Abstract
SRSF2 is a prototypical SR protein which plays important roles in the alternative splicing of pre-mRNA. It has been shown to be involved in regulatory pathways for maintaining genomic stability and play important roles in regulating key receptors in the heart. We report here the solution structure of the RNA recognition motifs (RRM) domain of free human SRSF2 (residues 9–101). Compared with other members of the SR protein family, SRSF2 structure has a longer L3 loop region. The conserved aromatic residue in the RNP2 motif is absent in SRSF2. Calorimetric titration shows that the RNA sequence 5′AGCAGAGUA3′ binds SRSF2 with a Kd of 61 ± 1 nM and a 1:1 stoichiometry. NMR and mutagenesis experiments reveal that for SFSF2, the canonical β1 and β3 interactions are themselves not sufficient for effective RNA binding; the additional loop L3 is crucial for RNA complex formation. A comparison is made between the structures of SRSF2–RNA complex with other known RNA complexes of SR proteins. We conclude that interactions involving the L3 loop, N- and C-termini of the RRM domain are collectively important for determining selectivity between the protein and RNA.
INTRODUCTION
Constitutive and alternative splicing are influenced by splicing factors such as the Serine Arginine (SR) family. SR proteins are made up of one or two N-terminal RNA recognition motifs (RRM) domains followed by a C-terminal RS domain. RNA binding domains or RRM (RBD) consists of a ∼90 amino acids domain comprising a 4-strand anti-parallel β-sheet connected to two α-helices (1).
SR proteins exhibit dual functionality in constitutive and alternative splicing. In constitutive splicing, the SR proteins appear to interact with the RNA in a non-specific manner. However, in alternative splicing, the SR protein family has differing RNA binding specificities that play an important role in splice site selection and regulation (2). Although the mechanisms behind the different functions of these proteins in the two splicing actions are not yet fully understood, it is known that the regulation of alternative splicing relies upon the interaction of SR proteins with RNA regulatory sequences. These sequences, known as ESEs, ISEs, ESSs and ISSs (exonic splicing enhancer, intronic splicing enhancers, exonic splicing silencers and intronic splicing silencers, respectively) provide the mechanism by which exon skipping is prevented, ensuring the correct order of exonic sequences in the spliced messenger RNA (mRNA) (3). Regulatory RNA sequences are involved in both constitutive and, to a greater extent, alternative splicing, to enable the assembly of a functional spliceosome at the correct splice site (4).
The SR protein Serine/Arginine-rich Splicing Factor 2 (SRSF2), previously known as SC35, is a prototypical SR protein, involved in splicing proteins essential for a number of pathways. In the thymus and pituitary glands, SRSF2 functions during organ development where it is an integral part of regulatory pathways maintaining genomic stability (5,6). In the heart, SRSF2 plays an important role in regulating key receptors essential for heart function (7) and hypoxic hearts have been shown to trigger SRSF2 phosphorylation, which is surmised to counteract heart damage (8). SRSF2 has also been shown to work antagonistically to SRSF1 (9) and also compete with SRSF6 (10). The recurring theme throughout the studies of SRSF2 is that SRSF2 is involved in pathways that require tight control and regulation and SRSF2 expression is self-regulated by a negative feedback loop. Self-regulation occurs by SRSF2 splicing its own pre-mRNA to introduce premature stop codons which bring about destruction of the pre-mRNA by the nonsense-mediated mRNA decay pathway (NMD) (11). Furthermore the antagonistic effect of SRSF2 is activated by several low-affinity exon–SRSF2 interactions (12).
The specificity of the differing SR proteins for distinct RNA sequences has been probed in detail by a number of groups. SRSF2 binding RNA sequences have been identified by Systematic Evolution of Ligands by Exponential Enrichment (SELEX) (13,14). Several sequences identified by this method were found to have high binding affinity although the Exon Splicing Enhancer (ESE) activity was low (13,15). Furthermore it is proposed that the binding of ESE is necessary but not sufficient to promote splicing and an additional cofactor is also required (12). It is apparent that understanding the molecular detail of the interactions between protein and RNA is essential to interpret some of the nuances between binding and activity.
The only known SR protein–RNA complex structure is of SRSF3 (previously known as Srp20) in complex with a four nucleotide RNA fragment (16). In the case of SRSF3 the consensus RNA sequence from SELEX yielded the CAUC sequence that was then used to determine the resultant structure. The nature of the complex was found to be semi-specific with only the 5′ cytosine selectively recognized by specific interactions to β-strand 4 whereas the other three nucleotides were shown to interact indiscriminately with aromatic amino acids on the exposed surface of the β-sheet.
Here we report the Nuclear Magnetic Resonance (NMR) structure of the RRM domain of SRSF2. Utilizing intermolecular Nuclear Overhauser Effects (NOEs) and chemical shift mapping in conjunction with mutagenesis and RNA–protein cross-linking, we also probed the RNA binding specificity of SRSF2. From the specific interactions identified we have determined that the long flexible loop between β-strands two and three (loop 3) plays an essential role in stabilizing the interaction of the 5′-end of the RNA (adenine 1 and guanidine 2) whilst the flexible C-terminus interacts with the RNA toward the 3′-end (Uridine 8).
MATERIALS AND METHODS
Cloning, expression and purification of SRSF2 RRM domain
The DNA encoding the RRM domain (amino acids 9–101) of human SRSF2 was subcloned into pET-24b containing the 58-amino acids GB1 solubility enhancement tag and a 6× His tag. Point mutations were performed by quick change mutagenesis using Pfu Turbo (Stratagene). Oligonucleotide sequences will be provided on request. GB1-His6-SRSF2 RRM (hereafter referred to as SRSF2 RRM) was expressed in Escherichia coli BL21 Acella (EdgeBio). For labeled samples, expression was carried out in M9 minimal media containing 15NH4Cl and [13C6]-D-Glucose. Cells were grown at 37°C from a single colony to an OD600 of 0.8 at which point the cultures were transferred to 4°C for 30 min, then returned to 30°C and allowed to equilibrate for a further 30 min. Expression of SRSF2 RRM was then induced by 1 mM IPTG followed by incubation at 30°C for 3 h.
SRSF2 RRM was purified by nickel affinity chromatography using a 6.4 ml HIS-Select column (Sigma-Aldrich), and eluted from the column with 50 mM sodium phosphate, pH 8, containing 0.3 M NaCl and 200 mM imidazole. The concentration and purity of the eluted protein were measured using a Bradford protein concentration assay and SDS–PAGE analysis. The fractions containing the eluted protein were dialyzed against H2O, lyophilized and stored at −80°C until required.
Structure determination of SRSF2 RRM and relaxation measurements
NMR spectra of 0.5 mM SRSF2 RRM in 25 mM sodium phosphate, pH 6.8, containing 100 mM NaCl, 2 mM DTT and 0.02% NaN3 were recorded at 305 K on Bruker Avance 600 and 800 MHz spectrometers equipped with [1H, 15N, 13C]-cryoprobes. Data were processed using TopSpin (Bruker) and analyzed using CCPN Analysis (17). Sequence-specific backbone and side-chain resonance assignment of SRSF2 RRM was made using 3D HNCA, HN(CA)CB, HN(CO)CA, HNCO, CBCA(CO)NH, HBHANH, HBHA(CO)NH and HCCH-TOCSY experiments. Assignment of aromatic side-chain residues was made using 2D [1H-13C] HSQC and homonuclear 1H NOESY and TOCSY spectra recorded in both D2O and H2O.
