directly (1). In contrast, HR is a high-fidelity mechanism reliant upon a homologous DNA sequence, such as a sister chromatid, as a template for retrieval of the lost genetic information (2).
The process of strand-exchange between homologous DNA molecules lies at the core of HR. The exchange of homologous DNA strands is preceded by the enzymatic resection of DNA ends, to generate the 3′ ssDNA tails that are utilized in the search for homology by recombinases of the RecA/Rad51/RadA family. A variety of genetic and biochemical studies have revealed that dedicated, functionally conserved helicase and nuclease activities act in concert at sites of DSB damage to produce the required 3′ overhangs (3–8). In eukaryotic organisms, DNA-end resection is performed by the Exo1 and Dna2 nucleases, working in combination with the yeast Sgs1 and human Bloom RecQ helicases (5,8–11), whereas in bacteria, resection is undertaken by the RecBCD, AddAB or AdnAB helicase–nuclease complexes (3,6,7,12). The highly conserved Mre11-Rad50 (MR) complex, an ATP-dependent molecular machine with a range of architectural and enzymatic roles at sites of DNA damage (13,14), appears to stimulate the activity of the nucleases and helicases responsible for DNA-end resection (9,10), and is likely to be required when the ends are blocked by covalent modifications or bulky adducts (14–16).
It is well-established that the DNA-information processing machineries of archaea and eukaryotes bear a strong similarity, presumably indicating a common evolutionary derivation of their molecular components (17,18). However, no orthologues of the eukaryotic DNA-end resection enzymes, such as RecQ helicases or Exo1/DNA2 nucleases, have been identified so far in archaea (19,20). In thermophilic archaea, the mre11 and rad50 genes reside within an operon that also encodes the NurA nuclease and the HerA helicase (21–24). Previous biochemical and bioinformatic characterization has indicated that NurA possesses a 5′ to 3′ exonuclease activity and that HerA belongs to the FtsK superfamily of hexameric translocases and helicases. The presence of the four genes within the same transcription unit suggests strongly that their gene products act in combination at sites of DNA damage to produce the 3′ tails necessary for homologous repair. Indeed, initial in vitro characterization of the mre11-rad50-herA-nurA operon from Pyrococcus furiosus provided biochemical support for this hypothesis (25).
In this study, we use biochemical and structural approaches to provide insights into the mechanism of DNA processing by the HerA helicase and NurA nuclease from the hyperthermophilic archaeon Sulfolobus solfataricus. We show that, in isolation, HerA and NurA possess little or no enzymatic activity and that efficient processing of DNA ends requires their reconstitution in a specific physical complex. Crystallographic analysis of NurA reveals an obligate toroidal dimer of RNAseH-like domains. The channel formed by NurA dimerization is too narrow for B-form, double-stranded (ds) DNA, but it can accommodate simultaneously one or two strands of an unwound DNA duplex. Furthermore, we identify by site-directed mutagenesis NurA residues that are important for enzymatic activity and for interaction with HerA. We combine our experimental observations with existing data to formulate possible models of DNA-end processing by the combined action of HerA and NurA. Our study suggests that the nature of the DNA ends might dictate the mode of activity adopted by the HerA–NurA complex.
MATERIALS AND METHODS
Protein purification
The herA and nurA genes were amplified by PCR from S. solfataricus P2 genomic DNA, cloned into the NdeI and XhoI sites of the pET30a vector (Novagen), and expressed in E. coli Rosetta (DE3) pLysS cells (Novagen). Cell lysates were heat-treated at 70°C for 20 min. Soluble proteins were subsequently purified by affinity chromatography, using a Hi-Trap heparin column, and size exclusion chromatography, using a Superdex 200 16/60 column. Mutant constructs were generated from the wild-type clones by site-directed mutagenesis (Stratagene). Full details are given in the Supplementary Data.
Helicase and nuclease assays
The catalytic activities of the HerA helicase and NurA nuclease were assayed on a variety of DNA substrates. Short oligo-based DNA substrates (50–110 nt) were 32P end-radiolabelled on one strand with T4 PNK (NEB), and after incubation with the HerA and NurA proteins, the assay products resolved on 12% SDS-PAGE using 1× TBE, and visualized by phosphoimaging. The unlabelled ϕX174 dsDNA assay products were resolved on a 1.0% agarose gel (after treatment with 0.4% SDS, 1.6 mg/ml proteinase K for 1 h at 37°C) and visualized by staining with SYBR-gold (Invitrogen). Full details of the assay conditions and DNA substrate preparation are provided in the Supplementary Data.
Holliday junction branch migration assays
The synthetic Holliday junction substrates were generated by annealing three oligonucleotides together to produce the cruciform structure: HH2 plus HH3, and either HH1a (5′ overhang substrate) or HH1b (blunt-ended substrate). Prior to the annealing, the HH1a and HH1b oligonucleotides were 32P end-radiolabelled with T4 PNK (NEB). Further details of the substrate construction are described in the Supplementary Data. After incubation with the HerA (or HerA K154A) and NurA D58A proteins at 65°C, the assay products were resolved on 12% SDS-PAGE, using 1× TBE, and visualized by phosphoimaging. Full details of the assay conditions are given in the Supplementary Data.
Triplex displacement assays
Triplex displacement assays were based on (26) but used a linear 500 bp PCR product containing a 22 bp triplex forming oligo (TFO) binding site. An alternative shorter oligo-based substrate was also designed that represented the 22 bp TFO binding site only. Triplex forming oligo was end labeled with 32 P using polynucleotide kinase. Triplexes were formed as previously described (Firman and Szczelkun, 2000). Proteins were preincubated in reaction buffer (25 mM MOPS, 50 mM NaCl, 10 mM MgCl2 and 0.5 mM Spermidine, pH 5.5) at 60°C for 30 min and then cooled to 50°C. A 5 nM triplex was added and incubated at 50°C for 0.5, 1, 1.5 or 5 min. Reactions were terminated by cooling on ice and addition of ×6 glycerol loading buffer (30% glycerol, 20 mM Tris pH 8, 0.25% bromophenol blue and 0.6% SDS). Resulting samples were analysed using either 6% non-denaturating PAGE or 1% agarose in (40 mM tris-acetate, 5 mM sodium acetate, 1 mM MgCl2, pH 5.5) at 120 V for 2.5 h at 4°C. Gels were analysed by PhosphorImager and ImageQuant software (GE Healthcare).
ATPase assays
ATP turnover of HerA and the HerA-NurA complex was determined through the monitoring of inorganic phosphate (Pi) release, utilizing a proprietary malachite dye kit (Pi ColorLockTM Gold—Innova Biosciences). Full experimental procedures are described in the Supplementary Data.
Physical interactions between HerA and NurA
HerA and NurA physical interactions between the wild-type and mutant proteins were examined by size exclusion chromatography using an analytical S200 HR 10/30 column, immunoprecipitation using rabbit polyclonal antibodies raised against purified HerA (CovalAb), or his-tagged pull-downs. Full experimental details are described in the Supplementary Data.
Crystallization and X-ray structure determination
The crystal structure of NurA was determined by single anomalous dispersion (SAD) using selenomethionine-labelled crystals and refined to a resolution of 2.5 Å. Full details of the structure determination and refinement are given in the Supplementary Methods. Coordinates and structure factors have been deposited in the Protein Data Bank under entry ID 2YGK.
Circular dichroism
Circular dichroism (CD) spectra between 250 and 185 nm were recorded on an AVIV 410 spectropolarimeter, at 25°C and 0.5 nm steps. The full methodology is provided in the Supplementary Data.
