Abstract
An AUG in an optimal nucleotide context is the preferred translation initiation site in eukaryotic cells. Interactions among translation initiation factors, including eIF1 and eIF5, govern start codon selection. Experiments described here showed that high intracellular eIF5 levels reduced the stringency of start codon selection in human cells. In contrast, high intracellular eIF1 levels increased stringency. High levels of eIF5 induced translation of inhibitory upstream open reading frames (uORFs) in eIF5 mRNA that initiate with AUG codons in conserved poor contexts. This resulted in reduced translation from the downstream eIF5 start codon, indicating that eIF5 autoregulates its own synthesis. As with eIF1, which is also autoregulated through translation initiation, features contributing to eIF5 autoregulation show deep evolutionary conservation. The results obtained provide the basis for a model in which auto- and cross-regulation of eIF5 and eIF1 translation establish a regulatory feedback loop that would stabilize the stringency of start codon selection.
INTRODUCTION
Eukaryotic translation initiation is a complex process requiring the activities of many factors (1). A distinctive feature of eukaryotic translation initiation is that the small ribosome subunit, including the initiator tRNA and assorted initiation factors constituting the 43 S pre-initiation complex (PIC), binds to the 5′ cap and scans downstream for a proper initiation codon. In most cases, initiation occurs at an AUG codon. In mammals, and perhaps most eukaryotes, strong bias exists for the nucleotides in the immediate vicinity of the initiation codon (2,3). The consensus initiation context in mammals is GCC(A/G)CCAUGG. This sequence is optimal for efficient initiation with the underlined nucleotides at positions −3 and +4 (relative to the +1 A of AUG, shown in italics) playing the most important role in providing an optimal context (3).
Both in vitro and in vivo results have demonstrated that the PIC component eIF1 is crucial for discrimination between poor and optimal initiation contexts (4,5). eIF1 binds near the P-site on the 40S ribosomal subunit (6,7). This is thought to result in the formation and maintenance of an open PIC conformation favoring scanning and inhibiting initiation of translation (8). Release of eIF1 from the PIC leads to a closed conformation of the small subunit which favors initiation and abrogates scanning (9).
In Saccharomyces cerevisiae, mutations in eIF1, eIF2 and eIF5, which are each factors associated with the PIC, affect start codon selection (10). Until recently it was thought that the role of eIF5 was promotion of hydrolysis of eIF2-bound GTP in response to initiation codon recognition (11,12). It is now known that eIF5 also stabilizes the binding of GDP to eIF2 and acts to inhibit the GDP–GTP exchange function of eIF2B (13). There is additional evidence for a distinct role for eIF5 as a competitor to eIF1 for binding at a critical site in the small ribosomal subunit such that successful competition leads to ejection of eIF1 (9,14). eIF1 ejection would then stimulate the formation of the closed conformation that favors initiation.
We recently showed that overexpression of eIF1 in mammalian cells led to reduced utilization of AUG start codons in poor context and of non-AUG start codons (e.g. CUG and AUU start codons) (5). Interestingly, the start codon of eIF1 is itself in a poor context in most eukaryotes for which sequence data is available (5,15). In addition, overexpression of eIF1 led to reduced utilization of its own poor context AUG in S. cerevisiae (16). These and other experimental data are consistent with a model in which eIF1 levels are controlled by an autoregulatory mechanism so that high eIF1 reduces translation initiation from its own start codon. This is analogous to the autoregulatory control of synthesis of bacterial initiation factor 3 (IF3), a protein which like eIF1 discriminates between initiation at AUG and near-cognate non-AUG codons. Initiation of IF3 mRNA translation is at an AUU codon and high IF3 levels result in reduced initiation at this codon, reducing IF3 synthesis (17,18). Autoregulation at the level of translation also controls the expression of other translation factors (19).
Here, we describe experiments that examined the consequences of eIF5 overexpression in human cells. eIF5 overexpression resulted in increased initiation at poor-context AUG codons and at non-AUG start codons. We discovered that many eukaryotic mRNAs encoding eIF5 contain one or more upstream open reading frames (uORFs) whose start codons are in poor contexts. This suggested a model for autoregulation in which an increased eIF5 level increases initiation at these uORF start codons and as a consequence decreases translation from the eIF5 start codon, reducing eIF5 synthesis. We tested this using reporter constructs and obtained results consistent with this model. Furthermore, using a series of reporters initiated by either AUGs in different contexts or by non-AUG start codons, eIF5 and eIF1 overexpression were observed to have opposite effects on the stringency of start codon selection. The data also suggest that eIF5 and eIF1 positively cross-regulate each other's expression at the level of translation initiation, providing additional means for a homeostatic cellular control mechanism to maintain stringency in start codon selection.
