Abstract
The chloroplast psbD and psbC genes encode the D2 and CP43 proteins of the photosystem II complex, and they are generally cotranscribed. We report studies on the basic translation process of tobacco psbD-psbC mRNAs using an in vitro translation system from tobacco chloroplasts. The primary transcript has an unusually long 5′-UTR (905 nt). We show that it is translatable. Processing of the 5′-UTR greatly enhances the translation efficiency of the psbD cistron. A striking feature is that psbD and psbC cistrons overlap by 14 nt. Removal of the psbD 5′-UTR plus the start codon and introduction of a premature termination codon in the psbD cistron considerably reduce the translation efficiency of the downstream psbC cistron. These results indicate that translation of the psbC cistron depends largely on that of the upstream psbD cistron and thus shows translational coupling; however, a portion is independently translated. These observations, together with the presence of monocistronic psbC mRNAs, suggest that the psbD and psbC cistrons are translated via multiple processes to produce necessary amounts of D2 and CP43 proteins.
INTRODUCTION
The chloroplast DNA in flowering plants is a circle ∼150 kb long, and includes approximately 80 different protein-encoding genes (1,2). Chloroplast gene expression is regulated in part at the transcriptional level (3,4), whereas posttranscriptional steps such as RNA editing, splicing, cleavage, trimming, translation and control of mRNA stability are more important for expression of most genes (5–11). Many chloroplast genes are clustered and are cotranscribed to polycistronic transcripts that are then processed into mature RNAs (12–15). Some gene clusters consist of functionally unrelated genes; for example, the psaC-ndhD operon encodes a photosystem I subunit and an NADH dehydrogenase subunit (16). To express functionally unrelated cistrons from a polycistronic mRNA, regulation at posttranscriptional steps for each cistron would be required to supply proper amounts of products during development and under different environmental conditions. The photosystem II complex consists of approximately 20 different subunits, of which 14 are encoded in the chloroplast genome of flowering plants (17,18). Many of these genes are also clustered. One example is the clustered psbD gene for D2 protein and psbC gene for CP43 protein. A striking feature is that these two genes overlap by 14 nt in flowering plants (19–21), algae (22) and cyanobacteria (23). A notable exception is that these two genes are separated in Chlamydomonas reinhardtii (24).
In flowering plants, the overlapping psbD and psbC genes (psbD/C) are cotranscribed with the downstream psbZ gene (formerly ORF62 or ycf9) to produce multiple overlapping transcripts (25,26). Transcription of monocot psbD/Cs starts from the preceding psbK and psbI gene cluster to give longer transcripts (27–29). The promoter for psbD/C is blue-light responsive (30,31) and is recognized by E. coli-type RNA polymerase with sigma factor 5 (32,33). However, little is known about the translation process of psbD/C mRNAs. In contrast, translation of separated psbD and psbC mRNAs in Chlamydomonas has been well studied using a biolistic chloroplast transformation system and heterologous reporter systems. Translation and stability of Chlamydomonas psbC and psbD mRNAs are controlled by interactions between 5′-UTRs and nucleus-encoded proteins (7–10).
We previously reported that the tobacco monocistronic psbC mRNA is translatable (34). However, translation of the psbC cistron from the dicistronic psbD/C mRNA has not been investigated. In addition to being overlapping, the primary psbD/C transcript has an unusually long 5′-UTR. A portion of the primary transcript is processed into mRNA with a shorter 5′-UTR (25). Hence, chloroplasts contain a mixture of primary and processed psbD/C mRNAs. In vitro translation systems are a useful tool to study the basic translation process for single mRNA species by eliminating possible effects of mRNA cleavage or stability, translation of other mRNAs, assembly-dependent control and levels of necessary components (e.g. ATP, GTP, amino acids, ribosomes).
Using our in vitro translation system from tobacco chloroplasts, we show here that the psbD cistron is translatable from the primary transcript. Processing of 5′-UTRs of both psbD/C dicistronic and psbC monocistronic mRNAs significantly enhances translation efficiency. We then show that translation of the psbC cistron occurs from the dicistronic mRNA and it depends highly on translation of the upstream psbD cistron (indicating translational coupling). We discuss how translation of the downstream cistron proceeds in the overlapping psbD/C mRNA.
MATERIALS AND METHODS
The coding region for Cerulean (35) was prepared from the EGFP-coding region (36) and that for Citrine was obtained as described (36). Procedures for plasmid construction and primers are in Supplementary Figures S1 and S2. Necessary regions from psbD/C were amplified by PCR from tobacco chloroplast DNA (37) and PCR products were confirmed by DNA sequencing. Synthesis and purification of test mRNAs were as described (38). Test mRNA sequences are shown in Supplementary Figure S3. Isolation of intact tobacco chloroplasts and preparation of chloroplast extracts (S30 fractions) were as described (38). In vitro translation was performed at 28°C for 1 h as described (38). Protein products were separated by 12.5% native polyacrylamide gel electrophoresis (PAGE). Fluorescent signals were detected and quantified with a Typhoon 9400 system (GE Healthcare, Little Chalfont, UK). The 526SP and 555BP20 filters were used to detect Cerulean and Citrine bands when excited by 457-nm and 532-nm actinic light, respectively.