Fluorescence anisotropy
Fluorescence anisotropy measurements were recorded in a PHERAstar FS plate reader (BMG Labtech; Germany) equipped with a fluorescence polarization optic module (excitation = 485 nm; emission = 520 nM). The anisotropy was measured at 25°C in black 96-well half area, flat bottom, NBS plates (Corning; USA). DNA substrates and the full experimental details are described in the Supplementary Data.
RESULTS
HerA and NurA are mutually dependent for their enzymatic activity
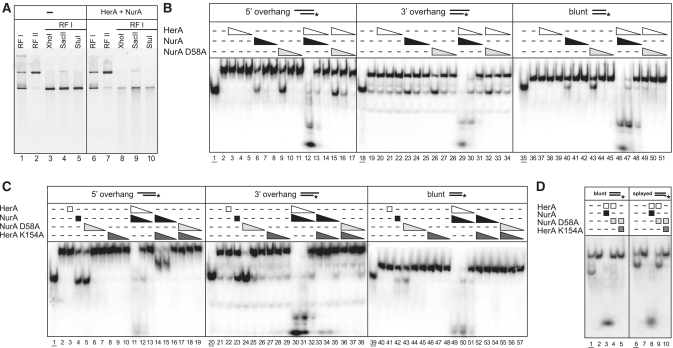
We commenced our functional characterization of the S. solfataricus HerA helicase and NurA nuclease by confirming that their enzymatic activity requires the presence of free DNA ends (Figure 1A). When a mixture of HerA and NurA proteins was added to either supercoiled or relaxed forms of a circular plasmid, the substrate remained refractory to digestion (Figure 1A; lane 6). In marked contrast, when the plasmid was linearized by digestion with either XhoI, SacII or StuI, addition of the HerA–NurA proteins produced extensive digestion of the template (Figure 1A; lanes 8–10). We then proceeded to analyse the effect of the nature of the DNA ends on the activity of the enzymes. The HerA and NurA proteins acted together to process efficiently linear dsDNA probes with 3′ or 5′ single-stranded overhangs or blunt ends (Figure 1B; lanes 12–14, 29–31, 46–48). Importantly, their catalytic activities displayed complete mutual dependency, in agreement with a previous report on the P. furiosus complex (25), but at variance with the studies on the S. acidocaldarius and S. todokaii HerA and NurA proteins (22,27). Thus, we were unable to detect HerA helicase activity in the absence of NurA (Figure 1B; lanes 3–5, 20–22, 37–39), or NurA nuclease activity in the absence of HerA (Figure 1B; lanes 6–8, 23–25, 40–42), on all three linear dsDNA substrates tested. We also observed that NurA displayed partial DNA unwinding at the highest enzyme to substrate ratio (Figure 1B; lanes 6, 23 and 40). A similar ability to induce strand separation in dsDNA has been described for the ssDNA-binding protein RPA (28).
Figure 1.
Enzymatic characterization of the HerA–NurA complex. (A) Activity of the HerA–NurA complex on ϕX174 DNA: supercoiled (RFI), nicked (RFII) or linearized with XhoI, SacII or StuI. Lanes 1–5: controls; lanes 6–10: DNA substrates following incubation with 200 nM HerA–NurA complex for 30 min at 60°C. (B) Activity of the HerA–NurA complex on a 54-bp dsDNA oligonucleotide. The DNA was either blunt-ended or with one 25 nt overhang at either 5′- or 3′-end. The position of the 32P-radiolabel is denoted by an asterisk. Substrates were incubated at 60°C for 30 min with HerA (lanes 3–5, 20–22 and 37–39), NurA (lanes 6–8, 23–25 and 40–42), NurA D58A (lanes 9–11, 26–28 and 43–45), HerA-NurA complex (lanes 12–14, 29–31 and 46–48) or HerA–NurA D58A complex (lanes 15–17, 32–34 and 49–51). Triangles depict decreasing concentrations of protein (450 nM, 45 nM or 4.5 nM of HerA hexamer or NurA dimer). Lanes 2, 19 and 36: no protein added. Lanes 1, 18 and 35: boiled substrates (underlined). (C) Activity of the HerA–NurA complexes in the presence of helicase-deficient (HerA K154A) and nuclease-deficient (NurA D58A) mutants. Reactions performed as described in B. Substrates were incubated at 60°C for 30 min with HerA (lanes 3, 22 and 41), NurA (lanes 4, 23 and 42), NurA D58A (lanes 5–7, 24–26 and 43–45), HerA K154A (lanes 8–10, 27–29 and 46–48), HerA–NurA complex (lanes 11–13, 30–32 and 49–51), HerA K154A–NurA complex (lanes 14–16, 33–35 and 52–54) or HerA K154A–NurA D58A complex (lanes 17–19, 36–38 and 55–57). Boiled substrates in Lanes 1, 20 and 39: boiled substrates (underlined). Lanes 2, 21 and 40: no protein added. (D) Activity of the HerA–NurA complex on a 6 nt splayed-end dsDNA substrate. Lanes 1–6: blunt-ended control substrate. Lanes 7–12: splayed substrate. Lanes 1 and 6: boiled substrates (underlined). Lanes 2 and 7: no protein added. Reactions performed as described in (B) using 450 nM HerA–NurA complex.
Although we could not detect DNA unwinding when HerA was used in isolation, clear HerA-dependent helicase activity was observed in the presence of NurA. This unwinding was best demonstrated with a catalytically inactive version of the nuclease, in which one of the three predicted catalytic residues (21) was mutated from aspartic acid to alanine (NurA D58A) (Figure 1B; lanes 15–17; 32–34). Importantly, unwinding was dependent on ATP hydrolysis, as no strand separation was observed in the presence of the ATPase-inactive HerA K154A mutant (Figure 1C; lanes 17–19, 36–38, 55–57). We also observed that, in the presence of the HerA K154A mutant, NurA was stimulated to digest the ssDNA region of the 5′ overhang substrate (Figure 1C; lanes 14–16). This enhanced activity was not detected, however, on either the 3′ overhang or blunt substrate molecule in the presence of magnesium (Figure 1C, lanes 14–16, 33–35, 52–54). In the case of NurA, it was also possible to compensate partially for the lack of HerA by replacing magnesium with manganese (Supplementary Figure S1). However, even in these buffer conditions, only the 5′ ssDNA overhang was processed by NurA alone, whereas dsDNA or 3′ ssDNA overhangs remained resistant to digestion. Taken together, the functional analyses indicate that stimulation of the HerA helicase activity by NurA is compatible with the production of ssDNA substrates, in agreement with the published evidence for the P. furiosus HerA–NurA complex (25).
The nature of the DNA ends modulates the activity of the HerA-NurA D58A complex
Interestingly, stimulation of HerA by the NurA D58A mutant resulted in strand separation for substrates with 5′ or 3′ ssDNA overhangs (Figure 1B; lanes 15–17, and 32–34, respectively), but little unwinding was apparent on the blunt substrate (Figure 1B; lanes 49–51). However, all three substrates were processed fully by the wild-type complex (Figure 1B; lanes 12–14, 29–31, and 46–48). This observation suggested that the HerA–NurA complex might adopt different modes of activity depending upon the nature of the DNA ends. To explore this possibility further, we examined the ability of the HerA–NurA D58A complex to unwind a blunt dsDNA substrate with the last 6 nucleotides mismatched to generate a splayed end (Figure 1D). Indeed, the HerA–NurA D58A complex unwound the mismatched substrate, but did not separate the two strands of the fully paired duplex in the control reaction (Figure 1D; lanes 4 and 9, respectively).