MATERIALS AND METHODS
Plasmids
Fusion of the wild-type 5′-UTR of eIF5 to firefly luciferase was accomplished as follows [Supplementary Table S1 lists the sequences for the oligonucleotides (1–54) used]: First, we replaced the large (1056 nt) intron within eIF5 uORF1 with the 133-nt intron obtained from the 5′-UTR of the mRNA specified by the Renilla luciferase reporter gene in phRL-CMV (Promega); the same intron/exon boundaries were retained. To accomplish this, the 5′ and 3′ regions of eIF5 5′-UTR were amplified with primers 1 and 2 (5′-end) and 3 and 4 (3′-end) by RT-PCR on RNA isolated from HEK-293 T cells. Next the 133-nt intron from the 5′-UTR of Renilla luciferase was amplified using primers 5 and 6 using phRL-CMV as template. Equimolar amounts of each of the three 1st step PCR products were mixed and used as template for the 2nd step of the PCR with primers 1 and 4 to generate wild-type 5′-UTR eIF5. Wild-type 5′-UTR eIF5 was then cloned using Pst1 and BamHI restriction sites into dual luciferase vector p2-Luc (20) to make pWT 5′-UTR eIF5-FFluc.
Constructs for overexpressing eIF5-WT, eIF5-AAA and eIF5-Perfect were synthesized by 2-step PCR. First, the eIF5 coding sequence was amplified using primers 7 and 8 by RT-PCR on RNA isolated from HEK-293 T cells. Wild-type eIF5 5′-UTR with the 133-nt intron was amplified by PCR using primers 9 and 10 on template pWT 5′-UTR eIF5-Fluc. eIF5-Perfect 5′-UTR was amplified by 2-step PCR using primers 9 and 11 (5′ end) and primers 12 and 10 (3′ end) again on template pWT 5′-UTR eIF5-Fluc. AAA eIF5 5′-UTR was amplified by PCR using primers 9 and 10 on a template synthesized commercially (Genscript Corp.) which had all of the uORF AUG-codons changed to AAA-codons. Equimolar amounts of each eIF5 5′-UTR amplicon were separately mixed with the eIF5 coding sequence amplicon and used as templates for PCR with primers 9 and 8 to generate eIF5-WT 5′-UTR, eIF5-perfect 5′-UTR and eIF5-AAA 5′-UTR. These three eIF5 constructs were then cloned using SacI and XbaI restriction sites into phRL to make peIF5-WT 5′-UTR, peIF5-Perfect 5′-UTR and peIF5-AAA 5′-UTR.
The construction of peIF1-good* (overexpressing eIF1), pSV40-eIF1-FFLuc, pSV40-Renilla and all of the non-AUG initiating FFLuc constructs shown in Figure 3B, has been described (5). For the synthesis of firefly luciferase reporters initiating with the first, second, third or main eIF5 AUG (Figure 3A) sense and antisense oligonucleotide pairs 13–20 (Table S1) were annealed and ligated into p2-Luc digested with PstI and BamHI. Firefly luciferase fusions initiating with AUGs in various contexts (Figure 3B) were made in a similar way using oligonucleotide pairs 21–54.
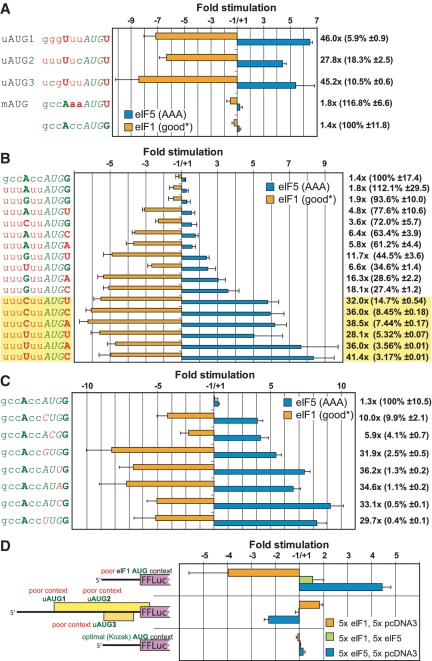
Figure 3.