RESULTS
Measurement of translation from psbD and psbC cistrons
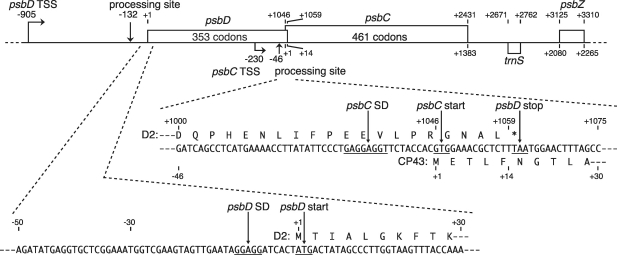
Figure 1 shows a schematic of the tobacco chloroplast psbD-psbC-psbZ operon (17,37). These genes are cotranscribed to produce various mRNAs and the psbD/C mRNA is one of the major transcripts accumulated in chloroplasts (25,26). In vitro capping assays have identified two transcription initiation sites: the main site is located 905-bp upstream from the psbD ATG start codon and the additional site is 230-bp upstream from the psbC GTG start codon (25,34). The primary psbD/C transcript has a 905-nt 5′-UTR with a processing site at position −132 relative to the A (+1) of the AUG start codon (25).
Figure 1.
The tobacco chloroplast psbD/C/Z operon. Positions relative to the psbD ATG start codon (A is +1) are shown above, and positions relative to the psbC GTG start codon (the first G is +1) are shown below. Bent arrows indicate transcription start sites, and vertical arrows indicate processing sites. mRNA sequences around the psbD and psbC start codons are shown below. SD-like sequences, start codons and a stop codon are underlined. Deduced amino acid sequences from the psbD and psbC cistrons (encoding the D2 and CP43 proteins) are respectively shown above and below the mRNA sequences.
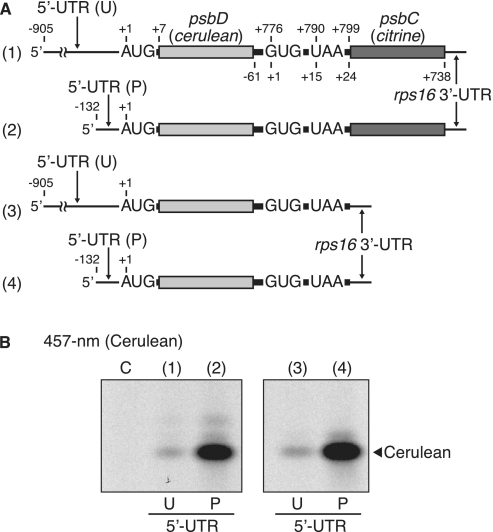
To differentially monitor translation of the psbD and psbC cistrons in psbD/C mRNA, we utilized Cerulean (a cyan-fluorescent protein) and Citrine (a yellow-fluorescent protein). A large portion of the psbD cistron was replaced with the Cerulean-coding sequence (cerulean, 236 codons without start and stop codons), and that of the psbC cistron with a Citrine-coding frame (citrine, 237 codons without start codon) (Figure 2A). We left the last 75 nt of the psbD cistron and the first 24 nt of the psbC cistron in the mRNA, because these sequences include the Shine–Dalgarno (SD)-like sequence (GAGGAGGU), the psbC start codon (GUG) and the psbD stop codon (UAA), and because sequences downstream from start codons are often required for efficient translation (39). The tobacco chloroplast rps16 3′-UTR was linked after citrine to standardize test mRNAs (38).
Figure 2.
Translation of the psbD cistron with primary and 5′-processed UTRs. (A) mRNA templates in which the psbD and psbC cistrons were respectively replaced with Cerulean (cerulean) and Citrine (citrine) coding regions. Positions are as in Figure 1. (template ‘1’) mRNA with the primary unprocessed (U) 5′-UTR. (template ‘2’) mRNA with processed (P) 5'-UTR. (templates ‘3’ and ‘4’) mRNAs lacking the downstream cistron with U and P 5′-UTRs. (B) Gel patterns of translation products. After in vitro translation, the synthesized Cerulean products were detected by fluorescence under 457-nm light. Lanes labeled ‘C’, no-mRNA control. Faint bands above the main Cerulean band are probably Citrine products (top, lane 2) or loosely folded Cerulean (middle, lanes 2 and 4).
Translation efficiencies of test mRNAs were measured using the in vitro translation system under template-limiting and linear progression (1 h) conditions, which enable estimation of the relative rate of translation (38). We confirmed by primer extension analysis that 5′-processing or cleavage of test mRNAs was hardly detectable during a 1-h incubation, probably due to the presence of an RNase inhibitor (38) (Supplementary Figure S3). After reaction, protein products were resolved by native PAGE. The fluorescent intensity of Cerulean and Citrine products was measured (see Figure 4B). Hence, this assay can monitor either psbD translation or psbC translation from the psbD/C mRNA by changing the excitation wavelength. The Cerulean product migrates faster in native gels than the Citrine product due to charge and size differences, which further helps differential monitoring (see Figure 4B).