The Hα, Cα, Cβ and CO chemical shifts were analyzed to give secondary structural information from the chemical shift index (CSI, 18). The structural analysis of SRSF2 RRM was performed using CYANA 2.1 software (19), with input data of shift lists derived from 15N- and 13C-HSQC spectra, along with un-assigned NOESY peak lists and additional restraints from 34 hydrogen bonds and 114 φ and ψ torsion angles produced by TALOS (20). CYANA 2.1 was run with standard protocols using seven cycles of automated NOE assignment and structural calculations, producing 100 structures per cycle. Of these 100, the 20 with the lowest target function were retained for analysis. The best 20 structures from CYANA 2.1 were further refined in ARIA 1.2 (21) using a total of 3406 unambiguous interproton distance restraints. A final ensemble of the best 20 water-refined structures was selected on the basis of lowest energies, and was characterized with PROCHECK-NMR (22) using the iCing interface (http://nmr.cmbi.ru.nl/icing/iCing.html). Atomic coordinates and NMR restraints of GB1-SRSF2 RRM have been deposited in the Protein Data Bank under the accession code 2KN4.
Structural analysis employed NACCESS (http://www.bioinf.manchester.ac.uk/naccess/) for identification of exposed hydrophobic residues, CCP4MG (23) for calculation and displaying electrostatic surface potentials, and Pymol (The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC) for secondary structure and side chain analysis. In addition comparative analysis of SR family employed promals3D (24) for secondary structure driven sequence alignment and Multiprot (25) for homologous structure alignment.
15N R1, R2 and 15N{1H}-NOE experiments were carried out on a Bruker Avance 600 MHz spectrometer at 298 K with a uniformly labeled 15N sample of SRSF2–RRM and conventional techniques with incorporation of gradient selection and sensitivity improvement (26). Heteronuclear 15N longitudinal (T1) and transverse (T2) relaxation rates were obtained by two-parameter fit of the experimental peak intensities to the equation I(t) = I0exp(−t/T). The [1H]-15N-heteronuclear NOEs were calculated from the ratio of peak intensities in 1H-saturated and unsaturated spectra.
Cross-linking of SRSF2 RRM with RNA
The RNAs GAGUA and AGCAGAGUA were synthesized, purified by PAGE and identified by HPLC by Dharmacon and Sigma-Aldrich (UK), respectively. Unless otherwise stated, all RNA and protein samples for NMR and ITC experiments were suspended in 25 mM N-(2-Acetamido)-2-aminoethanesulfonic acid (ACES), pH 6.8, containing 25 mM NaCl, 0.2 mM TCEP and 0.02% NaN3, and then dialyzed individually against the same buffer to ensure identical buffer conditions for all samples. RNA cross-linking was carried out as described previously (27).
Isothermal titration calorimetry
Isothermal Calorimetry was carried out on purified samples of protein exchanged into 25 mM ACES buffer contain 25 or 200 mM KCl using a NAP25 desalting column (GE Healthcare). In order to handle the RNA as little as possible HPLC purified lyophilized material was directly resuspended in the required buffer and pH adjusted where necessary. Control experiments whereby the RNA was titrated into buffer and buffer into protein exhibited undetectable heat exchange, confirming that there was appropriate match of buffer conditions with no evidence of dilution effects. To maintain consistency between titrations the same stock buffer was used in all protein and RNA preparations. Experiments were conducted at 25°C with an ITC200 (GE Healthcare) with a 60 µl syringe volume and 200 µl cell capacity. Titrations were carried out using between 2 µM and 200 µM of protein in the cell and a 10-fold concentration of RNA (between 20 µM and 2 mM). RNA was added into the cell in sequential 1 µl injections (at a rate of 0.5 µl/s) with a 180-s interval between each injection. One site (three parameters) and two site (six parameters) curve fitting was carried out using the MicroCal-supported ITC module within Origin version 7.
NMR experiments of SRSF2–RNA complex
15N-HSQC NMR titration experiments were carried out on a Bruker Avance 800 MHz spectrometer equipped with a 5 mm Cryoprobe at an experimental temperature of 305 K. Initial conditions indicated that lowering salt concentration improved binding and therefore optimal binding conditions were found to be 50 mM ACES buffer pH 6.8; negligible change to SRSF2 RRM spectra led us to conclude that these buffer modifications had minimal effects on protein structure. The RNA AGCAGAGUA was titrated from zero to a final concentration of 2.5 mM RNA in 0.5 mM SRSF2 RRM by mixing two protein samples of different concentrations of RNA in order to ensure no buffer mismatch or sample dilution. The peaks in each HSQC were assigned in the CCPN software.
RNA assignment
Natural abundance 1H 13C HSQC and homonuclear 2D TOCSY and NOESYs (300 ms mixing time) were collected to assign the 9-mer RNA in 50 mM ACES buffer pH 6.8 at 305 K through well-established methods (28). Lack of base-ribose H1′ sequential residues indicate that the RNA in isolation does not have a well defined structure.
Protein–RNA complex modeling
The assignments of both the RNA and SRSF2 RRM in the complexed forms were obtained by tracking chemical shifts during an RNA titration. Several types of spectra were collected: 1D 1H spectra, natural abundance 2D 1H 13C HSQC (for RNA shifts) and 1H 15N/1H 13C HSQC using labeled SRSF2. Intermolecular NOEs between 15N,13C labeled SRSF2 RRM and unlabeled RNA were collected using filtered NOESY experiments (29) with unlabeled RNA at a 5-fold excess and a mixing time of 300 ms. We modeled the SRSF2–RNA structure using two different approaches, HADDOCK (High Ambiguity Driven biomolecular DOCKing) (30,31) and CNS (32). For both protocols, ambiguous interaction restraints were defined by RRM residues which showed chemical shift perturbations of higher than 0.15 ppm or whose mutation led to loss of RNA binding. For the RNA, active residues were defined based on chemical shift changes upon binding to SRSF2. Experimental intermolecular NOEs between the RRM domains and RNA were also included. The RNA structure was poorly defined due to a lack of intramolecular NOEs. As the HADDOCK method works best when the individual interacting components have well-define structures, the HADDOCK-derived structures did not satisfy all the experimental intermolecular NOEs, this possibly due to the limited conformational space sampled by the ensemble of random coil RNA coordinates. Simulated annealing using the CNS software allowed a flexible treatment of the RNA coordinates and was performed with the protein restraints employed for the calculation of the GB1-SRSF2 structure, together with the six intermolecular RRM-RNA NOEs. After the first iteration of structures, it was apparent that a distance restraint between the conserved F57 and F59 to the RNA C3 could be included based on the proximity of the cystidine to both phenylalanines and the consensus of π stacking of these amino acids in homologous structures. The final ensemble of the best 20 water-refined structures was selected on the basis of lowest energies and without intermolecular NOE violations. The clusters were analyzed using criteria defined in the HADDOCK program. Pairwise RMSD analysis of these structures was carried out to define clusters of models with overall RMSD of 5 Å or less based on alignment of backbone atoms of the RNA 9-mer and the secondary structure elements of SRSF2.