eIF5 and eIF1 have opposing effects on the stringency of start codon selection. (A) Comparative effects of eIF1 and eIF5 overexpression on initiation at a reporter starting with AUG in the following different contexts: human eIF5 uAUG1, uAUG2, uAUG3 or the main eIF5 AUG. (B) The effects of eIF1 and eIF5 overexpression on AUG start codons with varied context at positions −3 and +4. All other positions between −6 and −1 contain the least favorable nucleotide U. (C) The effects of eIF1 and eIF5 overexpression on initiation at non-AUG start codons. In (B) and (C), the results are displayed in descending order with the most efficient initiation contexts (B) or codons (C) toward the top and the least efficient toward the bottom. In (A–C), ‘10×’ eIF1-overexpression, eIF5-overexpression or control vectors were co-transfected with the reporter vectors. Co-transfected Renilla luciferase was used for normalizing reporter activity; the fold-stimulation in response to overexpression of eIF1 or eIF5 was determined as in Figure 2. Negative ‘stimulation’ values indicate repression. In each case the firefly reporter was initiated by the codon and the context indicated on the left. In (B), the firefly luciferase reporters starting with contexts in which −3 is a pyrimidine and the +4 position is not G, are highlighted in yellow. In all cases, nucleotides matching the preferred initiation consensus in humans are indicated in green. Nucleotides deviating from the preferred context are in red. The fold-difference in translation of the reporter co-transfected with eIF1 compared to co-transfection with eIF5 is indicated on the right. The percentage of normalized firefly reporter activity is given relative to reporter activity from the construct whose AUG start is in optimal ‘Kozak’ context in the parentheses following this value. (D) Results from co-overexpressing eIF1 and eIF5. Schematic representations of the firefly luciferase reporter constructs used are on the left. (A–C): fold-stimulation of normalized firefly luciferase activity in cells co-transfected with (i) ‘10×’ vector expressing eIF1 in a near consensus context (eIF1 good*)—orange bars; or (ii) ‘10×’ vector expressing eIF5 in which its 5′-UTR is altered so that all uAUG codons are eliminated by substitution with AAA codons (eIF5 AAA)—blue bars. (D)—fold translation stimulation in cells co-transfected with (iii) ‘5×’ ‘eIF1 good*’ and ‘5×’ pcDNA3 vectors—orange bars; (iv) ‘5×’ ‘eIF1 good*’ and ‘5×’ ‘eIF5 AAA’ green bars; (v) ‘5×’ ‘eIF5 AAA’ and ‘5×’ pcDNA3—blue bars. The results in (A–D) each represent two independent experiments done it triplicate. Error bars or ‘±’ values represent SDs.
Cell culture and transfections
HEK-293 T cells (ATCC) were maintained in DMEM supplemented with 10% FBS, 1 mM l-glutamine and antibiotics. HEK-293 T cells were transfected in quadruplicate with Lipofectamine 2000 reagent (Invitrogen), using the 1-day protocol in which suspended cells are added directly to the DNA complexes in half-area 96-well plates. For each transfection the following were added to each well: 50 ng (or 25 ng each for mixing experiments shown in Figure 3D) peIF5-WT 5′-UTR, peIF5-Perfect 5′-UTR and peIF5-AAA 5′-UTR as indicated, 5 ng pSV40-firefly vector (with initiation contexts and/or codons as indicated in the figures), 0.2 ng pSV40-Renilla vector and 0.2 µl lipofectamine 2000 in 25 µl Opti-Mem (Gibco). The transfecting DNA complexes in each well were incubated with 4 × 104 cells suspended in 50 µl DMEM +10% FBS. Transfected cells were incubated overnight at 37°C in 5% CO2 for 16 h.
Dual luciferase assay
Firefly and Renilla luciferase activities were determined by dual-luciferase assay as described previously (5). Firefly luciferase activity was calculated relative to the activity of the co-transfected control plasmid expressing Renilla luciferase (pSV40-Renilla). All data points were averaged and the SD calculated. Data shown in Figure 2D represent the mean and SD from 3 independent experiments each done in quadruplicate (12 independent data points). Data shown in Figure 3 represent the mean and SD from two independent experiments each done in triplicate (six independent data points).
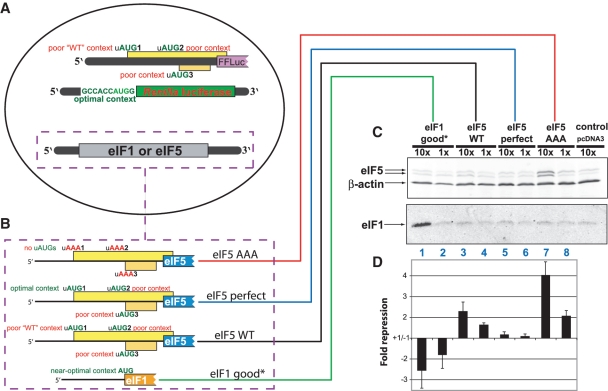
Figure 2.