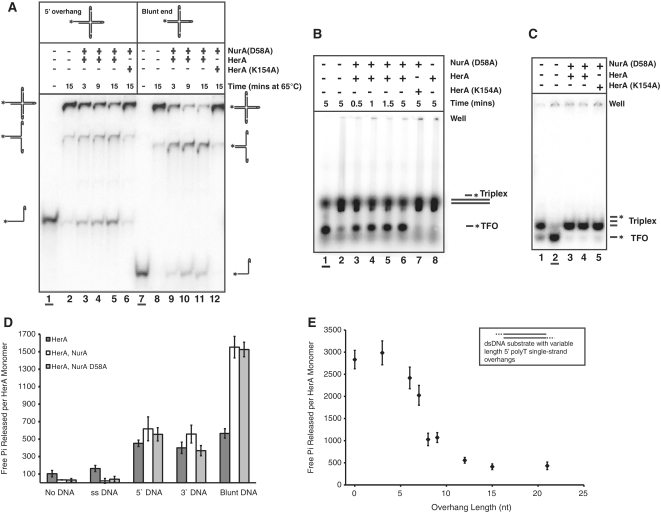
The apparent influence of DNA end on the outcome of the reaction was explored further by performing a branch migration assay, using a synthetic Holliday junction substrate (Figure 2A). Similar substrates have been used previously to characterize the activity of known translocases, such as DnaB (29). In our assays, the substrate was designed so that all but one of the four DNA ends were capped by hairpins, ensuring that protein loading could only occur on the remaining free-end (Figure 2A). The predominant DNA product resolved by the HerA–NurA D58A complex was dependent on the nature of the free DNA-end (Figure 2A). When the accessible end was blunt, the major resolved product was double-stranded, consistent with branch migration of the Holliday-junction (Figure 2A; lanes 9–11). In contrast, when the substrate possessed a 5′ overhang at the free-end, a smaller single-stranded product was generated most efficiently, indicating preferential unwinding of a single DNA strand (Figure 2A; lanes 3–5).
Figure 2.
The nature of the DNA end encountered by the complex affects the outcome of HerA–NurA processing. (A) Holliday junction branch migration, and strand unwinding by the HerA–NurA D58A complex. The 900 nM HerA hexamer and NurA D58A dimer were incubated with the Holliday junction substrate at 65°C for 3, 9 or 15 min. Lanes 1–6: substrate with a single 5′ overhang. Lanes 7–12: blunt-ended substrate. Boiled substrates in lanes 1 and 7 (underlined). Lanes 2 and 8: no protein added (65°C for 15 min). Lanes 6 and 12: HerA K154A–NurA D58A controls. (B) Triplex displacement assay. Lane 1 (underlined), boiled triplex (TFO, triplex forming oligonucleotide); lane 2, no protein added; lanes 3–6, time course of HerA–NurA D58A (230 nM HerA hexamer and NurA D58A dimer); lane 7, HerA K154A–NurA D58A; lane 8, HerA alone. Assays were performed at 50°C using 5 nM triplex DNA. Reactions were resolved on a 1% agarose gel. (C) Triplex displacement control experiment, with the dsDNA shortened to the length of the TFO. Lane 1, no protein added; lane 2 (underlined) boiled sample; lanes 3 and 4 wild-type (230 or 94 nM HerA hexamer and NurA D58A dimer, respectively) HerA–NurA D58A complex after incubation at 50°C for 3 min, respectively; lane 5, HerA K154A–NurA D58A control. Reactions were resolved on a 6% polyacrylamide gel. (D) ATPase assays demonstrating free Pi released per HerA monomer over 20 min (mean value for three independent experiments). Reactions in the absence or presence of DNA for HerA, HerA–NurA and HerA–NurA D58A (40 nM hexamer or dimer in the absence of DNA, or 20 nM for reactions containing both DNA and protein). Error bars depict the standard deviation (SD). (E) HerA ATP turnover versus 5′ overhang length. HerA–NurA complex (20 nM hexamer and dimer) was incubated with dsDNA substrates possessing varying length 5′ polyT overhangs. Data points represent the mean value for five independent experiments.
To characterize further the mode of action of the HerA–NurA complex, we performed a DNA triplex displacement assay, a technique commonly used to demonstrate translocation on dsDNA (26). In these experiments, a DNA triplex forming oligonucleotide (TFO) was annealed to a preformed DNA duplex under conditions that promote Hoogstein base pairing of the shorter third strand (further details in Supplementary Data). We observed that the TFO was displaced from a blunt-ended duplex after addition of the HerA–NurA D58A complex in an ATP-dependent manner (Figure 2B). Protein loading on the ends of the DNA duplex was necessary for TFO displacement since it was not observed on a control triplex assembled from three strands of identical length (Figure 2C).
The rate of ATP hydrolysis of the HerA–NurA complex is dependent upon the nature of the DNA ends
Our functional assays indicated that the activity of the HerA–NurA complex was modulated by the blunt or overhang nature of the DNA end. We decided to investigate the activity of the HerA–NurA complex further, by measuring the ability of HerA to hydrolyse ATP, using a malachite green phosphate-detection system that monitors release of free inorganic phosphate (Pi). In the absence of DNA, HerA alone displayed a low level of ATPase activity, releasing ∼100 free Pi molecules per monomer during the course of a 20-min experiment. Addition of ssDNA had no significant effect on this basal ATPase activity. However, ATP hydrolysis increased ∼5-fold in the presence of linear dsDNA (Figure 2D).
In the presence of NurA, stimulation of HerA ATPase activity became dependent on the nature of the DNA ends. In the absence of dsDNA, or in the presence of ssDNA, the complex was inactive. However, addition of dsDNA with either a 5′ or 3′ overhang increased ATP hydrolysis by the HerA–NurA complex to levels comparable to those measured for HerA alone on dsDNA (Figure 2D). Notably, a further 3-fold increase in the ATPase activity of the complex was observed in the presence of blunt DNA, compared to the overhang substrates (Figure 2D). This observation prompted us to measure the ATPase activity of the HerA–NurA complex on a set of dsDNA substrates that were either blunt-ended, or possessed 5′ overhangs of increasing length, ranging from 3 to 21 nucleotides. ATPase activity was highest on the blunt-ended and 3-nucleotide overhang substrates and declined as the size of the overhang increased (Figure 2E). The correlation was not linear: a sharp drop in ATP turnover was observed at overhang sizes of between 5 and 10 nucleotides, reaching a minimum for overhangs that were 12 nucleotides in length or longer (Figure 2E).
A stable physical interaction is observed between HerA and NurA
The functional assays revealed that the enzymatic activities of HerA and NurA had an absolute dependency on the presence of their protein partner. We therefore set out to investigate whether their functional interdependency reflected a physical interaction. Earlier studies had failed to identify a physical association between HerA and NurA of S. acidocaldarius and S. tokodaii (26,30), whereas HerA and NurA from P. furiosus have previously been shown to interact (25).