RESULTS
SRSF2 RRM domain is completely independent of GB1 solubility tag
The RRM structure of SRSF2 was determined in the presence of the N terminally tethered GB1 fusion protein, the latter being essential to keep the protein in solution (33). The fusion protein is monomeric in solution with a molecular mass of 16 kDa as assessed by size-exclusion chromatography and multi-angle light scattering analysis (data not shown). The RMSD (using backbone atoms Cα, NH, C′) for the RRM over the 20 lowest energy structures was 0.5 Å (Table 1 and Figure 1). T1 T2 and heteronuclear NOE analysis of the SRSF2 RRM construct demonstrated the lack of interaction between the two domains (Supplementary Figure S1); the T1/T2 values for the RRM and GB1 domain indicate different rotational correlation times for the two domains, with the T1/T2 values of GB1 fused to SFSR2 being similar to those of isolated GB1, which suggests that the RRM domain has very little effect on the solution reorientation of the GB1 domain. The lack of interdomain NOEs between the two domains also corroborates the autonomy of the two separate domains. Comparison of the structure of GB1 determined here and in isolation (PDB number 2IGG; BMRB Accession number 1639) showed that the two structures are similar, further confirming that the presence of the RRM domain had negligible influence on the GB1 tag. The chemical shifts of GB1 alone and fused to the RRM are identical under the same buffer conditions. We, therefore, conclude that the two folded domains are completely independent of each other and do not have any artefactual interactions.
Table 1.
NMR statistics for the structure of SRSF2 RRM
| Experimental restraints | |
|---|---|
| Restraints | |
| Unique/Ambiguous NOEs | 3406/145 |
| Intraresidue | 1441/43 |
| Sequential | 735/29 |
| Short range (1< [i − j] <5) | 379/23 |
| Long range ([i − j] >4) | 851/50 |
| φ/ψ dihedral anglesa | 114 |
| Energies (kcal mol−1)b | |
| Total | −5948 ± 80 |
| Van Der Waals | −1370 ± 17 |
| NOE | 78 ± 23 |
| RMS deviationsb | |
| NOEs (Å) (no violations >0.5 Å) | 0.02 ± 0.003 |
| Dihedral restraints (°) (no violations >0.5 Å) | 0.44 ± 0.09 |
| Bonds (Å) | 0.0040 ± 0.0001 |
| Angles (°) | 0.51 ± 0.02 |
| Impropers (°) | 1.55 ± 0.07 |
| Ramachandran map analysisc (%) | |
| Allowed regions | 80.2 |
| Additional allowed regions | 15.5 |
| Generously allowed regions | 2.6 |
| Disallowed regions | 1.7 |
| Pairwise rms difference (Å)d | |
| Residues 70–146 | 1.31 (2.13) |
| 2o Structure | 0.40 (0.99) |
aFrom chemical shifts using Talos.
bCalculated in ARIA 1.2 for the 20 lowest energy structures refined in water.
cObtained using PROCHECK-NMR.
dFor backbone atoms; value for all heavy atoms in brackets.
Figure 1.
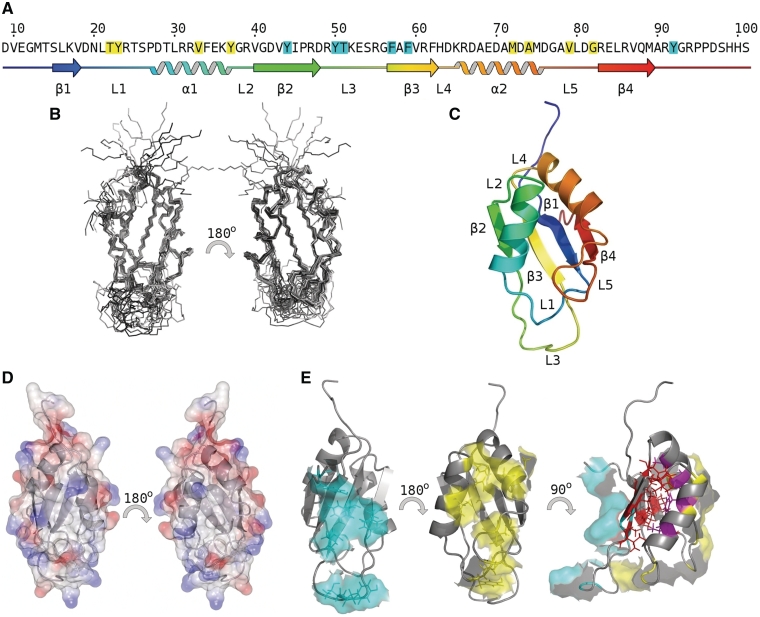
(A) SRSF2 RRM sequence and secondary structure. (B–E) Structure of SRSF2 RRM: (B) ensemble structures. (C) cartoon representation and schematic colored blue to red N-terminus to C-terminus, loops, strands and helices labeled according to RRM consensus (1). (D) electrostatic surface (red −5, blue +5). (E) hydrophobic residue analysis of SRSF2 identified two surface exposed patches, one on the helical face (yellow) the other on the β-sheet face (cyan); the hydrophobic core of the molecule comprises residues from both helices (magenta) and strands (red), for clarity flexible C-terminus residues 94–101 are omitted.
The structure of SRSF2 RRM domain comprises a four strand anti-parallel β-sheet and two α-helices. The highly ordered secondary structure elements are apparent in the ensemble of structures (Figure 1B) with a significant degree of flexibility in the apical loop 3 between strands 2 and 3 as inferred from 1H{15N} heteronuclear relaxation, Random Coil Index (34) data (Supplementary Figure S1), and a lack of intramolecular proton NOEs for this region of the protein. The N- and C-termini are flexible as evident from the chemical shift values and the lack of long-range NOE correlations. Due to significant resonance overlap of resonances for the C-terminal region, reliable 15N relaxation data could only be obtained for the N-terminus residues, which show low-frequency motions that are associated with conformational exchange. The compact, well-ordered structure is maintained by extensive hydrogen bonds in the β-sheet together with a buried, internal hydrophobic core formed by residues L30, F34, A68, A71 and M75 of the two α-helices and residues on the inward face of the β-sheet, L16, V18, V40, V43, I45, A58, V60, F62, L85 and V87. In addition to the hydrophobic core, NACCESS analysis revealed two surface-exposed hydrophobic patches on opposite sides of the protein. On the outward face of the β-sheet Y44 of strand 2 and F57, F59 of strand 3 create a hydrophobic patch with C-terminus residue Y92 and loop 3 residues Y50 and T51. On the helical side of the protein, a second hydrophobic patch comprises helical residues V33 Y37 M72 and A74 together with loop 1 residues T22, Y23 and loop 5 residues V79 and G82.
Structure of SRSF2 RRM domain is typical of the SR family
The structure of the SRSF2 RRM domain resembles the classical fold for RRM domains with the C terminal residues exhibiting a greater degree of flexibility. In comparison with the homologous SR RRM domains, SRSF2 aligns well, in particular, with the single or first RRM domains (Figure 2A). When RRMs occur in tandem in SR proteins the second RRM domain has an extended loop 5 and is shown to bind RNA in a different manner to the first/single RRM domains (27,35). Therefore, a comparison of existing SR RRM domains has been carried out exclusively on single/first RRM structures (Figure 2B). The aligned RRM domains adopt a highly homologous structure with a RMSD of the structured regions of 1.21 Å (over the 58 backbone residues indicated in the alignment). However, differences are apparent in the flexible N and C termini and also the hairpin loop L3 between β2 and β3, which varies in length between the RRM domains.