Overexpression of eIF5 is autoregulatory. (A) Schematic representation of the triple transfections used in these experiments. The firefly luciferase reporter is fused downstream of the wild-type 5′-UTR of human eIF5 mRNA. Its initiation context matches the context of the eIF5 start codon, which is near optimal (see Figure 3A for its sequence). The translation of the Renilla luciferase reporter is initiated with an AUG codon in optimal ‘Kozak’ context. The third plasmid used in the triple transfection encoded one of the four eIF5 or eIF1 expression constructs shown in (B). (B) Schematic representation of the constructs used to overexpress eIF5 or eIF1 (see text for details). (C) Western blots of protein lysates from cells transfected with the overexpression eIF5 or eIF1 constructs indicated in (B). The eIF1 overexpression construct is the same as ‘eIF1 good*’ described previously (5). In lanes marked ‘10×’, 10-fold more vector with insert was transfected compared with lanes marked ‘1×,’ where the difference in the amount of transfecting DNA is made up with the inert plasmid pcDNA3. The control cells are transfected with ‘10×’ amount of pcDNA3. The blot shown was probed with anti-eIF5 and anti-β-actin antibodies and separately with anti-eIF1 antibodies. The corresponding detected proteins are indicated by arrows. Anti-β-actin antibody is used to control for loading differences. (D) Fold repression of firefly luciferase activity in response to eIF5 or eIF1 overexpression. The ratio from dual luciferase measurements from the same cells for which western blots were performed in (C) was calculated. The firefly luciferase measurements were normalized to those from Renilla luciferase. The Renilla and firefly reporters are those illustrated in (A). The ratios in test cells were then compared to the luciferase ratio in control cells transfected with pcDNA3 and the fold-repression was calculated from this comparison. Negative ‘repression’ values indicate stimulation.
Western analysis
Cells were transfected in 6-well plates essentially as described in (5) with either 3 µg (10×) or 0.3 µg (1×) peIF5-WT 5′-UTR, peIF5-perfect 5′-UTR or peIF5-AAA 5′-UTR vectors as indicated plus 0.3 µg pWT 5′-UTR eIF5-FFluc and 12 ng pSV40-Renilla vector. To maintain identical levels of transfecting DNA, those transfections with 0.3 µg vector also included 2.7 µg pcDNA3 plasmid DNA (control cells were transfected with 3 µg pcDNA3). Transfected cells (2.4 × 106 HEK-293 T/well) were incubated at 37°C in 5% CO2 for 16 h then lysed and assayed by dual luciferase assay and western blotting as described in (5). Immunoblots were incubated at 4°C overnight in 2% powdered milk with a 1:50 dilution of goat anti-eIF1 (Santa Cruz D-15), 1:1000 dilution of rabbit anti-eIF5 (Abcam AB85913) and a 1:10 000 dilution of mouse anti-β-actin (Sigma). Immunoreactive bands were detected on membranes after incubation with appropriate fluorescently labeled secondary antibodies using a LI-COR Odyssey® Infrared Imaging Scanner (LI-COR Biosciences). Quantification of eIF5, eIF1 and β-actin protein levels was accomplished using IMAGEQUANT software (Molecular Dynamics).
RESULTS
eIF5 mRNAs contain multiple upstream AUGs present in conserved poor context
While examining the mRNA sequences of eIF5 from Neurospora crassa and human we noticed that upstream of the main open reading frame (mORF) there were three AUG codons (uAUGs). A schematic representation of the uAUGs in the single human gene for eIF5 is shown in Figure 1A. These uAUGs are each in a poor initiation context. The possibility that eIF5 might compete with eIF1 for the same binding site in the PIC (9) suggested to us that these uAUGs, and the uORFs they initiate, might be involved in translational autoregulation of eIF5. If a high level of eIF5 protein enables it to out-compete eIF1 from the PIC, then initiation at poor-context AUGs would increase. Ribosomes would, therefore, initiate more frequently at the inhibitory uORFs of the eIF5 mRNA, reducing translation from the downstream eIF5 start codon. To investigate this hypothesis further, the sequences of over 300 homologs of eIF5 were compiled and analyzed. This analysis revealed that the presence of uAUGs (and uORFs) is a near universal feature in eukaryotic eIF5 genes. Most eIF5 mRNAs have multiple uAUGs, except for the fungi where in the majority of cases (except Pezizomycotina) a single uAUG (and uORF) is present. eIF5 uORFs were previously noted in seven plant homologs (21); we examined 140 plant homologs of eIF5 and all contained uORFs. The 337 eIF5 eukaryotic homologs analyzed have a total of 867 uAUGs (2.6 uAUGs/mRNA on an average).
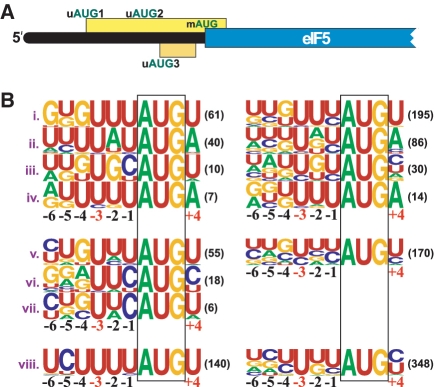
Figure 1.