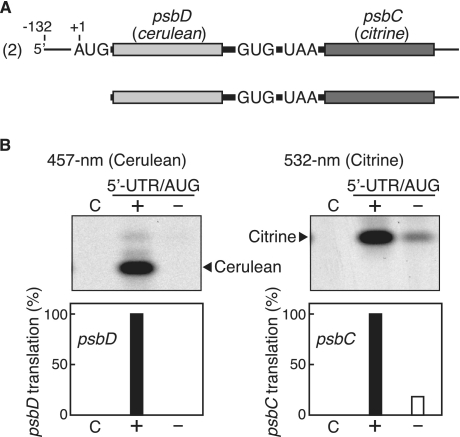
Figure 4.
Effect of psbD 5′-UTR/AUG deletion on psbC translation. (A) mRNA templates. The processed dicistronic mRNA is as in Figure 2A (template ‘2’), and the mRNA after deletion of its 5′-UTR and the following AUG. (B) Gel patterns of translation products. Cerulean products were detected by fluorescence under 457-nm light (left panel) and Citrine products under 532-nm light (right panel). Lanes labeled ‘C’, no-mRNA control. Lanes labeled ‘+’ and ‘−’ respectively represent mRNAs with and without the sequence for the psbD 5′-UTR and the following AUG. Faint bands are as in the legend for Figure 2. Quantified efficiencies of psbD and psbC translation are shown in the bar graphs below (‘2’ mRNA value defined as 100%, mean values from ≥3 independent assays).
Roles of 5′-processing on translation efficiency of the psbD cistron
The tobacco psaC-ndhD transcript is processed into functional mRNA species (16). It is hence possible that the primary psbD/C transcript is simply a precursor and not functional in translation because it has an extremely long 5′-UTR. We first examined whether translation of the psbD cistron occurs from the primary psbD/C mRNAs. Two test psbD/C mRNAs were prepared: one contains the primary (unprocessed) 5′-UTR of 905 nt and the other the processed 5′-UTR of 132 nt (Figure 2A, lanes 1 and 2). These mRNAs are expected to synthesize Cerulean products when translation starts from the psbD start codon. As shown in Figure 2B (lane 1), 457-nm excitation gave a single major band, indicating that the primary transcript is translatable. However, processing at position −132, removing 85% of the primary 5′-UTR, greatly enhanced its translation efficiency (10-fold, lane 2).
The long 5′-UTR of the primary transcript may interact with the downstream cistron, thereby interfering with psbD translation. To exclude this possibility, we prepared additional test mRNAs that lack the psbC (citrine) cistron, namely, artificial monocistronic psbD mRNAs (Figure 2A, lanes 3 and 4), and assayed their translation. Figure 2B (lanes 3 and 4) shows that their translation efficiencies were similar to the original dicistronic mRNAs, indicating that the downstream cistron does not affect translation of its upstream cistron.
The tobacco psbD mRNA contains a typical SD-like sequence, GGAGG, in its 5′-UTR (see Figure 1). The distance between the SD-like sequence and the AUG codon is 6 nt, a proper spacing (40). Changing GGAGG to AAGAA nearly abolished psbD translation, indicating a crucial role of the SD-like sequence on psbD translation initiation (Supplementary Figure S5).
Translation of the psbC cistron from dicistronic and monocistronic mRNAs
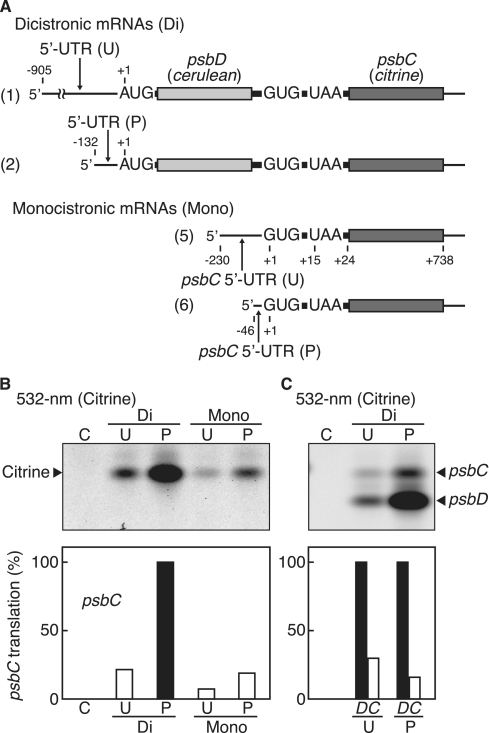
The second promoter located within the upstream psbD cistron produced 1.5- to 2.6-kb transcripts, of which two are monocistronic psbC mRNAs (25). We previously showed that the monocistronic psbC mRNA is translatable and its start codon is GUG, not the upstream in-frame AUG (34). The psbC start codon is located 14 nt upstream from the preceding psbD stop codon, and this feature may interfere with psbC translation initiation in the dicistronic psbD/C mRNA. We then examined whether translation of the psbC cistron occurs from the dicistronic mRNA. We again used the primary unprocessed and 5′-processed mRNAs (Figure 3A). As shown in Figure 3B (labeled ‘Di’), excitation with 532-nm light enabled detection of Citrine products from the psbC start codon in both mRNAs, indicating that the psbC cistron can be translated from the dicistronic mRNA. Again, the translation efficiency of the psbC cistron was much higher with the 5′-processed mRNA than with the primary transcript. This difference parallels that observed in translation efficiencies of the upstream psbD cistron (Figure 2B), which suggests that translation of the downstream psbC cistron depends on translation of the preceding cistron.