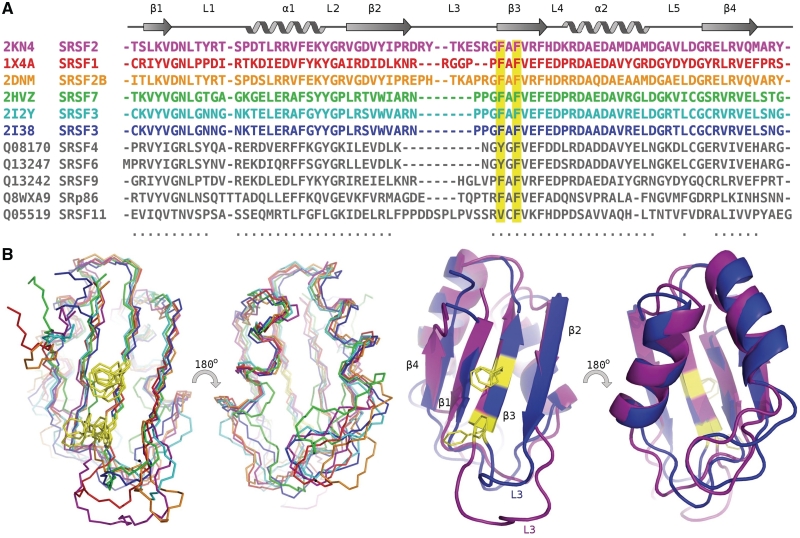
Figure 2.
Alignment of the SR protein RRM domains. (A) PDB deposited structures colored according to overlay and identified by PDB number and name, all other RRMs colored grey and identified by uniprot number and name. Conserved residues for RNA binding are highlighted in yellow (B) Left: Overlay of backbone atoms of the molecular structures of all known SR RRM domains, backbone alignment using 57 residues (indicated in by dots beneath the sequence) with an RMSD of 1.18 Å. Structures shown of human SR family RRM domains; SRSF1 (1X4A–RSGI), SRSF2B (2DNM–RSGI), SRSF7 [2HVZ (16)] and SRSF3 [2I2Y, 2I38 (16)]. Right: Cartoon representation of structures; for clarity only two SR–RRM domains are shown; SRSF2 and SRSF3, the SRSF3 structure used for this alignment is from PDB ID 2I2Y, the only SR–RRM structure determined in the presence of RNA.
RRMs of SR proteins possess classic ribonucleoprotein (RNP) recognition motifs, RNP1 and RNP2, on β3 and β1 strands, which are essential for RRM–RNA interactions (1), although the degree of hydrophobicity in β1 is reduced in SRSF2 and SRSF3. The RNP1 sequence is conserved in SRSF2, with core conserved amino acids on β3 being F57 and F59 (Figure 2A). However, the aromatic residue that is normally found in β1, and which is important for RNA binding, is missing in SRSF2, this being replaced by a lysine residue (K17).
Comparison of electrostatic surface potential and hydrophobic surfaces show non-polar areas differ between SR-RRMs, with SRSF2 having a marginally greater area of exposed hydrophobic residues in L3 (Supplementary Figure S2). In the SRSF3 RRM structure, the corresponding residues for the β-sheet outward face hydrophobic patch are Y13, G15, W40, A41, F48, F60, L80, with P45, G47, of loop 3 and G83 of the C-terminus region; Y50 and T51 found in SRSF2 RRM do not exist in SRSF3 RRM. On the helical side, the corresponding residues in SFSR3 are T24, G31, Y32, P35 P56, A60 G68, T70, L71, G73 (Supplementary Figure S2).
Binding of SRSF2 RRM to RNA using NMR and mutagenesis
Various RNA constructs were used to probe the nucleotide specific interactions of SRSF2. SELEX analysis has previously identified several sequences which preferentially bind SRSF2 (13). For the purpose of this study we focused primarily on the GAGUA SELEX motif and found this 5-mer bound preferentially over non-specific sequences such as AUAUA (Supplementary Figure S3). However, NMR chemical shift mapping yielded an approximate Kd in the order of 0.5 mM. By extending the 5′-end of GAGUA to give a 9-mer construct, AGCAGAGUA, the RNA bound with higher affinity to SRSF2, as assessed initially by NMR. This complex was taken forward for further studies.
The 9-mer RNA lacks internucleotide NOEs, suggesting that the RNA is largely unstructured in the unbound state. Titration of RNA into 15N-labeled protein causes large chemical shift changes and/or line-broadening to many resonances in the 1H-15N HSQC spectrum, suggesting that a large number of residues in the protein are affected by RNA binding (Figure 3 and Supplementary Figure S4). The resonances of the RNA bases also show significant shift changes and/or line broadening upon interaction with SRSF3.
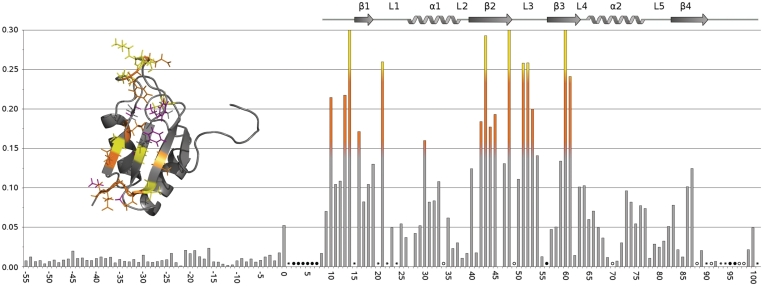
Figure 3.
Histogram of chemical shift changes—residues with combined shifts greater than 0.15 ppm (orange) and 0.25 ppm (yellow) marked on structure inset. Filled circle represents NH peak not assigned, open circles represent peaks that broaden or cannot be tracked upon titration and asterisks represent overlapping NH peaks. Significant shift changes/line-broadening can be seen for residues in the β-strands as well and residues in L3, in particular residues Y50 and T51, and the N-termini.
The mixture of the resonance characteristics (shifts and line-broadening) over the course of the RNA titration is not unusual, as often observed for many protein–ligand titrations. Typically, for a given equilibrium, while the overall exchange rate constants are indeed constant, different resonances will show different exchange behavior on the NMR timescale, as revealed in the differences in the degree of line-broadening, depending on the total chemical shift change (Δvtotal) between the free and bound states. Deriving dissociation constant values (Kd) from NMR titrations is only reliable under conditions of extreme fast exchange on the NMR timescale (and hence very weak binding). For SRSF2, since fast exchange was not universally observed for all the resonances, dissociation constants for the 9-mer could not be reliably extracted from the NMR titrations. However, despite severe attenuation for some of the peaks, it was possible to obtain the resonance assignment of the bound protein and RNA since chemical shift changes could be followed over the course of the titration.