The mRNAs of eIF5 homologs from eukaryotes have one or more uAUGs in poor contexts preceding the AUG for the main ORF. (A) Schematic representation of the eIF5 mRNA from mammals. The position of the uAUGs and the AUG of the main ORF are indicated. The uORFs initiated by the uAUGs are shown as yellow-hued rectangles. The main ORF is shown as a blue rectangle. Representation of the 3′-end of the eIF ORF and mRNA 3′-UTR is omitted. (B) Weblogo representation of initiation contexts of the uAUGs in eIF5 mRNAs from diverse eukaryotes. Letter heights are proportional to the frequency of conservation of each nucleotide at each position. Each line represents a different eukaryotic branch. (i–iv Animalia): (i) Vertebrata; (ii) Arthropoda; (iii) Nematoda; (iv) Mollusca. (v–vii Fungi): (v) Pezizomycotina; (vi) Basidiomycota; (vii) Zygomycota, Glomeromycota, Neocallimastigomycota and Chytridiomycota. (viii) Plantae. The column on the left represents the contexts of uAUG1 which almost invariably initiates the longest uORF. The column on the right represents the contexts of all uAUGs. The number of AUGs used to generate each representation is indicated in parentheses on its right. The nucleotide position relative to the A of the AUG start codon is indicated below (crucial positions −3 and +4 are in red). The AUG is boxed. Alignments of the sequences used to generate the logogram are shown in Supplementary Figure S1.
Almost invariably, uORF1 is the longest uORF. In vertebrate homologs of eIF5, this longest uORF is more than 90 codons long and invariably overlaps with the mORF. In mammalian mRNAs, uORFs longer than 30 codons or uORFs overlapping the mORF greatly inhibit reinitiation at the mORF (22). In invertebrate and plant homologs, the longest uORF is at least 50 codons long. In fungi, its length is smaller but still greater than 21 codons and ribosomes that translate it would not be expected to resume scanning downstream following termination (23). These features suggest that ribosomes that initiate translation of the longest uORF of eIF5 mRNA would not be expected to translate the mORF. uAUG1's poor context is conserved (Figure 1B and Supplementary Figure S1). In 98.8% of cases, the −3 nt is U (which is the least favorable −3 nt for initiation) and it is never a favorable purine. The +4 nt is never the most-favored G. There is also a strong selection for highly unfavorable U in positions −1 and −2. Considering all eIF5 uAUGs together, strong selection against purines in the −3 position and against G in the +4 position is apparent (Figure 1B).
The longest eIF5 uORF-encoded peptide sequences show some phylum-specific conservation (Supplementary Figure S2). Conservation of the eIF5 uORF1 peptide sequence among plant homologs has already been noted (21); however, there is little conservation between more distantly related eukaryotic branches suggesting that, if the uORF plays a regulatory role, it is not strictly dependent on a conserved peptide sequence.
Overexpression of eIF5 protein leads to autoregulatory repression
If initiation at the uAUGs of eIF5 mRNA leads to reduced translation of the mORF, and if the uAUGs are sensors for autoregulation, then overexpression of eIF5 protein would result in reduced translation of a downstream ORF preceded by the 5′-UTR of eIF5 mRNA. This prediction was tested in human cells (HEK-293 T) using a triple-transfection strategy (Figure 2A). One vector overexpressed eIF5 mRNA or alternatively eIF1 (empty vector was used as a negative control). Three different versions of the eIF5 gene were placed into the overexpression vector (Figure 2B)—one with the wild-type eIF5 5′-UTR, one with eIF5 uAUG1 placed in optimal initiation context, and one in which all eIF5 uAUG codons are changed to AAA (non-initiating) codons. The endogenous 3′-UTR of eIF5 was omitted from the constructs to avoid potential 3′-UTR effects on regulation. Cells were also co-transfected with two luciferase reporter vectors. In one, the wild-type 5′-UTR of eIF5, up to and including the AUG codon and context of the mORF, was fused to firefly luciferase. In the other reporter, Renilla luciferase was initiated from AUG in optimal context. Renilla luciferase activity was used for normalizing firefly luciferase activity.
Western blots from these experiments showed that overexpressing the eIF5 wild-type construct results in only a modest increase (2-fold and no statistically significant increase with ‘10×’ ‘1×’ transfected DNA, respectively) in eIF5 protein expression (Figure 2C, lanes 3 and 4; Supplementary Figure S3). In contrast, overexpressing the eIF5 construct in which all uAUG-codons were changed to AAA-codons led to substantially increased (2- and 8-fold with ‘1×’ and ‘10×’ transfected DNA, respectively) eIF5 protein expression (Figure 2C, lanes 7 and 8; Supplementary Figure S3). This result was consistent with an inhibitory function for the uORFs and with a model of eIF5 autoregulation mediated by translation of the eIF5 uORFs.