Figure 3.
Translation of the psbC cistron. (A) mRNA templates. Dicistronic mRNAs (labeled ‘Di’) with primary/unprocessed (labeled ‘U’) and processed (labeled ‘P’) 5′-UTRs as shown in Figure 2A (template ‘1’) and (template ‘2’). Monocistronic mRNAs (labeled ‘Mono’) with primary/unprocessed (‘U’) and processed (‘P’) 5′-UTRs. (B) Gel patterns of translation products. Synthesized Citrine products were detected by fluorescence under 532-nm light. Lanes labeled ‘C’, no-mRNA control. Faint bands above the main Citrine bands could be loosely folded Citrine. (C) Templates are the dicistronic mRNAs above with cerulean replaced with citrine. Gel patterns of translation products. Synthesized Citrine products from both cistrons were detected by fluorescence under 532-nm light. Lanes labeled ‘C’, no-mRNA control. Quantified efficiencies are shown in the bar graphs below (‘P’-psbC value defined as 100% in (B), and psbD value as 100% in (C), mean values from ≥3 independent assays).
We prepared two monocistronic psbC mRNAs: one from the second transcription initiation site (the primary 230-nt 5′-UTR) and another from the processed site (the processed 46-nt 5′-UTR), as shown in Figure 2A, and assayed as above. Unexpectedly, the translation efficiency of the psbC cistron was much higher from the dicistronic mRNA than the monocistronic mRNA (Figure 3B, compare lanes labeled ‘Di’ and ‘Mono’). The 5′-processed mRNA was again high in translation efficiency (lanes labeled ‘Mono’). Both the primary and 5′-processed mRNAs accumulate to fairly high levels in tobacco chloroplasts (25). Hence, the results in Figures 2 and 3 strongly suggest that processing of the 5′-UTRs has a role in controlling translation efficiency.
To compare translation efficiencies for the psbD and psbC cistrons, we modified the above test mRNAs (labeled ‘Di’), replacing cerulean with citrine in the psbD cistron) to minimize variables, because quantitative assays are difficult to compare between different fluorescent proteins. The Citrine products from the psbD start codon migrate faster in native gels than those from the psbC start codon due to charge and size differences; however, minor cross-contamination is inevitable between the upper and lower bands. We estimated that the translation efficiency of the psbC cistron is roughly one-third that of the psbD cistron from the primary mRNA, and roughly one-sixth that of the 5′-processed mRNA (Figure 3C, compare lanes labeled ‘U’ and ‘P’). The reason for such a difference remains unclear.
Translational coupling of psbD and psbC in psbD/C mRNA
If translation of the psbC cistron is coupled with that of the upstream psbD cistron, the translation efficiency of the psbC cistron should depend on translation of the upstream cistron. To examine this possibility, we removed the 5′-UTR and the following AUG start codon from the processed psbD/C mRNA (Figure 4A). As expected, this deletion led to no psbD translation (Figure 4B, lane labeled ‘−’ in the left panel). Consequently, the translation efficiency of the downstream psbC cistron decreased dramatically (to ∼20% of the wild-type), though did not disappear completely (lane ‘−’ in the right panel).
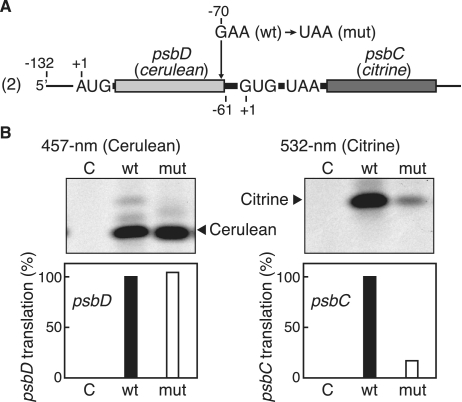
Next, we introduced a G-to-A point mutation at position −70 relative to the G (+1) of the psbC GUG codon, creating a premature stop codon (UAA) in the 3′-region of cerulean (Figure 5A). Consequently, the psbD and psbC cistrons no longer overlap and are separated by a 70-nt spacer, which prevents translational coupling (41). As shown in Figure 5B (left panel), no significant change in psbD translation was observed, whereas the translation efficiency of the psbC cistron was greatly reduced, to 20% (the right panel). On the basis of these results, we concluded that translation of the psbC cistron largely depends on translation of the preceding psbD cistron, though there is some independent psbC translation.