The resonance perturbation of the protein spectrum enabled the RNA-binding region of SRSF2 RRM to be mapped and key residues identified. When compared with GAGUA (and the non-specific control AUAUA), more extensive shift changes and of larger magnitudes are observed (Supplementary Figure S3), suggesting that the extra nucleotides increase the number of protein–RNA contacts. This is corroborated by the intermolecular NOE data (see later). Analyses of the resonance perturbations show that amino acids from three regions of the RRM domain are significantly affected by RNA binding: the N-terminus leading into β1, namely residues V10, M13 and T14; residues D48, T51, K52 and E53 of the long flexible L3 loop; and residues L16, D42, V43, Y44, I45, V60 and R61, comprising the β-sheet formed by β1, β2 and β3 (Figure 3 and Supplementary Figure S4). In addition, line broadening upon RNA titration of the L3 loop residue R49, supports the notion that these regions are interacting with the RNA. In addition to probing the backbone NH resonances of SRSF2 RRM, the 13C side chain resonances were also monitored throughout the RNA titration. Large side chain shifts were identified for the aromatic rings of F57 and F59 in β3, the methyl groups of V10, M13 at the N-terminus and also V60 in the L3 loop (Supplementary Figure S5). The chemical shift data show that SRSF2 RRM domain, like most RRM domain, binds RNA using the β-sheet. In addition, however, resonances from the L3 region appear to be significantly perturbed (Figure 3).
We attempted to obtain information on intermolecular contacts between the RRM domain and the RNA by acquiring 13C, 15N-filtered NOE data using a 13C, 15N-labeled SRSF3-RNA complex sample, although only a limited number of contacts are observed. From the SRSF2:AGCAGAGUA complex, four intermolecular NOEs could be assigned to specific residues; these involved NOEs from residues in the L3 loop, the N and C-termini, namely between V10-Ade6, D48-Gua2, Y50-Gua2 and Y92-Uri8 (Figure 6A). The intermolecular NOEs agree well with the chemical shift mapping data (Figure 3). From the specific interactions identified, we determine that the long flexible loop L3, between β2 and β3, plays an essential role in stabilizing the interaction with the RNA 5′-end (Adenine 1 and Guanidine 2), whereas the flexible C-terminus interacts with the RNA toward the 3′-end (Uridine 8).
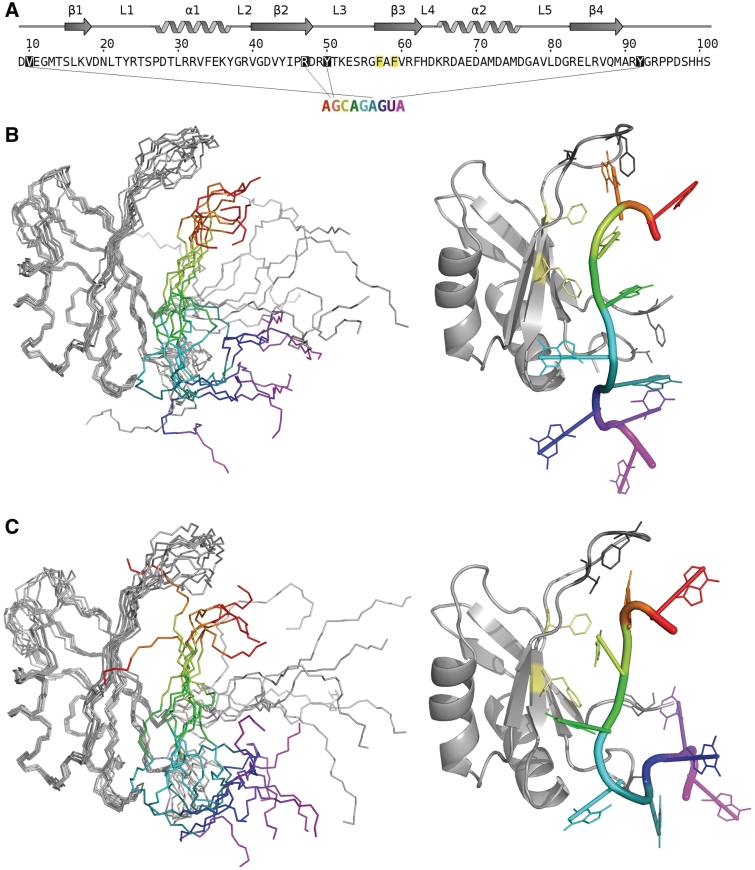
Figure 6.
(A) NOEs used to derive the model of SRSF2–RRM bound to 9-mer AGCAGAGUA RNA; V10-Ade6, D48-Gua2, Y50-Gua2 and Y92-Uri8. (B) Left: Ensemble of 10 structures from CNS calculations that contribute to the lowest energy cluster. (C) Left: Ensemble of five structures from CNS calculations that contribute to the second cluster. In both (B) and (C) the mobility of loop 3 and terminal regions afford a great degree of freedom to the orientation of the RNA. Right: representative structure (closest to mean) from each cluster with side chain residues shown for the incorporated intermolecular NOEs. In addition conserved hydrophobic residues F57 and F59 (pale yellow) are found to be involved in the binding.
The number of intermolecular contacts is small. One possible explanation is the severe chemical exchange line-broadening of some of the residues at the protein–RNA binding interface. Of those observed, most are from regions of high flexibility in the SRSF2 structure—the L3 loop region, and the N- and C-termini. Detection of these intermolecular NOEs suggests that the interactions involving these regions are significantly long-lived rather than transient. However, the limited number of intermolecular contacts, plus the fact that these are between poorly structured, flexible regions of both the SRSF2–RRM and the RNA, precluded the calculation of a high-resolution structure of the complex.
To probe the importance of the L3 region, several mutants were generated. Being in the loop region, these mutations have minimal effects on the integrity of the RRM fold, as confirmed by the 1H-15N HSQC spectra of the mutants. These spectra show that the mutant proteins are folded, with minimal shift changes compared to the wild-type spectrum, and these confined mainly to the sites of mutation (Supplementary Figure S6). Hence, any effects on the affinity of the RNA to the mutant proteins are the results of the specific mutation.
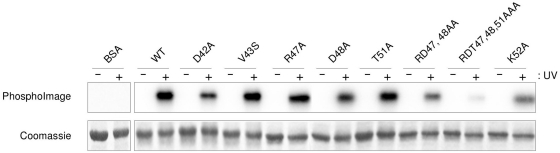
RNA binding was assayed by UV cross-linking (Figure 4). The mutated amino acids with the most pronounced effect were K52, the double mutant R47-D48 and the triple mutant R47, D48, T51. These results unambiguously demonstrate the importance of the mutated residues in mediating RNA binding. That the loop mutations cause the most dramatic decrease in RNA binding affinity compared to the wild-type protein suggests that, in the case of SRSF2–RRM, the canonical β1 and β3 interactions (1) found in typical RRM:RNA binding are themselves not sufficient for effective RNA binding; the additional loop L3 is crucial for RNA complex formation.
Figure 4.
SRSF2:RNA UV cross-linking. Various purified proteins (BSA, GB1-6His-SRSF2 9-101 wild-type and point mutants) were incubated with 32P-labelled 9-mer RNA (AGCAGAGUA) before binding reactions were irradiated with UV (+) or not (−) and analysed by 15% SDS–PAGE stained with Coomassie and autoradiography.