The role of the eIF5 uORFs in autoregulation was strongly supported by expression data for the co-transfected luciferase reporters. The firefly luciferase reporter with the wild-type eIF5 5′-UTR was repressed 4-fold (Figure 2D, column 7) when the expression of the co-transfected eIF5 gene was highest (i.e. using eIF5 vector in which the uAUGs were mutated to AAA). Even with the construct expressing the eIF5 with the wild-type 5′-UTR, which in western blots showed only a slight increase in eIF5 protein over endogenous levels, reporter expression was repressed more than 2-fold (Figure 2D, column 3). In contrast, the eIF5 construct in which the first uAUG is placed in optimal context, which is expected to strongly reduce translation from the downstream start codon for eIF5, did not induce appreciable repression of the reporter (Figure 2D, columns 5 and 6).
Overexpression of eIF1 is known to increase the stringency of start codon selection and thus to reduce initiation at AUG start codons in suboptimal contexts (5). Consistent with our hypothesis that the uAUGs in the 5′-UTR of eIF5 are suboptimal for initiation, and that initiation at these uAUGs inhibits expression of the mORF, overexpression of eIF1 protein led to ∼2.5-fold induction of the firefly reporter containing the wild-type eIF5 5′-UTR (Figure 2D, column 1).
Overexpression of eIF5 protein leads to induced initiation from AUG codons in poor contexts and from non-canonical initiation codons
We next investigated whether eIF5 overexpression has a direct effect on the stringency of start codon selection. For this purpose, a triple-transfection scheme similar to the one illustrated in Figure 2A was used except that the firefly luciferase reporters were different. In this series of firefly luciferase reporters, ‘inert’ 5′-UTR (not related to eIF5 and lacking any uAUGs) was followed by an AUG that initiates synthesis of the luciferase reporter. This AUG was placed in a variety of initiation contexts. Figure 3A compares the results of placing this AUG in the context of human eIF5 uAUG1, uAUG2, uAUG3 or the AUG of the main eIF5 ORF to the results obtained with an AUG in an optimal context. As expected, uAUG1, uAUG2 and uAUG3 contexts were inefficient for initiation—they were 5.9%, 18.3% and 10.5% as efficient, respectively, relative to AUG in an optimal context. In contrast, and also as expected, the mORF AUG was as efficient as AUG in optimal context. Importantly, overexpression of eIF5 substantially increased initiation from uAUG1, uAUG2 and uAUG3 (by ∼6-, 4- and 5-fold, respectively), but did not have a corresponding effect on initiation from the mORF or the optimal-context AUGs (<1.2-fold increase, Figure 3A), indicating that eIF5 overexpression affected the stringency of start codon selection.
While eIF5 overexpression increased initiation at the eIF5 uAUGs which are in poor-contexts, eIF1 overexpression, which increases stringency (5), should correspondingly decrease expression from these uAUGs. Consistent with this, in triple-transfection experiments, eIF1 overexpression had the opposite effect of eIF5 overexpression (Figure 3A). Thus, expression of the uAUG1-, uAUG2- and uAUG3-initiated reporters were reduced by ∼7-, 6- and 8-fold, respectively. As expected, there was little effect on initiation from the mORF or optimal-context AUGs (<1.6-fold decrease, Figure 3A).
We next assessed the importance of nucleotide-substitutions at two critical positions that define start codon context in modulating the effects of eIF5 and eIF1 on start codon selection. The AUG codon of firefly luciferase was placed in the context uuuNuuAUGN where the two underlined positions (−3 and +4), which are most important for determining the relative strengths of the initiation context (3), were varied to all 16 possible permutations. The suboptimal nucleotide U was placed in positions −6, −5, −4, −2 and −1 to make the dependence on positions −3 and +4 more pronounced (24). The results of triple-transfection experiments using this series of firefly luciferase reporters are shown in Figure 3B; the results were ranked with the least efficient initiators at the bottom. There was a striking correlation between the inefficiency of a given initiation context to support initiation and the increase in its utilization when eIF5 was overexpressed. That is, the least efficient context (uuuUuuAUGC) showed the highest stimulation following eIF5 overexpression, while the most efficient context (gccAccAUGG) showed the lowest stimulation. These data are entirely consistent with eIF5 overexpression leading to a relaxation in the stringency of start codon selection.
eIF1 overexpression generally had an effect inverse to eIF5 overexpression (Figure 3B). However, for start codon contexts in which there was a −3 purine and which also lacked a +4 G, repression by eIF1 overexpression was much greater than stimulation by eIF5 overexpression. The greatest effects of eIF5 and eIF1 overexpression were observed for start codon contexts containing a pyrimidine at −3 and lacking G at +4 (highlighted in yellow in Figure 3B). For all of these, the difference in reporter activity when eIF1 was overexpressed compared to when eIF5 was overexpressed was greater than 28-fold while it was not greater than 19-fold for any context having either a purine at −3 or a G at +4.