Figure 5.
Effect of premature termination of the upstream cistron on psbC translation. (A) The mRNA template is as in Figure 2 (template ‘2’). Arrow indicates the point mutation from G to U, creating a premature termination codon. (B) Gel patterns of translation products. Cerulean products were detected by fluorescence under 457-nm light (left panel) and Citrine products under 532-nm light (right panel). Lanes labeled ‘C’, no-mRNA control. Lanes labeled ‘wt’ and ‘mut’ represent wild-type and mutant mRNAs, respectively. Faint bands are as in the legend for Figure 2. Quantified efficiencies of psbD and psbC translation are shown in the bar graphs below (‘wt’ value defined as 100%, mean values from ≥3 independent assays).
DISCUSSION
In psbD/C transcripts of tobacco, spinach and pea (25,26,31), a striking feature is that the primary transcript has an unusually long 5′-UTR; for example, it is 905 nt in tobacco psbD, 75 nt in Chlamydomonas psbD (24) and 85 nt in tobacco psbA (encoding D1 protein) (38). This study was the first to demonstrate that the psbD cistron with its long 5′-UTR is translatable, indicating that the primary transcript is not merely a precursor for functional mRNAs. We then showed that the 5′-processed mRNA with a 132 nt 5′-UTR has very high translation efficiency for the psbD cistron and that the SD-like sequence is essential for its translation. Both primary and processed mRNAs are abundant in chloroplasts of mature tobacco leaves (25). Hence, we hypothesize that psbD/C expression is regulated at least in part via 5′-processing. This hypothesis could be applied for translation of the monocistronic psbC mRNA, because its 5′-processing greatly increases psbC translation efficiency. The 5′-UTR processing of psbD/C and psbC transcripts is presumably catalyzed or mediated by nucleus-encoded proteins. The amount or activity of these proteins could be modulated in response to light or developmental cues. Specific RNA-binding proteins are involved in mRNA processing and mRNA stabilization or translation (7–10,42–46). For example, maize PPR10 defines the processing site of atpH mRNAs and thereby might enhance atpH translation (46). Our previous in vitro study indicated that the effect of 5′-UTR processing on translation varies from mRNA to mRNA (38). Translation efficiencies from primary and processed rbcL and atpH 5′-UTRs were comparable, whereas processing of atpB and psbB 5′-UTRs enhanced translation efficiency (38). It is interesting to note that 5′-processing of tobacco psbD/C mRNAs leads to a 10-fold increase in translation efficiency (Figure 2B).
Another striking feature is the partial overlap of psbD and psbC cistrons. We demonstrated that translational coupling does occur between psbD and psbC cistrons. As expected, the translation efficiency of the psbC cistron was much lower than that of its upstream cistron (Figure 3C). Blocking psbD translation still supported 20% of psbC translation (Figure 4B), indicating a contribution from independent psbC translation, probably in a manner similar to translation of the monocistronic psbC mRNA.
Four pairs of overlapping genes have been reported in the tobacco chloroplast: ndhC-ndhK, atpB-atpE, psbD-psbC and rpl22-rps3 (37). The atpB and atpE cistrons have an overlap (AUGA) of the atpB stop codon (UGA) with the atpE start codon (AUG). We recently revealed that translation of the downstream atpE cistron is independent of that of the upstream atpB from the dicistronic atpB/E mRNA and that the atpE cistron is translated via its own cis-element (36). In contrast, we found that the ndhC and ndhK cistrons overlap by 7 nt (the ndhK start codon is located 4 nt upstream from the ndhC stop codon) and ndhK translation is strictly dependent on the upstream termination codon (47). It has been suggested that the ndhK cistron is exclusively translated by the ribosomes that complete translation of the upstream cistron; that is, some proportion of the ribosomes participating in ndhC translation move 4 nt upstream after termination and resume translation of the ndhK cistron; the remainder are released at the termination codon. It has been suggested that in E. coli, the ribosome could undergo bidirectional diffusion for a very short time following termination (48). Generally, increasing the distance between the stop codon of an upstream cistron and the start codon of the downstream cistron reduces or arrests translation of the downstream cistron. When the distance between the ndhC stop codon and the ndhK start codon is increased artificially, translation of the downstream ndhK cistron is reduced or abolished; for example, a separation of 11 nt between the stop and start codons arrests ndhK translation (47). Because the distance between the psbD stop codon and the psbC start codon is 11 nt, it seems unlikely that the psbC cistron is translated exclusively by ribosomes participating in psbD translation. We favor the hypothesis that the psbC cistron becomes capable of being translated by ribosomes from the free ribosome pool through the action of the ribosome translating the psbD cistron.