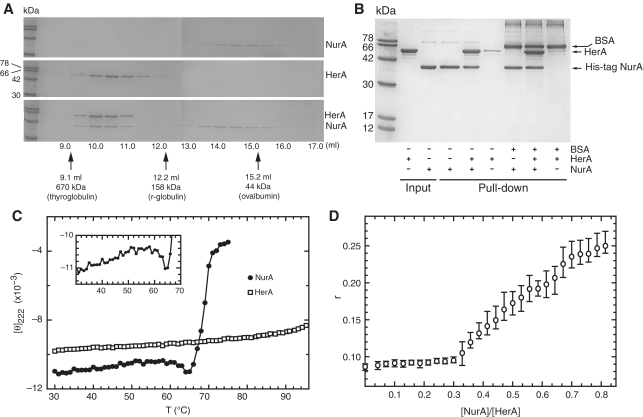
Analytical gel filtration chromatography of the HerA helicase showed a hydrodynamic behaviour consistent with an hexameric arrangement of protomers, in agreement with earlier reports for the HerA in Methanobacter thermoautotrophicus (24) (Figure 3A). When HerA and NurA were mixed together, the two proteins coeluted at an earlier volume than HerA alone (Figure 3A), indicative of the formation of a stable complex. The interaction was verified by pull-down using C-terminal His-tagged NurA and untagged HerA (Figure 3B). Reconstitution of a stable complex was promoted by preincubation of HerA and NurA at 60°C for 20 min (Supplementary Figure S2).
Figure 3.
Analysis of the physical interaction between S. solfataricus HerA and NurA. (A) Analytical gel filtration over an S200 Superdex HR 10/30 column. A total of 150 µg HerA (0.44 nmol of hexamer) and 90 µg NurA (1.15 nmol of dimer) were heated individually (top and middle panel) or together (bottom panel) at 60°C for 20 min before gel filtration. Protein fractions were separated by SDS–PAGE. Migration of molecular markers is indicated by arrows. (B) Pull-down assay confirming the HerA–NurA interaction. A total of 30 µg His-tagged NurA (0.38 nmol of dimer) was mixed with 50 µg untagged HerA (0.15 nmol of hexamer) at 60°C for 20 min. The complex was retrieved on nickel-agarose beads, and eluted by boiling. (C) Circular dichroism thermal melting profile. Mean residue elipticity at [θ]222 (×10−3) in degrees cm2 dmol−1 residue−1) as a function of temperature. Closed circles represent NurA (0.40 mg·ml−1), and HerA (0.47 mg·ml−1) is depicted by open squares. The insert is a detailed view of the low enthalpy transition for NurA between 30°C and 64°C. (D) Determination of the stoichiometry of the HerA–NurA complex by fluorescence anisotropy. HerA (0.62 µM) in buffer B plus 10 nM dsDNA with 5′ single-stranded overhangs (construct F2, Supplementary Data) were titrated with increasing concentrations of NurA. The abscissa axis shows the monomeric ratio of NurA to HerA. The open circles are the measurements mean (n = 3) and the vertical bars are the range of the error.
To investigate further the physical determinants of the HerA-NurA interaction, we measured the thermal stability of both proteins by circular dichroism (CD). The thermal melting profile of HerA (Figure 3C) showed no apparent unfolding in the temperature range 30–95°C indicating that its Tm is greater than 95°C. In strong contrast, NurA had a much lower Tm of 68.5°C (Figure 3C). Inspection of the melting profile of NurA between 30 and 64°C (Figure 3C; inset and Supplementary Figure S3) showed a departure from the linear decrease in ellipticity, which is typically attributed to fraying of α-helices (31). The ‘hump’ in the CD melt profile suggests that NurA undergoes a small enthalpic structural change before cooperative unfolding takes place. The combined evidence that heating is necessary for the optimal association of HerA and NurA, and that a conformational rearrangement is induced in NurA at around 60°C, suggests that complex formation depends on a specific structural alteration in one or both protein partners.
HerA and NurA interact with 6:2 stoichiometry
Fluorescence anisotropy measurements demonstrated that NurA binds to fluorescein-labelled DNA with a lower apparent equilibrium dissociation constant than HerA or the HerA–NurA complex. Under identical conditions, the difference of fluorescence anisotropy magnitude (Δr) for NurA binding to a dsDNA construct with 5′ single-stranded overhangs (substrate F2 in the Supplementary Data) was observed to be much larger than the Δr for HerA (Supplementary Figure S4). We exploited this experimental observation to investigate the stoichiometry of the HerA–NurA interaction. Titration of NurA against a fixed concentration of HerA (0.62 µM monomer) in the presence of 10 nM DNA substrate resulted in an inflection in the anisotropy plot (Figure 3D), which marks the point at which all of the NurA protein is sequestered in a complex with HerA. At larger values of the NurA:HerA ratio, the excess NurA binds to the DNA with high affinity, resulting in a large increase in Δr. The inflection point occurs at a NurA:HerA (monomer) ratio of 0.33, indicating that the complex contains at least 1 monomer of NurA to 3 monomers of HerA. Considering the known hexameric state of HerA, we conclude that the complex is composed of a HerA hexamer bound to a NurA dimer. A complex of three HerA chains bound to one NurA molecule is ruled out by the observation that the HerA–NurA complex elutes earlier than the HerA hexamer in analytical gel filtration (Figure 3A; lower and middle panel).
NurA folds in a toroidal dimer of RNAseH-like domains
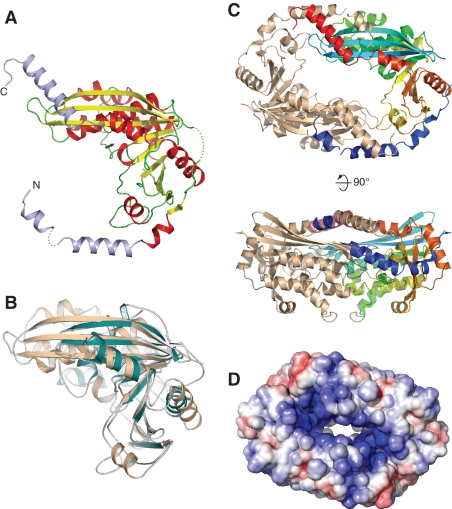
Our combined biochemical and biophysical approaches support the notion that formation of a specific HerA–NurA complex is a prerequisite for efficient processing of DNA ends. To investigate further the nature of the interaction between HerA and NurA and the mechanism of action of the HerA–NurA complex, we determined the crystal structure of S. solfataricus NurA at 2.5 -Å resolution (Table S2, refinement and model statistics). The crystallographic analysis revealed that NurA adopts an RNAseH-like fold (Figure 4A). The closest, functionally characterized, structural relative of NurA is endonuclease V from Thermotoga maritima (PDB id: 2W36) (33), with an rmsd of 3.0 Å over 143 Ca positions (Figure 4B). In the NurA crystal, two L-shaped chains dimerize in a head-to-tail fashion, via the reciprocal intertwining of helical N- and C-terminal extensions that protrude from the RNAseH domain (Figure 4C). The large surface area buried at the interface between NurA chains (∼2800 Å2/protomer) indicates that this dimeric arrangement reflects a biologically relevant, constitutive interaction. Our conclusion is corroborated further by the presence of a similar dimeric arrangement in the crystal structure of an RNAseH-like fold protein from T. maritima (TM1739; PDB id: 1ZUP). Thus, the crystallographic analysis of NurA supports our proposed stoichiometry of six HerA and two NurA subunits in the complex.
Figure 4.
Crystal structure of NurA. (A) NurA belongs to the RNAseH-like family of nucleases. The structure is shown as a ribbon, coloured according to secondary structure elements (alpha helices are red and beta sheets yellow). The N- and C-terminal helical extensions that are not part of the RNAseH fold are coloured in light blue. The position of the active site is indicated by an arrow. (B) Structural superposition of NurA and Endo V from T. maritima (PDB ID: 2w36). NurA is shown in light brown and Endo V in green. (C) Dimeric arrangement of NurA chains, as observed in the crystal. In the dimer, one protomer is coloured from the N- (blue) to the C-end (red), the other is coloured light brown. Two views are shown, related by a 90° rotation. (D) Electrostatic properties of the NurA protein. The electrostatic potential was calculated with APBS (32) and mapped onto the solvent-accessible surface of the structure, at contouring levels of ±5 kT (blue/red).