Binding of SRSF2 RRM to RNA by isothermal titration calorimetry
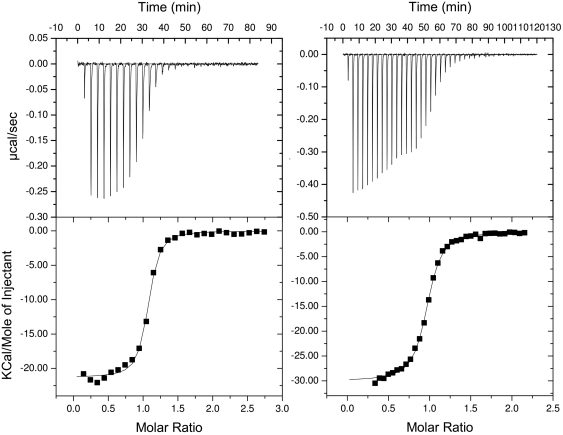
The 5-mer GAGUA SELEX motif bound too weakly and was unsuitable for ITC investigations. Isothermal calorimetric titrations using the 9-mer give a good binding curve and showed that the binding is exothermic (Figure 5). Fitting the calorimetry curve to a one site model yielded good thermodynamics parameters The 9-mer binds with a stoichiometry of 1:1 and a dissociation constant, Kd, value of ∼61 nM (Figure 6), with a large negative enthalpy, ΔH, of −21 kcal/mol and change in entropy, ΔS, of approximately −38.4 cal mol−1 K−1 (at 298 K). The same experiments repeated at 200 mM KCl gave a Kd value of ∼1.36 µM, ΔH, of −26 kcal/mol and change in entropy, ΔS, of −60.6 cal mol−1 K−1, with a reduction in the minor non-specific initial interactions observed at low salt concentrations (36) (Supplementary Figure S7). The enthalpy-driven interactions accompanied by large heat of associations are not dissimilar to many protein–RNA interactions. The salt dependence of the RNA binding suggests the presence of electrostatic interactions between SRSF2 and the 9-mer.
Figure 5.
Isothermal titration calorimetry curves for WT and K52A mutant fit to a one-site model. (A) WT SRSF2 with AGCAGAGUA (25 mM PO43-, 25 mM KCl, 25°C) curve fitting to a one-site 1:1 model yields fit parameters: N (stoichiometry ratio) = 1.03, Kd = 6.17 × 10−8M, ΔH = −21.3 kcal/mol and ΔS = −38.4 cal mol−1 K−1. (B) K52A SRSF2 with AGCAGAGUA (25 mM PO43-, 25 mM KCl, 25°C), curve fitting to a one-site model yields fit parameters: N = 0.953, Kd = 1.63 × 10−7M, ΔH = −30 kcal/mol and ΔS = −69.5 cal mol−1 K−1.
A comparable binding curve, albeit weaker binding, is obtained for the single point mutant K52A, with a Kd value of ∼170 nM at 25 mM KCl, and a binding stoichiometry of 1:1, a negative enthalpy of −29.4 kcal/mol and ΔS of −67.7 cal mol−1 K−1 at 298 K (Figure 5). The same experiments repeated at 200 mM KCl gave a Kd value of ∼3.6 µM. The effects of KCl on the mutant protein interactions are similar to those of the wild-type protein. This suggests that apart from electrostatic interactions, the other types of interactions such as aromatic ring stacking and hydrogen bonds are also likely to be important in the SRSF2–RNA complex. The dissociation constant for the R47-D48 double mutant was too weak to be measured by ITC.
Comparison of SRSF2–RNA interactions with SRSF3 RRM:CAUC
The binding of SRSF3 to 4-mer CAUC relies on π stacking interactions between amino acids Y13, F50 and F48 across the β-sheet to C1, A2 and U3, respectively (16); in this complex, the aromatic side-chains of these residues and the RNA nucleotide bases formed a very compact network of hydrophobic interactions. In the case of SRSF2 these positions on the β-sheet are occupied by aromatic residues F57 and F59 and are involved in binding, as evident from the 1H-13C-chemical shift; however, the residue corresponding to Y13 is the basic residue K17. The NMR chemical shift mapping data show K17 not to be significantly affected by the binding of the RNA. Given that the structure of SRSF3:CAUC is already known, it is highly likely that replacing Y13 (in SRSF3) with K17 in SRSF2 will have a significant effect on the affinity of SRSF2 for RNA.
The amino acids present in the β-sheets are thought to be non-selective as they are common to all RRM (1). However, in nature, alternative splicing via SR protein are known to proceed via selective interaction between specific RNA sequences and SR RRM domains. Along with the β-sheet interactions, the loop region L3 of both SRSF2 and SRSF3 interact with the RNA. In the case of SRSF3, the L3 loop comprises four residues including two prolines that contribute to a relatively well constrained short loop region. SRSF2, however, consists of nine amino acids which are shown here by NMR relaxation studies to be relatively flexible. The flexibility and length of SRSF2 L3 may explain why it was difficult to observe a high number of intermolecular NOEs. In addition the longer length of the RNA used here in SRSF2 binding was necessary to obtain a high-affinity complex, compared to the much shorter 4-mer RNA for SRSF3 which was chosen for the quality of the resultant NMR spectra rather than for its affinity to SRSF3 (16). The C-terminus residues of both SRSF2 and SRSF3 provide binding interactions to the 3′-end of the RNA (G7) through residue N82 of SRSF3 and Y92 of SRSF2. The N-termini interactions found in SRSF2 namely V10-RNA (G7) are also consistent with K11–RNA interaction identified in SRSF3-CAUC complex although this interaction was not satisfied in the final structure reported for the SRSF3–CAUC complex, due possibly to the truncated nature of the RNA used. It is possible that with a longer RNA sequence might make more contacts with the RRM domain of SRSF3, similar to the ones observed here.
In summary, our results show that the flexible L3 loop of SRSF2 together with its flexible N- and C-termini collectively provide the necessary binding sites for the RNA interaction
DISCUSSION
Roles of loop regions of SRSF2 key to RNA interaction
The structure of SRSF2 exhibits the classic RRM-SR protein fold comprising a four-strand anti-parallel β-sheet and two α-helices. The L3 loop region between β-stands 2 and 3 of all the SR-RRM domains shown in Figure 2 are of variable lengths, with L3 in SRSF2 being somewhat longer and highly flexible. RNA binding to SRSF2 was initially probed by interaction with a 5-mer RNA identified by SELEX. Although this sequence bound favorably when compared to control 5-mers selected on purine/pyrimidine composition, the resultant interaction was of low affinity with Kd of the order of 10−4 M. The extension of the sequence to the 9-mer, again based on SELEX, increased the affinity to the order of 10−8 M. Changes in chemical shifts of SRSF3 upon binding the 9-mer AGCAGAGUA RNA showed that SRSF3 binds RNA with the expected features, involving the β-sheet and loop regions.
The importance of the L3 loop is most interesting; this appears to be a primary site since it is the region whose chemical shifts are most affected upon addition of both the weak-binding 5-mer GAGUA and the 9-mer AGCAGAGUA. The mutagenesis studies also demonstrate that L3 residues such as R47, D48 and K52 are responsible for mediating SRSF2 binding to the RNA. Other structural and mutagenesis studies of RRM–RNA interactions have previously highlighted that the loop regions can play important roles in RNA recognition although which and how many loops are important is protein specific (37). Focusing on the L3 loop (β2–β3 loop), in human RBMY this loop is required for the recognition of the shape of the RNA, based on the fact that all the loop residues contact the phosphate backbone (38). In the case of SRSF2, the side chains D48 and Y50 form intermolecular NOEs with the G2 nucleotide base; this together, with the significant effects of mutagenesis, suggests that L3 has a role in nucleotide recognition.