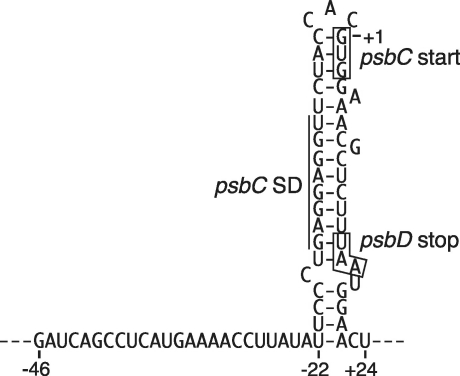
A rigid stem–loop structure can be predicted from the region including the SD-like sequence, the psbC start codon and the psbD stop codon (Figure 6). Because the SD-like sequence and the psbC start codon are embedded in the hairpin, loading of ribosomes (or 30S subunits) could interfere with initiation of psbC translation. The ribosome translating the upstream cistron could unwind the hairpin to facilitate initiation of translation of the psbC cistron. Both primary and processed psbC 5′-UTRs are also predicted to form this hairpin, which could be one reason why translation of the monocistronic psbC mRNAs is inefficient (Figure 3B). The translation efficiency of the psbC cistron from the dicistronic mRNA is apparently insufficient to assemble a functional photosystem II complex. Chloroplasts accumulate a considerable amount of the monocistronic psbC mRNAs in tobacco (25) and other plants (27,49,50). Translation of monocistronic psbC mRNAs may compensate for a shortage of CP43.
Figure 6.
Predicted secondary structure of the mRNA region around the psbC start site. Positions are relative to the GUG start codon. The SD-like sequence, psbC start codon and psbD stop codon are marked.
In conclusion, D2 and CP43 are produced via multiple translation processes, from primary and 5′-processed mRNAs and by translational coupling and uncoupling pathways. The sequences encompassing psbC start/psbD stop codons are well conserved among various land plants, algae and cyanobacteria (Supplementary Figure S6), suggesting that the basic translation process of psbD/C mRNAs is also conserved. Information on the basic translation process, such as reported here, should help to understand more about the translational control mechanisms operating within the chloroplast.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6.
FUNDING
New Energy and Industrial Technology Development Organization (to M.S.); and Grants-in-Aid for Scientific Research (19370021 and 22570050 to M.S., 20570043 to H.K.). Funding for open access charge: Grant-in-Aid for Scientific Research (22570050 to M.S.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
This work was performed in part in Sugiyama Jogakuen University, Nagoya.
REFERENCES
- 1.Sugiura M. The chloroplast genome. Plant Mol. Biol. 1992;19:149–168. doi: 10.1007/BF00015612. [DOI] [PubMed] [Google Scholar]
- 2.Bock R. Structure, function, and inheritance of plastid genomes. In: Bock R, editor. Cell and Molecular Biology of Plastids. Berlin Heidelberg: Springer; 2007. pp. 29–63. [Google Scholar]
- 3.Shiina T, Tsunoyama Y, Nakahira Y, Khan MS. Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int. Rev. Cytol. 2005;244:1–68. doi: 10.1016/S0074-7696(05)44001-2. [DOI] [PubMed] [Google Scholar]
- 4.Liere K, Börner T. Transcription and transcriptional regulation in plastids. In: Bock R, editor. Cell and Molecular Biology of Plastids. Berlin Heidelberg: Springer; 2007. pp. 121–174. [Google Scholar]
- 5.Gruissem W, Tonkyn JC. Control mechanisms of plastid gene expression. Crit. Rev. Plant Sci. 1993;12:19–55. [Google Scholar]
- 6.Sugiura M, Hirose T, Sugita M. Evolution and mechanism of translation in chloroplasts. Annu. Rev. Genet. 1998;32:437–459. doi: 10.1146/annurev.genet.32.1.437. [DOI] [PubMed] [Google Scholar]
- 7.Rochaix J-D. The role of nucleus- and chloroplast-encoded factors in the synthesis of the photosynthetic apparatus. In: Wise RR, Hoober JK, editors. The Structure and Function of Plastids. Dordrecht: Springer; 2006. pp. 145–165. [Google Scholar]
- 8.Marín-Navarro J, Manuell AL, Wu J, Mayfield SP. Chloroplast translation regulation. Photosynth. Res. 2007;94:359–374. doi: 10.1007/s11120-007-9183-z. [DOI] [PubMed] [Google Scholar]
- 9.Peled-Zehavi H, Danon A. Translation and translational regulation in chloroplasts. In: Bock R, editor. Cell and Molecular Biology of Plastids. Berlin Heidelberg: Springer; 2007. pp. 249–281. [Google Scholar]
- 10.Wobbe L, Schwarz C, Nickelsen J, Kruse O. Translational control of photosynthetic gene expression in phototrophic eukaryotes. Physiol. Plant. 2008;133:507–515. doi: 10.1111/j.1399-3054.2008.01091.x. [DOI] [PubMed] [Google Scholar]
- 11.Tangphatsornruang S, Birch-Machin I, Newell CA, Gray JC. The effect of different 3′ untranslated regions on the accumulation and stability of transcripts of a gfp transgene in chloroplasts of transplastomic tobacco. Plant Mol. Biol. 2011;76:385–396. doi: 10.1007/s11103-010-9689-1. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka M, Obokata J, Chunwongse J, Shinozaki K, Sugiura M. Rapid splicing and stepwise processing of a transcript from the psbB operon in tobacco chloroplasts: determination of the intron sites in petB and petD. Mol. Gen. Genet. 1987;209:427–431. doi: 10.1007/BF00331145. [DOI] [PubMed] [Google Scholar]