The mode of NurA dimerization leads to formation of a toroidal shape, with a central channel lined by diametrically opposed active sites. The inner surface of the channel is predominantly positively charged, consistent with the requirement to interact with the phosphodiester chains of DNA (Figure 4D). Importantly, the size of the channel is too narrow to fit a B-form DNA duplex, but it could accommodate in principle one or two ssDNA molecules, such as the strands of an unwound DNA helix. In agreement with the crystallographic model, the affinity of NurA for ssDNA is greater than for dsDNA by approximately one order of magnitude (Supplementary Figure S4). Thus, the structure of the NurA nuclease is compatible in principle with nucleolytic mechanisms that can result alternatively in the generation of ssDNA overhangs or wholesale digestion of dsDNA.
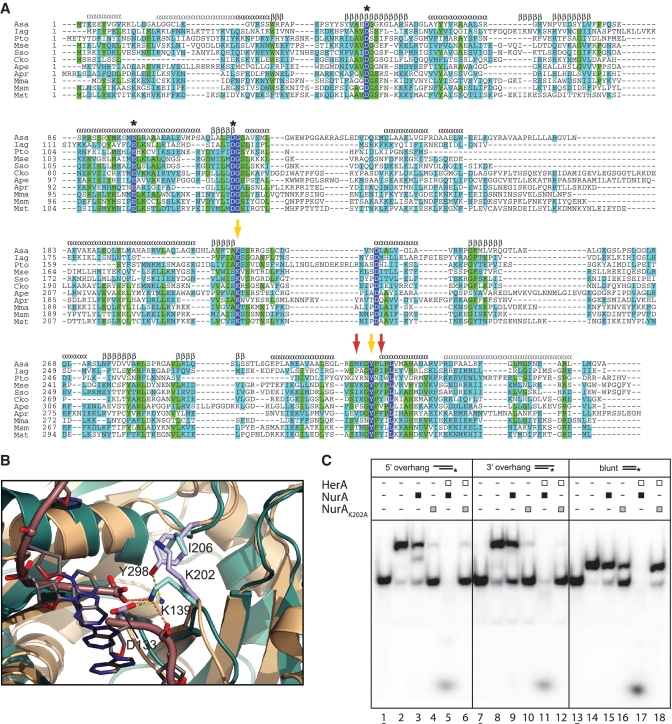
Absolute conservation of amino acids among NurA orthologues was low beyond the catalytic residues D58, D133 and E116, observed at the active site of the crystal structure, as predicted by Constantinesco et al. (21). Inspection of a multiple sequence alignment for diverse archaeal NurA proteins revealed further conservation of Y298 and K202 (Figure 5A). The conserved tyrosine residue Y298 had been noted in a previous study, although the function of this key residue was unknown at the time (21). In the structure, both residues are situated near the active site of the enzyme. Y298 is located on the loop preceding the last helix of the RNAseH fold, from where it projects its side chain towards the active site, contacting D133 via a water-mediated hydrogen bond (Figure 5B). The side chain of K202 rises from the inner edge of the central beta sheet, to pack against the aromatic ring of the tyrosine (Figure 5B). Analysis of the interaction by the program CaPTURE (http://capture.caltech.edu/) identifies the contact between K202 and Y298 as a strong cation-pi interaction at −4.4 kcal/mol (30,34). The high degree of conservation of K202 and its location adjacent to the active site points to its functional involvement in catalysis. A clue to the role of K202 was obtained by structural superposition of NurA with the structure of T. maritima endo V–DNA complex (33). The equivalent lysine in endo V (K139) interacts simultaneously with the phosphate groups flanking the scissile phosphodiester bond (Figure 5B). It seems likely that the highly conserved K202 performs an analogous functional role in NurA, by binding to ssDNA and selecting the correct orientation of the DNA strand for optimal nucleolytic processing. Indeed, mutation of K202 to alanine yielded a viable NurA mutant that retained its ability to interact with HerA, as identified by analytical gel filtration (Supplementary Figure S5), but was inactive in the functional assays (Figure 5C).
Figure 5.
Sequence conservation of NurA residues important for function and HerA binding. (A) Multiple sequence alignment of NurA sequences (Asa, Acidilobus saccharovorans; Iag, Ignisphaera aggregans; Pto; Picrophilus torridus; Mse, Metallosphaera sedula; Sso, Sulfolobus solfataricus; Cko, Candidatus Korarchaeum cryptofilum; Ape, Aeropyrum pernix; Apr, Archaeoglobus profundus; Mma, Methanococcus maripaludis; Msm, Methanosphaera stadtmanae; Mst, Methanobrevibacter smithii). Absolutely conserved residues are coloured white in a blue background, identical residues are highlighted in green and conserved residues highlighted in light blue. Secondary structure elements are identified as α for alpha helix and β for beta sheet on top of the alignment, coloured black for the elements of the RNaseH-like fold and grey for the N- and C-helical extensions. The known catalytic residues D58, E116 and D133 are marked by asterisks. Yellow arrows indicate the position of the K202 and Y298 residues, and red arrows mark the location of the I295 and F300 hydrophobic residues. (B) Cation–pi interaction between side chains of K202 and Y298 at the active site of NurA. The structure of the T. maritima Endo V - DNA complex (PDB: 2W35) at the post-incision stage is also shown superimposed. NurA is shown in light brown and Endo V in green. The side chain of conserved K139 in Endo V contacts the two phosphate groups on either side of the DNA backbone cleavage. The phosphodiester backbone of the DNA is shown as a tube and the bases as sticks. (C) Loss of nucleolytic activity in the NurA K202A mutant. Helicase–nuclease assays using the radiolabelled substrates as described in Figure 1B. Assays performed as described in Figure 1B using 450 nM HerA hexamer or NurA dimer. The 5 mM MnCl2 was included in all reactions. Lanes 1, 7 and 13 (underlined); boiled samples. Lanes 2, 8 and 14; no protein added. Lanes 3, 9 and 15; NurA. Lanes 4, 10 and 16; NurA K202A. Lanes 5, 11 and 17; HerA–NurA complex. Lanes 6, 12 and 18; HerA–NurA K202A complex.
Mapping of the HerA–NurA interaction surface
The arrangement of secondary structure elements in the quaternary structure of NurA is best appreciated when observed from a viewpoint perpendicular to the 2-fold symmetry element of the dimer (bottom view in Figure 4C). The central beta sheets harbouring the catalytic aspartates D58 and D133 separate two outer layers of mainly alpha helical structure. The two outer layers of the toroidal dimer have drastically different shapes: on one side of the molecule, the helical N- and C-terminal extensions intermesh to form a flat surface, whereas the alpha helices of the opposite layer are packed in a globular arrangement (Figure 4C).