Another class of suboptimal start codons are near-cognate non-AUG codons (25,26). We examined the effect of overexpressing eIF5 on initiation at these start codons (Figure 3C). Consistent with the results obtained with other suboptimal start codons, overexpression of eIF5 led to stimulation of initiation at these non-AUG codons. The greatest stimulation was seen with the least efficient codons. Again, eIF1 overexpression had the opposite effect of eIF5 overexpression (Figure 3C).
When eIF1 and eIF5 are co-overexpressed in the same cells they cancel each other's effect on stringency of start codon selection
The opposing effects of overexpressing eIF1 and eIF5 on stringency of start codon selection raised the question of whether the co-overexpression of eIF1 and eIF5 would neutralize each other's effect on the stringency of start codon selection or whether one is epistatic. A quadruple transfection experiment, similar to the triple transfection scheme used to test the overexpression of each factor individually, was used to examine co-overexpression. Three different firefly luciferase reporters were used (Figure 3D). In one, the firefly luciferase AUG start codon was in the poor eIF1 context. In another, the luciferase AUG was in an optimal context. In the third, the luciferase mORF was preceded by the wild-type 5′-UTR of eIF5 (this reporter was also used in Figure 2). Overexpression of eIF1 and eIF5 separately had the expected effects on the stringency of start codon selection. eIF1 overexpression repressed the reporter starting with AUG in poor context and stimulated the reporter preceded by the eIF5 5′-UTR. Conversely, eIF5 overexpression stimulated the reporter starting with AUG in poor context and repressed the reporter preceded by the eIF5 5′-UTR. Neither one, when overexpressed, had much effect on the reporter starting with AUG in optimal context (Figure 3D). Co-overexpression of eIF1 and eIF5 (green bars in Figure 3D) nearly cancelled each other's effect on the stringency of start codon selection. These data indicated that there was no epistatic relationship when these factors are overexpressed. The absence of epistasis in the eIF1 and eIF5 co-overexpression experiment is consistent with the previously suggested hypothesis that the two factors compete for binding to a crucial position on the scanning small ribosome subunit (9,27,28).
DISCUSSION
The results presented earlier provide strong evidence that the overexpression of eIF5 relaxes the stringency of start codon selection in human cells. They are also consistent with the hypothesis that this phenomenon is used for autoregulation of eIF5 expression. The conservation of structural features in the eIF5 mRNA indicates that autoregulation could be present in most eukaryotes.
The structural features in the human eIF5 mRNA responsible for conferring autoregulation are three uAUGs in poor contexts. Relaxed stringency that occurs with a high level of eIF5 would increase initiation at these uAUGs, resulting in reduced translation initiation at the eIF5 start codon. Consistent with this interpretation, the expression of transfected DNA specifying eIF5 was highest when all uAUGs were removed and lowest when uAUG1 was in optimal-context. This was established by western analysis of eIF5 levels and by assay of reporters that were sensitive to the level of eIF5 expression (Figure 2C and D).
The effect of eIF5 overexpression was opposite to the effect of eIF1 overexpression with regard to the stringency of start codon selection. While overexpression of either led to repression of its own synthesis, it led to derepression of a reporter with the mRNA architecture of the other. Importantly, co-overexpression of eIF1 and eIF5 cancel each other's effect on the stringency of start codon selection. The opposing effects of eIF5 and eIF1 on stringency, and the fact that eIF1 translation initiation increases and eIF5 translation initiation decreases when stringency is relaxed—while eIF1 translation initiation decreases and eIF5 translation initiation increases when stringency is strengthened—provide the basis for a set of auto- and cross-regulatory interactions through these factors that would stabilize the stringency of start codon selection (Figure 4). Since the cis-acting features in the mRNAs for eIF1 and eIF5 are conserved in different eukaryotic kingdoms, it appears that this regulatory loop is both ancient and widespread.
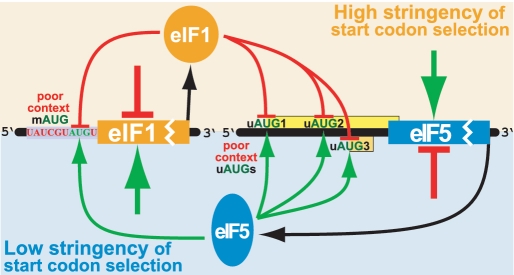
Figure 4.
Model for auto- and cross-regulation of eIF1 and eIF5 translation. eIF1 overexpression increases the stringency of start codon selection (upper half of the figure), resulting in reduced initiation at the mAUG of eIF1 and at the uAUGs of the eIF5. As a consequence, eIF1 translation decreases and eIF5 translation increases. eIF5 overexpression decreases the stringency of start codon selection (lower half of the figure), resulting in increased initiation at the mAUG of eIF1 and at the uAUGs of eIF5. As a consequence, eIF1 translation increases and eIF5 translation decreases.