- 13.Barkan A. Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 1988;7:2637–2644. doi: 10.1002/j.1460-2075.1988.tb03116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westhoff P, Herrmann RG. Complex RNA maturation in chloroplasts. The psbB operon from spinach. Eur. J. Biochem. 1988;171:551–564. doi: 10.1111/j.1432-1033.1988.tb13824.x. [DOI] [PubMed] [Google Scholar]
- 15.Stern DB, Goldschmidt-Clermont M, Hanson MR. Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 2010;61:125–155. doi: 10.1146/annurev-arplant-042809-112242. [DOI] [PubMed] [Google Scholar]
- 16.Hirose T, Sugiura M. Both RNA editing and RNA cleavage are required for translation of tobacco chloroplast ndhD mRNA: a possible regulatory mechanism for the expression of a chloroplast operon consisting of functionally unrelated genes. EMBO J. 1997;16:6804–6811. doi: 10.1093/emboj/16.22.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, Mori M, Kondo C, Honji Y, Sun C-R, Meng B-Y, et al. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Gen. Genet. 1989;217:185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- 19.Holschuh K, Bottomley W, Whitfeld PR. Structure of the spinach chloroplast genes for the D2 and 44 kd reaction-centre proteins of photosystem II and for tRNASer (UGA) Nucleic Acids Res. 1984;12:8819–8834. doi: 10.1093/nar/12.23.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alt J, Morris J, Westhoff P, Herrmann RG. Nucleotide sequence of the clustered genes for the 44 kd chlorophyll a apoprotein and the “32 kd”-like protein of the photosystem II reaction center in the spinach plastid chromosome. Curr. Genet. 1984;8:597–606. doi: 10.1007/BF00395705. [DOI] [PubMed] [Google Scholar]
- 21.Rasmussen OF, Bookjans G, Stummann BM, Henningsen KW. Localization and nucleotide sequence of the gene for the membrane polypeptide D2 from pea chloroplast DNA. Plant Mol. Biol. 1984;3:191–199. doi: 10.1007/BF00029654. [DOI] [PubMed] [Google Scholar]
- 22.Wakasugi T, Nagai T, Kapoor M, Sugita M, Ito M, Ito S, Tsudzuki J, Nakashima K, Tsudzuki T, Suzuki Y, et al. Complete nucleotide sequence of the chloroplast genome from the green alga Chlorella vulgaris: the existence of genes possibly involved in chloroplast division. Proc. Natl Acad. Sci. USA. 1997;94:5967–5972. doi: 10.1073/pnas.94.11.5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golden SS, Stearns GW. Nucleotide sequence and transcript analysis of three photosystem II genes from the cyanobacterium Synechococcus sp. PCC7942. Gene. 1988;67:85–96. doi: 10.1016/0378-1119(88)90011-x. [DOI] [PubMed] [Google Scholar]
- 24.Rochaix J-D, Dron M, Rahire M, Malnoe P. Sequence homology between the 32K dalton and the D2 chloroplast membrane polypeptides of Chlamydomonas reinhardtii. Plant Mol. Biol. 1984;3:363–370. doi: 10.1007/BF00033383. [DOI] [PubMed] [Google Scholar]
- 25.Yao WB, Meng BY, Tanaka M, Sugiura M. An additional promoter within the protein-coding region of the psbD-psbC gene cluster in tobacco chloroplast DNA. Nucleic Acids Res. 1989;17:9583–9591. doi: 10.1093/nar/17.23.9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swiatek M, Kuras R, Sokolenko A, Higgs D, Olive J, Cinque G, Müller B, Eichacker LA, Stern DB, Bassi R, et al. The chloroplast gene ycf9 encodes a photosystem II (PSII) core subunit, PsbZ, that participates in PSII supramolecular architechture. Plant Cell. 2001;13:1347–1367. doi: 10.1105/tpc.13.6.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sexton TB, Christopher DA, Mullet JE. Light-induced switch in barley psbD-psbC promoter utilization: a novel mechanism regulating chloroplast gene expression. EMBO J. 1990;9:4485–4494. doi: 10.1002/j.1460-2075.1990.tb07899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawaguchi H, Fukuda I, Shiina T, Toyoshima Y. Dynamical behavior of psb gene transcripts in greening wheat seedling. I. Time course of accumulation of the psbA through psbN gene transcripts during light-induced greening. Plant Mol. Biol. 1992;20:695–704. doi: 10.1007/BF00046454. [DOI] [PubMed] [Google Scholar]
- 29.Chen S-CG, Lu J-H, Cheng M-C, Chen L-FO, Lo P-K. Four promoters in the rice plastid psbK-psbI-psbD-psbC operon. Plant Sci. 1994;99:171–182. [Google Scholar]