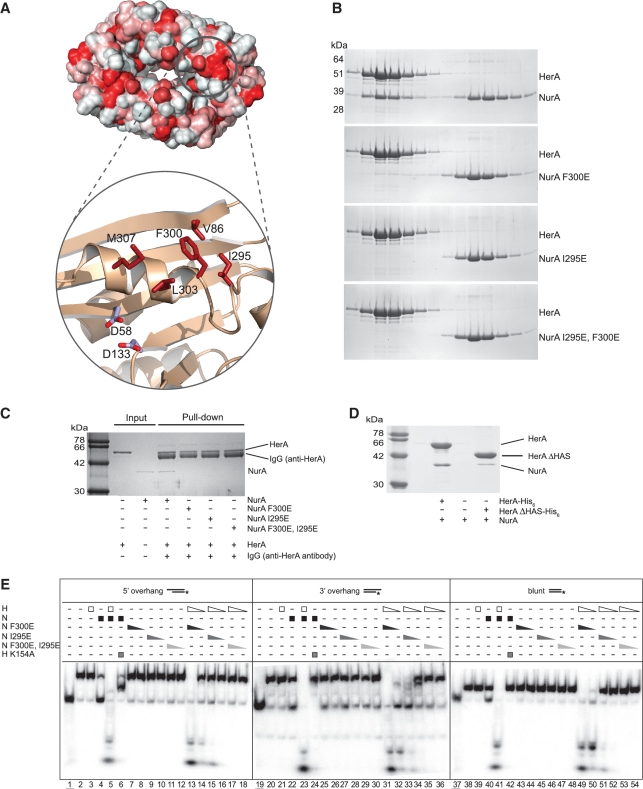
Protein surfaces that mediate constitutive association with interacting partners usually display a predominant hydrophobic character in the chemical nature of their constituent amino acids. Inspection of the flat surface in the proximity of the active site highlighted the presence of a solvent-exposed hydrophobic patch centred around Phe 300 and including Val 86, Ile 295, Leu 303 and Met 307 (Figure 6A). With the exception of Val 86, the residues are located in a segment of the NurA polypeptide that forms an essential architectural component of the active site. The hydrophobic nature of the exposed patch and its proximity to the active site of NurA made it an attractive candidate as a potential HerA-interacting epitope.
Figure 6.
Characterization of the HerA–NurA interaction. (A) Hydrophobic nature of the NurA surface, plotted red (most hydrophobic) to white (least hydrophobic) on the solvent-accessible surface of the protein. A hydrophobic patch formed by V86, I295, F300, L303 and M307 adjacent to the active site (marked by the side chains of D58 and D133) is shown in close-up view. (B) Gel-filtration analysis of the interaction between HerA and NurA and the NurA single mutants F300E, I295E and double mutant I295E, F300E. SDS–PAGE analysis of eluted fractions from a Superdex 200 HR 10/30 column. (C) Immunoprecipitations between HerA and NurA or the NurA F300E, NurA I295E or NurA I295E/F300E mutant proteins using HerA-specific polyclonal antibodies. Complexes were retrieved on protein-A sepharose beads, before separation by SDS–PAGE. (D) Pull-down analysis demonstrating the loss of interaction between NurA and HerA ΔHAS. 35 µg of HerA his-tagged HerA (0.10 nmol of hexamer) was mixed with 15 µg untagged NurA (0.19 nmol of dimer) at 60°C for 20 min. The complex was retrieved on nickel-agarose beads and eluted by boiling. (E) Helicase–nuclease assays using the radiolabelled substrates as described in Figure 1B demonstrating reduced nucleolytic activity in the HerA–NurA F300E, HerA–NurA I295E and HerA–NurA F300E/I295E mutant complexes. Triangles depict decreasing concentrations of proteins (450 nM and 45 nM HerA hexamer or NurA dimer). Lanes 1, 19 and 37 (underlined); boiled samples. Lanes 2, 20 and 38; no protein added. Lanes 3, 21 and 39; HerA. Lanes 4, 22 and 40; NurA. Lanes 5, 23 and 41; HerA–NurA complex. Lanes 6, 24 and 42; HerA K154A–NurA. Lanes 7–8, 25–26 and 43–44; NurA F300E. Lanes 9–10, 27–28 and 45–46; NurA I295E. Lanes 11–12, 29–30 and 47–48; NurA F300E/I295E. Lanes 13–14, 31–32 and 49–50; HerA–NurA F300E. Lanes 15–16, 33–34 and 51–52; HerA–NurA I295E. Lanes 17–18, 35–36 and 53–54; HerA–NurA F300E/I295E.
We set out to investigate our hypothesis by site-directed mutagenesis. We reasoned that if the hydrophobic residues formed part of the interface with HerA upon complex formation, reversing their chemical nature by mutation to a hydrophilic, charged amino acid would weaken or abolish the HerA–NurA interaction. Indeed, single-point mutations to glutamic acid resulted in a substantial decrease in the interaction with HerA for the NurA F300E mutant and complete loss for the NurA I295E mutant, as determined by analytical gel filtration (Figure 6B). The analytical gel-filtration also revealed that the dimerization of NurA was unaffected by the F300E, I295E or F300E/I295E mutations. Coimmunoprecipitation (Co-IP) using polyclonal antibodies specific to S. solfataricus HerA (Figure 6C) confirmed the impairment of the NurA mutants in their ability to interact with HerA. Functional assays of the NurA mutants showed a reduced, but still substantial level of nucleolytic activity for the NurA F300E mutant and a clear reduction for the NurA I295E mutant (Figure 6E). A F300E, I295E double mutant of NurA lost completely the ability to interact physically with HerA (Figure 6B and C) and failed to show any nucleolytic activity (Figure 6E). The result of the biophysical and biochemical characterization of NurA supports the identification of the hydrophobic patch centred on I295E and F300E as part of the NurA–HerA interface.
A previous bioinformatics analysis reported that the HerA family of helicases possesses an N-terminal beta-barrel domain, named the HAS-barrel due to its homology to the N-terminal domain of ATP synthase (standing for HerA and ATP synthase) (23). On the basis of the known roles of this domain in ATP synthase, the authors predicted that the HAS barrel region of HerA would be important for multimerization as well as interactions with partner proteins. We therefore generated an N-terminally truncated S. solfataricus HerA protein (HerA ΔHAS) that lacked the HAS domain. The oligomeric state of the HerA ΔHAS protein did not appear to have been altered when analysed by analytical gel filtration (Supplementary Figure S6). Conversely, removal of the HAS domain abolished complex formation as the HerA ΔHAS and NurA failed to coelute. The results of the gel filtration analysis were supported by pull-down assays, which indicated that the ability of HerA ΔHAS to interact with NurA was severely impaired (Figure 6D; Supplementary Figure S6). In summary, the association between HerA and NurA appears to be mediated by the HAS domain of HerA and the exposed hydrophobic patch identified on the flat surface of NurA.
DISCUSSION
In this article, we have investigated the mechanism of DNA-end resection by the S. solfataricus HerA helicase and NurA nucleases, encoded in a conserved operon of thermophilic archaea that includes the mre11 and rad50 genes. We have confirmed that HerA and NurA display a high degree of mutual dependency for their enzymatic activity and that assembly of the HerA–NurA complex is a prerequisite for processing DNA ends. The stable physical interaction that we observe between the HerA and NurA proteins is consistent with a previous study of the P. furiosus proteins (25), whereas earlier investigations in Sulfolobus species had failed to identify this association (26,35). In our hands, optimal reconstitution of the HerA–NurA complex required incubation at 60°C.
The molecular basis for the mutual dependency of the enzymatic activities of HerA and NurA is currently unknown. It is reasonable to assume that the interaction of the two protein partners induces a reciprocal conformational rearrangement that is required for activity. An indirect clue that this might be the case comes from the thermal melting analysis of NurA, which indicates a conformational rearrangement in the nuclease during heating to 60°C. Because the putative HerA-binding site of NurA is juxtaposed to the active site of the nuclease, we envisage that upon complex formation HerA might guide the ssDNA towards the active site of NurA, thus promoting DNA digestion. A critical role in determining the path of the ssDNA in the complex might be played by the essential K202, which is oriented by the strong cation–pi interaction with Y298 in the NurA structure.