In many RRM–RNA structures, an aromatic residue present in loop L1 (β1–α1 loop) is crucial for RRM–RNA interactions. For example, the F126 of Fox-1 RRM is important for binding the 5′-end of the RNA (39). The equivalent residue in SRSF2 is Y23. Only a modest reduction is affinity for RNA was observed when Fox-1 F126 was mutated to a tyrosine residue, implying that Y23 in SRSF2 could, in principle, play a similar role in RNA binding as F126 in Fox-1. Surprisingly, the NMR resonances of Y23 of SRSF2 were not affected upon RNA binding and no intermolecular NOEs involving Y23 were observed (Supplementary Figure S5). In addition, SRSF3, like many other SR proteins, have no equivalent aromatic residues in loop 1. This suggests that for the SR family of RRM domains, loop 1 is not involved in RNA binding.
The results here show that SRSF2 binds RNA using features which are found in other RRM–RNA interactions, namely, via the canonical β-sheet binding interface and the crucial involvement of one loop region, that is, the L3 loop.
This change from an aromatic to a basic residue between the SR proteins could potentially be one of the factors which determine RNA sequence selectivity. A comparison between the low-resolution SRSF2-9-mer AGCAGAGUA RNA model structure from the cluster, with the structures of SRSF3:CAUC (and Fox-1:UGCAUGU) supports the variability of RRM–RNA interactions that are known to exist (Supplementary Figure S8).
Non-specific standard RRM interactions are present
A comparison with the structure of SRSF3 bound to a 4-nt RNA highlight that non-specific standard RRM interactions are present on the solvent-exposed face of the β-sheet of SRSF2. In particular, in both the SRSF2- and SRSF3–RNA complexes, the well-conserved F57 and F59 are shown to be involved in interactions with the counterpart RNA. However, these interactions alone are insufficient. In the case of SRSF3-CAUC, the Y13 in β1 provides that additional stabilizing interactions with C1. In SRSF2, Y13 is replaced by a lysine in the equivalent position (K17). The loss of one of the most conserved aromatic residues in the RNP2 motif provides the possible explanation as to why SRSF2 is only able to bind a 5-mer RNA weakly, and that longer RNA fragments such as the 9-mer are necessary in order to provide additional protein–RNA contacts to stabilize the SRSF2–RNA complex.
We characterized the thermodynamics of the 9-mer interactions with SRSF3 using isothermal titration calorimetry. This interaction represents an interaction between a RRM domain and an unstructured single stranded RNA. There are very few examples in the literature of the thermodynamics analyses of the interactions between RRM domains and unstructured RNAs. In several reported cases, these interactions have been accompanied by very large favorable enthalpy changes (−30 to −60 kcal mol−1) and unfavorable entropy changes, and these have been confirmed to be of physiological significance (40). The enthalpic and entropic changes for SRSF2–RNA interactions reported here are more modest (ΔH = −21 kcal/mol −TΔS = 11.43 cal mol−1 K−1) although still significant and larger than average protein–protein interactions. The large enthalpy and entropy changes observed for many of the RRM–RNA interactions are attributed to the extensive π stacking interactions involving aromatic residues in β1 and β3 whose positions are structurally conserved to afford these hydrophobic interactions with the nucleotides. In SRSF2, as discussed above, the β1 aromatic residue is missing, hence, providing a possible explanation for the smaller ΔH and ΔS.
Restraint-driven model of SRSF2-9-mer AGCAGAGUA RNA complex suggests different mode of binding
The limited number of intermolecular contacts, plus the fact that these are between poorly structured, flexible regions of both the SRSF2–RRM and the RNA, precluded the calculation of a high-resolution structure of the complex. However, models could be obtained from the limited intermolecular NOE data and chemical shift perturbations using the CNS software which produced an ensemble of the water-refined structures in which all the experimental restraints were satisfied. Pairwise RMSD analysis of the ensemble structures showed that the ensemble could be split into two clusters, using a cutoff of 5 Å (Figure 6B and C). These two clusters are quite similar with variation between them being <7 Å. In these models, the backbone of the nucleotides 2–4 of the RNA are aligned parallel to the β-strands with multiple orientations for the 5′ (proximal to loop 3) and 3′ (proximal to the N- and C-termini) end nucleotides. The variation between the two clusters is minimal (RMSD of <7 Å) for structured regions. The two clusters resolve below 5 Å and appear to arise due to differing local environments for G2 and A4. In the first cluster (Figure 6B), the orientations for loop 3 seem restricted due to G2 in close proximity to T51 (and restrained by G2-Y50 NOE). In the second cluster (Figure 6C) positioning of G2 appears more varied with orientation of A4 more restricted in close proximity to Y44 of β-strand 2. In both clusters, it is evident that A1 and G2 interact with L3, and the residues of the N- and C-termini (namely V10 and Y92) are in close contacts with 5′-end of the RNA. In addition, nucleotides C3 and A4 are located adjacent to the β-sheet.
In summary, it is possible, even with these low-resolution models, to discern the orientation of the RNA with respect to the RRM, which highlight a different orientation of the RNA relative to the protein when compared with SRSF3 and other RRM–RNA complexes. The models show A1 and G2 interacting with L3, and the residues V10 and Y92 in close contacts with 5′-end of the RNA. It is posited that the mode of interaction obtained here is due to the longer length of the RNA forming more points of contacts with the SRSF2 RRM domains (involving loop 3, and also the C- and N-termini) leading to the different RNA orientation relative to the RRM domain.
The results here show that the flexible L3 loop of SRSF2 together with its flexible N- and C-termini collectively provide the necessary binding sites for the RNA interaction. The flexibility and variability of loop 3 residues and C- and N-termini between SR family members could provide the selectivity required for the alternative splicing pathways targeted by different family members. Many structural RRM:RNA binding studies use small RNA fragments to facilitate ease of analysis; however, the results here show that longer RNA fragments are necessary in the case of SRSF2 in order to obtain better affinity, with binding afforded by the collaborative effects of two binding areas. Therefore, studies involving both longer RNA fragments and the N/C residues beyond the consensus RRM domain may provide further insights into the selectivity of the RRM binding in SR proteins.
ACCESSION NUMBER
PDB 2KN4.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–8.
FUNDING
Biotechnology and Biological Sciences Research Council (grant number BB/D012716/1 to S.A.W. and L-Y.L.); Wellcome Trust (grant number 086391 to L-Y.L.). Funding for open access charge: Wellcome Trust (grant number 086391).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Dr Igor Barsukov for help and useful discussions regarding protein–RNA intermolecular NOE experiments and ITC. The University of Liverpool is thanked for its support of the NMR Centre for Structural Biology. The authors also wish to thank Mark Tully and Phillip Widdowson for support in the laboratory.