- 30.Gamble PE, Mullet JE. Blue light regulates the accumulation of two psbD-psbC transcripts in barley chloroplasts. EMBO J. 1989;8:2785–2794. doi: 10.1002/j.1460-2075.1989.tb08424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christopher DA, Kim M, Mullet JE. A novel light-regulated promoter is conserved in cereal and dicot chloroplasts. Plant Cell. 1992;4:785–798. doi: 10.1105/tpc.4.7.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsunoyama Y, Ishizaki Y, Morikawa K, Kobori M, Nakahira Y, Takeba G, Toyoshima Y, Shiina T. Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. Proc. Natl Acad. Sci. USA. 2004;101:3304–3309. doi: 10.1073/pnas.0308362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagashima A, Hanaoka M, Shikanai T, Fujiwara M, Kanamaru K, Takahashi H, Tanaka K. The multiple-stress responsive plastid sigma factor, SIG5, directs activation of the psbD blue light-responsive promoter (BLRP) in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:357–368. doi: 10.1093/pcp/pch050. [DOI] [PubMed] [Google Scholar]
- 34.Kuroda H, Suzuki H, Kusumegi T, Hirose T, Yukawa Y, Sugiura M. Translation of psbC mRNAs starts from the downstream GUG, not the upstream AUG, and requires the extended Shine-Dalgarno sequence in tobacco chloroplasts. Plant Cell Physiol. 2007;48:1374–1378. doi: 10.1093/pcp/pcm097. [DOI] [PubMed] [Google Scholar]
- 35.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki H, Kuroda H, Yukawa Y, Sugiura M. The downstream atpE cistron is efficiently translated via its own cis-element in partially overlapping atpB-atpE dicistronic mRNAs in chloroplasts. Nucleic Acids Res. 2011;39:9405–9412. doi: 10.1093/nar/gkr644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yukawa M, Tsudzuki T, Sugiura M. The 2005 version of the chloroplast DNA sequence from tobacco (Nicotiana tabacum) Plant Mol. Biol. Rep. 2005;23:359–365. [Google Scholar]
- 38.Yukawa M, Kuroda H, Sugiura M. A new in vitro translation system for non-radioactive assay from tobacco chloroplasts: effect of pre-mRNA processing on translation in vitro. Plant J. 2007;49:367–376. doi: 10.1111/j.1365-313X.2006.02948.x. [DOI] [PubMed] [Google Scholar]
- 39.Kuroda H, Maliga P. Sequences downstream of the translation initiation codon are important determinants of translation efficiency in chloroplasts. Plant Physiol. 2001;125:430–436. doi: 10.1104/pp.125.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirose T, Sugiura M. Functional Shine-Dalgarno-like sequences for translational initiation of chloroplast mRNAs. Plant Cell Physiol. 2004;45:114–117. doi: 10.1093/pcp/pch002. [DOI] [PubMed] [Google Scholar]
- 41.Jackson RJ, Kaminski A, Pöyry TAA. Coupled termination-reinitiation events in mRNA translation. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 197–223. [Google Scholar]
- 42.Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15:1480–1495. doi: 10.1105/tpc.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sane AP, Stein B, Westhoff P. The nuclear gene HCF107 encodes a membrane-associated R-TPR (RNA tetratricopeptide repeat)-containing protein involved in expression of the plastidial psbH gene in Arabidopsis. Plant J. 2005;42:720–730. doi: 10.1111/j.1365-313X.2005.02409.x. [DOI] [PubMed] [Google Scholar]
- 44.Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson X, Wostrikoff K, Finazzi G, Kuras R, Schwarz C, Bujaldon S, Nickelsen J, Stern DB, Wollman F-A, Vallon O. MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell. 2010;22:234–248. doi: 10.1105/tpc.109.066266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl Acad. Sci. USA. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yukawa M, Sugiura M. Termination codon-dependent translation of partially overlapping ndhC-ndhK transcripts in chloroplasts. Proc. Natl Acad. Sci. USA. 2008;105:19550–19554. doi: 10.1073/pnas.0809240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adhin MR, van Duin J. Scanning model for translational reinitiation in eubacteria. J. Mol. Biol. 1990;213:811–818. doi: 10.1016/S0022-2836(05)80265-7. [DOI] [PubMed] [Google Scholar]
- 49.Meurer J, Berger A, Westhoff P. A nuclear mutant of Arabidopsis with impaired stability on distinct transcripts of the plastid psbB, psbD/C, ndhH, and ndhC operons. Plant Cell. 1996;8:1193–1207. doi: 10.1105/tpc.8.7.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gamble PE, Sexton TB, Mullet JE. Light-dependent changes in psbD and psbC transcripts of barley chloroplasts: accumulation of two transcripts maintains psbD and psbC translation capability in mature chloroplasts. EMBO J. 1988;7:1288–1297. doi: 10.1002/j.1460-2075.1988.tb02943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.