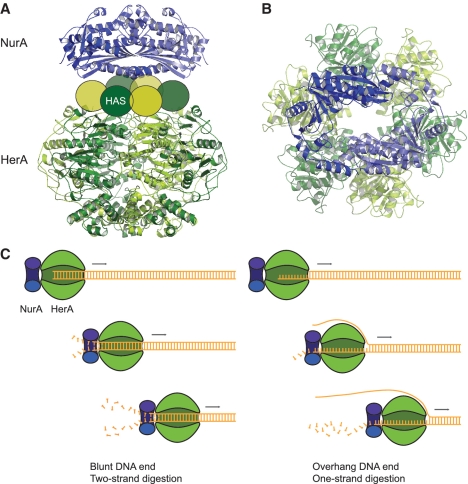
The crystal structure of the NurA nuclease reveals a constitutive homodimer of RNAseH-like domains, which are clamped together by intertwining N- and C-helical extensions. It seems plausible that this closed toroidal arrangement may significantly enhance the processivity of the HerA–NurA complex. The structure also provides a suitable template for rationalizing the ssDNA nuclease activity of NurA, as the central channel of the toroidal dimer is too narrow for dsDNA but can easily accommodate ssDNA. The dimeric arrangement of NurA makes it well suited in principle for digestion of one or two strands of an unwound duplex DNA, as would be conceivably produced by the action of the HerA helicase. Our combined biophysical and crystallographic evidence indicates that a NurA dimer associates with a hexameric ring of HerA subunits. No evidence is currently available on the reciprocal spatial arrangement of HerA and NurA in the complex. The simplest way to position the two protein partners is to arrange them so that the 2-fold symmetry axis of the NurA dimer coincides with the 6-fold axis of the HerA ring (Figure 7A). In the complex, the flat surface of NurA would contact the side of HerA harbouring the HAS barrel, in agreement with our mutagenesis study.
Figure 7.
Models of DNA end resection by the HerA–NurA complex. (A) 3D model of the interaction between the HerA helicase and NurA nuclease. The subunits of the HerA hexamer are coloured in alternating hues of dark and light green. The NurA subunits are coloured in light and dark blue. The HAS barrel portion of each HerA subunit is represented as an oval. The HerA hexamer and NurA dimer are arranged coaxially, with the flat side of NurA contacting the HAS barrel ring of HerA. The HerA structure is a homology model calculated in Modeller (37), based on the hexameric structure of the bacterial conjugation protein TrwB (PDB ID: 1E9R) (38). (B) Top-down view of the complex, highlighting the continuous channel formed by the proposed coaxial arrangement. The relative rotational setting of NurA and HerA is arbitrary. (C) The nature of the DNA end might influence the mode of DNA end resection adopted by the HerA–NurA complex. In the presence of a blunt or short (less than five bases) DNA overhang the complex would digest both DNA strands, leading to dsDNA degradation. Conversely, a larger (10 bases or more) DNA overhang would lead to degradation of one strand and generation of a ssDNA tail. The undigested strand might be entirely excluded from complex (as illustrated here), or conceivably could pass through the HerA ring, exiting at the interface between HerA and NurA.
Such coaxial arrangement generates a continuous tunnel traversing the hexameric helicase and the toroidal nuclease (Figure 7B). An appealing model of DNA processing would postulate that the DNA duplex translocates through the HerA hexamer towards the nuclease component on the opposite side of the complex. dsDNA unwinding could be performed by helicase activity inside HerA itself, or alternatively in concert with a ‘ploughshare’ action of NurA (36), separating the DNA strands and guiding them toward the active sites of each dimer subunit. In this model, both strands would be nucleolysed concurrently, albeit possibly at unequal rates on account of the different polarity of the DNA strands (Figure 7C). The apparent inability of the nuclease-inactive complex to separate blunt DNA substrates might reflect a dual-strand mode of processing, which would lead to the rapid reannealing of DNA strands after unwinding, as they emerge undigested from the exit channel on the opposite side of the NurA dimer.
Comprehensive degradation of linear dsDNA is not consistent with the generation of 3′ overhangs needed for homologous recombination. To account for such an outcome, a second mode of action of the HerA–NurA complex must be invoked, in which only one strand of the DNA duplex is digested (Figure 7C). A clue to the existence of this dual mode of action comes from the observation that the HerA–NurA complex has a higher rate of ATP hydrolysis on linear dsDNA substrates with blunt ends, or short overhangs, and a lower rate on substrates with overhangs above a certain size. Indeed, the nuclease-inactive complex can efficiently unwind a DNA substrate with 3′ or 5′ overhangs or one with frayed ends.
In this scenario, the type of DNA end presented to the HerA–NurA complex would determine how the complex interfaces with the DNA and hence the digestion mode: the presence of a blunt end or a short overhang (less than five bases) would activate the dual-strand digestion mode, whereas longer overhangs would direct activity towards the one-strand mode. Both modes of HerA–NurA action outlined here would correspond to a specific biological requirement: the dual-strand mode might be adopted as a defence mechanism against foreign DNA, whereas the single-strand mode would generate the ssDNA tails needed for DNA repair by homologous recombination. It is important to consider that viral integration into archaeal genomes is a common occurrence, even in thermophilic organisms such as S. solfataricus (39), and repair systems such as CRISPR/cas act to guard against the invasion of foreign DNA (40). It is therefore plausible that the HerA–NurA complex may protect the genome from insertion of extrachromosomal elements, as has been observed for the RecBCD complex in bacteria (41,42). This mode of DNA processing would need to be tightly regulated to ensure that vast regions of the chromosome are not illegitimately destroyed. We note that the bacterial helicase-nuclease RecBCD complex fulfils both the dsDNA destruction and single-strand resection functions. In the case of RecBCD, the Chi sequence elements, encoded within the bacterial genome, regulate the switch between the two modes (6,43). We speculate that, in archaeal recombination, selection of the appropriate activity mode might be determined by the initial processing of the DNA ends by the MR complex: preliminary cleavage of the DNA end by the 3′ to 5′ exonuclease activity of Mre11 would generate a 5′ overhang of sufficient size to direct the activity of the complex towards generation of 3′ overhangs suitable for recombination. At present, there is no mechanistic evidence to suggest how the HerA–NurA helicase–nuclease works in conjunction with the Mre11-Rad50 complex. Additional structural and functional analyses will provide fundamental insights into how accessory helicases and nucleases, such as HerA–NurA in archaea, or Sgs1-Exo1/Dna2 in eukaryotes, associate with the core Mre11-Rad50 components during DNA metabolism. The use of biochemically tractable model organisms, such as S. solfataricus, should prove instrumental to the further elucidation of these essential Mre11-Rad50-dependent DNA repair pathways.
ACCESSION NUMBER
The coordinates and structure factors of the S. solfataricus NurA crystal structure have been deposited in the Protein Data Bank under accession code 2YGK.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6, Supplementary Tables 1 and 2, Supplementary Methods and Supplementary References [26,44–50].
FUNDING
Research in the N.P.R. laboratory is funded by the Medical Research Council [Career Development Award (G0701443)]. Research in L.P. laboratory is funded by a Wellcome Trust Senior Fellowship Award in Basic Biomedical Sciences (08279/Z/07/Z). Funding for open access charge: the Medical Research (G0701443 to N.P.R.) and the Wellcome Trust (08279/Z/07/Z to L.P.) and the Department of Biochemistry, University of Cambridge.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
N.P.R. would like to thank Ron Laskey and the Hutchison-MRC Cancer Cell Unit for support and advice during the preliminary stages of this study. N.P.R. would also like to thank Adam McGeoch and Victoria Marsh for helpful discussion and experimental suggestions. J.K.B. would like to thank Mark Szczelkun and Kara van Aelst for technical assistance with the triplex experiments and the kind gift of plasmid, pLKS5.