REFERENCES
- 1.Maris C, Dominguez C, Allain FH-T. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 2.Bourgeois CF, Lejeune F, Stevenin J. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:37–88. doi: 10.1016/S0079-6603(04)78002-2. [DOI] [PubMed] [Google Scholar]
- 3.Ibrahim EC, Schaal TD, Hertel KJ, Reed R, Maniatis T. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc. Natl Acad. Sci. USA. 2005;102:5002–5007. doi: 10.1073/pnas.0500543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen H, Kan JLC, Green MR. Arginine–serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 5.Wang HY, Xu X, Ding JH, Bermingham JR, Jr, Fu XD. SC35 plays a role in T cell development and alternative splicing of CD45. Mol. Cell. 2001;7:331–342. doi: 10.1016/s1097-2765(01)00181-2. [DOI] [PubMed] [Google Scholar]
- 6.Xiao R, Sun Y, Ding JH, Lin S, Rose DW, Rosenfeld MG, Fu XD, Li X. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol. Cell. Biol. 2007;27:5393–5402. doi: 10.1128/MCB.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding J-H, Xu X, Yang D, Chu P-H, Dalton ND, Ye Z, Yeakley JM, Cheng H, Xiao R-P, Ross J, Jr, et al. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. EMBO J. 2004;23:885–896. doi: 10.1038/sj.emboj.7600054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cataldi A, Zingariello M, Rapino M, Zara S, Daniele F, Di Giulio C, Antonucci A. Effect of hypoxia and aging on PKC d-mediated SC-35 phosphorylation in rat myocardial tissue. Anat. Rec. 2009;292:1135–1142. doi: 10.1002/ar.20936. [DOI] [PubMed] [Google Scholar]
- 9.Solis AS, Peng R, Crawford JB, Phillips JA, Patton JG. Growth hormone deficiency and splicing fidelity - Two serine/arginine-rich proteins, ASF/SF2 and SC35, act antagonistically. J. Biol. Chem. 2008;283:23619–23626. doi: 10.1074/jbc.M710175200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandradas S, Deikus G, Tardos JG, Bogdanov VY. Antagonistic roles of four SR proteins in the biosynthesis of alternatively spliced tissue factor transcripts in monocytic cells. J. Leukocyte Biol. 2010;87:147–152. doi: 10.1189/jlb.0409252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dreumont N, Hardy S, Behm-Ansmant I, Kister L, Branlant C, Stevenin J, Bourgeois CF. Antagonistic factors control the unproductive splicing of SC35 terminal intron. Nucleic Acid. Res. 2006;38:1353–1366. doi: 10.1093/nar/gkp1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tacke R, Manley JL. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu HX, Chew SL, Cartegni L, Zhang MQ, Krainer AR. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell. Biol. 2000;20:1063–1071. doi: 10.1128/mcb.20.3.1063-1071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavaloc Y, Bourgeois CF, Kister L, Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, Lian L-Y, Wilson SA, Allain FHT. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO J. 2006;25:5126–5137. doi: 10.1038/sj.emboj.7601385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: Development of a software pipeline Proteins: Struct. Func. Bioinformatics. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 18.Wishart DS, Sykes BD. The C-13 chemical shift index - a simple method for the identification of protein secondary structure using C-13 chemical shift data. J. Biom. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 19.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 20.Cornilescu G, Delaglio F, Bax AJ. Backbone angle restraints from searching a database for chemical shift and sequence homology. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 21.Rieping W, Habeck M, Bardiaux B, Bernard A, Malliavin TE, Nilges M. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- 22.Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–496. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 23.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta. Cryst. D. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 24.Pei J, Kim B-H, Grishin NV. PROMALS3D: a tool for multiple sequence and structure alignment. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shatsky M, Nussinov R, Wolfson HJ. A method for simultaneous alignment of multiple protein structures. Proteins: Struct. Func. Bioinformatics. 2004;56:143–156. doi: 10.1002/prot.10628. [DOI] [PubMed] [Google Scholar]
- 26.Kay LE, Nicholson LK, Delaglio F, Bax A, Torchia DA. Pulse sequences for removal of the effects of cross-correlation between dipolar and chemical shift anisotropy relaxation mechanism on the measurement of heteronuclear T1 and T2 values in proteins. J. Magn. Reson. 1992;97:359–375. [Google Scholar]
- 27.Tintaru AM, Hautbergue GM, Hounslow AM, Lian LY, Craven CJ, Wilson SA. Structure of SF2/ASF RNA recognition motif 2 reveals a novel RNA binding interface, (2007) EMBO Reports. 2007;8:756–762. [Google Scholar]
- 28.Cromsigt J, van Buuren B, Schleucher J, Wijmenga S. Resonance assignment and structure determination for RNA. Meth. Enzymol. 2001;338:371–399. doi: 10.1016/s0076-6879(02)38229-6. [DOI] [PubMed] [Google Scholar]
- 29.Lee W, Arrowsmith C, Kay LE. A pulsed field gradient isotope-filtered 3D 13C HMQC-NOESY experiment for extracting intermolecular NOE contacts in molecular complexes. FEBS Lett. 1994;350:87–90. doi: 10.1016/0014-5793(94)00740-3. [DOI] [PubMed] [Google Scholar]
- 30.de Vries SJ, van Dijk ADJ, Krzeminski M, van Dijk M, Thureau A, Hsu V, Wassenaar T, Bonvin AMJJ. HADDOCK versus HADDOCK: New features and performance of HADDOCK2.0 on the CAPRI targets. Proteins: Struc. Funct. Bioinformatics. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 31.Dominguez C, Boelens R, Bonvin AMJJ. HADDOCK: a protein–protein docking approach based on biochemical and/or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 32.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR System (CNS), A new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Zhou P, Lugovskoy AA, Wagner G. A solubility-enhancement tag (SET) for NMR studies of poorly behaving proteins. J. Biomol. NMR. 2001;20:11–14. doi: 10.1023/a:1011258906244. [DOI] [PubMed] [Google Scholar]
- 34.Berjanskii MV, Wishart DS. A simple method to predict protein flexibility using secondary chemical shifts. J. Am. Chem. Soc. 2005;127:14970–14971. doi: 10.1021/ja054842f. [DOI] [PubMed] [Google Scholar]
- 35.Ngo JCK, Giang K, Chakrabarti S, Ma CT, Huynh N, Hagopian JC, Dorrestein PC, Fu X-D, Adams JA, Ghosh G. A sliding docking interaction is essential for sequential and processive phosphorylation of an SR protein by SRPK1. Mol. Cell. 2008;29:563–576. doi: 10.1016/j.molcel.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holbrook JA, Tsodikov OV, Saecker RM, Record MT. Specific and non-specific interactions of integration host factor with DNA: thermodynamic evidence for disruption of multiple IHF surface salt-bridges coupled to DNA binding. J. Mol. Biol. 2001;310:379–401. doi: 10.1006/jmbi.2001.4768. [DOI] [PubMed] [Google Scholar]
- 37.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not Quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 38.Skrisovka L, Bourgeois CF, Stefl R, Grellscheid SN, Kister L, Wenter P, Elliot DJ, Stevenin J, Allain FH. The testis specific human protein RBMY recognizes RNA through a novel mode of interaction. EMBO Rep. 2007;8:372–379. doi: 10.1038/sj.embor.7400910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S, Allain FH. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLaughlin JJ, Jenkins JL, Kielkopf CL. Large favorable enthalpy changes drive specific RNA recognition by RNA recognition motif proteins. Biochemistry. 2011;50:1429–1431. doi: 10.1021/bi102057m. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.