REFERENCES
- 1.Critchlow SE, Jackson SP. DNA end-joining: from yeast to man. Trends Biochem. Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 2.Holthausen JT, Wyman C, Kanaar R. Regulation of DNA strand exchange in homologous recombination. DNA Repair. 9:1264–1272. doi: 10.1016/j.dnarep.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Chedin F, Kowalczykowski SC. A novel family of regulated helicases/nucleases from gram-positive bacteria: insights into the initiation of DNA recombination. Mol. Microbiol. 2002;43:823–834. doi: 10.1046/j.1365-2958.2002.02785.x. [DOI] [PubMed] [Google Scholar]
- 4.Kooistra J, Venema G. Cloning, sequencing, and expression of Bacillus subtilis genes involved in ATP-dependent nuclease synthesis. J. Bacteriol. 1991;173:3644–3655. doi: 10.1128/jb.173.12.3644-3655.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 7.Sinha KM, Unciuleac MC, Glickman MS, Shuman S. AdnAB: a new DSB-resecting motor-nuclease from mycobacteria. Genes Dev. 2009;23:1423–1437. doi: 10.1101/gad.1805709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G, Lee SE. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 29:3370–3380. doi: 10.1038/emboj.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeeles JT, Dillingham MS. The processing of double-stranded DNA breaks for recombinational repair by helicase-nuclease complexes. DNA Repair. 9:276–285. doi: 10.1016/j.dnarep.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 13.Lammens K, Bemeleit DJ, Mockel C, Clausing E, Schele A, Hartung S, Schiller CB, Lucas M, Angermuller C, Soding J, et al. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 145:54–66. doi: 10.1016/j.cell.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams GJ, Williams RS, Williams JS, Moncalian G, Arvai AS, Limbo O, Guenther G, Sildas S, Hammel M, Russell P, et al. ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat. Struct. Mol. Biol. 18:423–431. doi: 10.1038/nsmb.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connelly JC, de Leau ES, Leach DR. Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair. 2003;2:795–807. doi: 10.1016/s1568-7864(03)00063-6. [DOI] [PubMed] [Google Scholar]
- 17.Barry ER, Bell SD. DNA replication in the archaea. Microbiol. Mol. Biol. Rev. 2006;70:876–887. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelman Z, White MF. Archaeal DNA replication and repair. Curr. Opin. Microbiol. 2005;8:669–676. doi: 10.1016/j.mib.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Fujikane R, Komori K, Shinagawa H, Ishino Y. Identification of a novel helicase activity unwinding branched DNAs from the hyperthermophilic archaeon, Pyrococcus furiosus. J. Biol. Chem. 2005;280:12351–12358. doi: 10.1074/jbc.M413417200. [DOI] [PubMed] [Google Scholar]
- 20.Guy CP, Bolt EL. Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands. Nucleic Acids Res. 2005;33:3678–3690. doi: 10.1093/nar/gki685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Constantinesco F, Forterre P, Elie C. NurA, a novel 5'-3' nuclease gene linked to rad50 and mre11 homologs of thermophilic Archaea. EMBO Rep. 2002;3:537–542. doi: 10.1093/embo-reports/kvf112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Constantinesco F, Forterre P, Koonin EV, Aravind L, Elie C. A bipolar DNA helicase gene, herA, clusters with rad50, mre11 and nurA genes in thermophilic archaea. Nucleic Acids Res. 2004;32:1439–1447. doi: 10.1093/nar/gkh283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manzan A, Pfeiffer G, Hefferin ML, Lang CE, Carney JP, Hopfner KP. MlaA, a hexameric ATPase linked to the Mre11 complex in archaeal genomes. EMBO Rep. 2004;5:54–59. doi: 10.1038/sj.embor.7400037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hopkins BB, Paull TT. The P. furiosus mre11/rad50 complex promotes 5' strand resection at a DNA double-strand break. Cell. 2008;135:250–260. doi: 10.1016/j.cell.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Firman K, Szczelkun MD. Measuring motion on DNA by the type I restriction endonuclease EcoR124I using triplex displacement. EMBO J. 2000;19:2094–2102. doi: 10.1093/emboj/19.9.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang S, Wei T, Hou G, Zhang C, Liang P, Ni J, Sheng D, Shen Y. Archaeal DNA helicase HerA interacts with Mre11 homologue and unwinds blunt-ended double-stranded DNA and recombination intermediates. DNA Repair. 2008;7:380–391. doi: 10.1016/j.dnarep.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 28.Georgaki A, Strack B, Podust V, Hubscher U. DNA unwinding activity of replication protein A. FEBS Lett. 1992;308:240–244. doi: 10.1016/0014-5793(92)81283-r. [DOI] [PubMed] [Google Scholar]
- 29.Kaplan DL, O'Donnell M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol. Cell. 2002;10:647–657. doi: 10.1016/s1097-2765(02)00642-1. [DOI] [PubMed] [Google Scholar]
- 30.Dougherty DA. Cation-pi interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 31.Dragan AI, Privalov PL. Unfolding of a leucine zipper is not a simple two-state transition. J. Mol. Biol. 2002;321:891–908. doi: 10.1016/s0022-2836(02)00699-x. [DOI] [PubMed] [Google Scholar]
- 32.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalhus B, Arvai AS, Rosnes I, Olsen OE, Backe PH, Alseth I, Gao H, Cao W, Tainer JA, Bjoras M. Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair. Nat. Struct. Mol. Biol. 2009;16:138–143. doi: 10.1038/nsmb.1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc. Natl Acad. Sci. USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quaiser A, Constantinesco F, White MF, Forterre P, Elie C. The Mre11 protein interacts with both Rad50 and the HerA bipolar helicase and is recruited to DNA following gamma irradiation in the archaeon Sulfolobus acidocaldarius. BMC Mol. Biol. 2008;9:25. doi: 10.1186/1471-2199-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi TS, Wigley DB, Walter JC. Pumps, paradoxes and ploughshares: mechanism of the MCM2-7 DNA helicase. Trends Biochem. Sci. 2005;30:437–444. doi: 10.1016/j.tibs.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 37.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 38.Gomis-Ruth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, de la Cruz F, Coll M. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature. 2001;409:637–641. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- 39.Prangishvili D, Garrett RA, Koonin EV. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res. 2006;117:52–67. doi: 10.1016/j.virusres.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 40.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 41.Enquist LW, Skalka A. Replication of bacteriophage lambda DNA dependent on the function of host and viral genes. I. Interaction of red, gam and rec. J. Mol. Biol. 1973;75:185–212. doi: 10.1016/0022-2836(73)90016-8. [DOI] [PubMed] [Google Scholar]
- 42.Myers RS, Stahl FW. Chi and the RecBC D enzyme of Escherichia coli. Annu. Rev. Genet. 1994;28:49–70. doi: 10.1146/annurev.ge.28.120194.000405. [DOI] [PubMed] [Google Scholar]
- 43.Spies M, Amitani I, Baskin RJ, Kowalczykowski SC. RecBCD enzyme switches lead motor subunits in response to chi recognition. Cell. 2007;131:694–705. doi: 10.1016/j.cell.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, et al. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 47.Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 48.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 49.John DM, Weeks KM. van't Hoff enthalpies without baselines. Protein Sci. 2000;9:1416–1419. doi: 10.1110/ps.9.7.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinberg RL, Veprintsev DB, Fersht AR. Cooperative binding of tetrameric p53 to DNA. J. Mol. Biol. 2004;341:1145–1159. doi: 10.1016/j.jmb.2004.06.071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.