A potential consequence of increased initiation at the eIF5 uORFs is that the eIF5 mRNA could be destabilized through the nonsense mediated mRNA decay (NMD) pathway (29,30). If the first (and often only) in-frame AUG of the eIF1 ORF, which is in a poor context, is skipped due to leaky scanning, then initiation at an internal ORF could potentially trigger NMD of the eIF1 mRNA. In both cases this could reinforce autoregulation.
While the homeostatic mechanism described earlier should stabilize the stringency of start codon selection, physiological conditions can change the balance of initiation events at poor versus optimal start codons. For example, ribosome-profiling experiments in S. cerevisiae showed that amino acid starvation can lead to relaxed stringency of start codon selection (31). In mammalian cells methionine-starvation can increase initiation from a non-AUG start codon (32). It was also proposed that polyamines could affect stringency of start codon selection based on studies of initiation at an evolutionarily conserved non-AUG start codon in the human antizyme inhibitor 1 mRNA 5′-UTR (33).
uORFs have wide-ranging, most often inhibitory, effects on expression of the downstream main ORF (34,35). The fact that stringency of start codon selection controls expression of the uORF-containing eIF5 mRNA raises the possibility that stringency of start codon selection regulates the expression of many uORF containing mRNAs. It is already known that uAUGs are disproportionately present in poor start contexts (36) and their utilization, and effects on the expression of the downstream gene ORF, would, therefore, be strongly influenced by changes in the cell that alter the stringency of start codon selection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table S1 and Supplementary Figures S1–S3.
FUNDING
Science Foundation Ireland (grant number 08/IN.1/B1889 to J.F.A.) and National Institutes of Health (grant number R01 GM079523 to J.F.A., R01 GM47498 to M.S.S.). Funding for open access charge: Science Foundation Ireland (grant number 08/IN.1/B1889).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the Atkins and Sachs labs for discussions.
REFERENCES
- 1.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984;12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 4.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivanov IP, Loughran G, Sachs MS, Atkins JF. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1) Proc. Natl Acad. Sci. USA. 2010;107:18056–18060. doi: 10.1073/pnas.1009269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 8.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Nanda JS, Cheung YN, Takacs JE, Martin-Marcos P, Saini AK, Hinnebusch AG, Lorsch JR. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J. Mol. Biol. 2009;394:268–285. doi: 10.1016/j.jmb.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donahue TF. In: Translational Control of Gene Expression. Sonenberg N, Hershey JWB, Mathews MB, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 595–614. [Google Scholar]
- 11.Das S, Ghosh R, Maitra U. Eukaryotic translation initiation factor 5 functions as a GTPase-activating protein. J. Biol. Chem. 2001;276:6720–6726. doi: 10.1074/jbc.M008863200. [DOI] [PubMed] [Google Scholar]
- 12.Paulin FE, Campbell LE, O'Brien K, Loughlin J, Proud CG. Eukaryotic translation initiation factor 5 (eIF5) acts as a classical GTPase-activator protein. Curr. Biol. 2001;11:55–59. doi: 10.1016/s0960-9822(00)00025-7. [DOI] [PubMed] [Google Scholar]
- 13.Jennings MD, Pavitt GD. eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation. Nature. 2010;465:378–381. doi: 10.1038/nature09003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valásek L, Phan L, Schoenfeld LW, Valaskova V, Hinnebusch AG. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J. 2001;20:891–904. doi: 10.1093/emboj/20.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyasaka H, Endo S, Shimizu H. Eukaryotic translation initiation factor 1 (eIF1), the inspector of good AUG context for translation initiation, has an extremely bad AUG context. J. Biosci. Bioeng. 2010;109:635–637. doi: 10.1016/j.jbiosc.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 16.Martin-Marcos P, Cheung YN, Hinnebusch AG. Functional elements in initiation factors 1, 1A, and 2beta discriminate against poor AUG context and non-AUG start codons. Mol. Cell. Biol. 2011;31:4814–4831. doi: 10.1128/MCB.05819-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butler JS, Springer M, Grunberg-Manago M. AUU-to-AUG mutation in the initiator codon of the translation initiation factor IF3 abolishes translational autocontrol of its own gene (infC) in vivo. Proc. Natl Acad. Sci. USA. 1987;84:4022–4025. doi: 10.1073/pnas.84.12.4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sacerdot C, Chiaruttini C, Engst K, Graffe M, Milet M, Mathy N, Dondon J, Springer M. The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli. Mol. Microbiol. 1996;21:331–346. doi: 10.1046/j.1365-2958.1996.6361359.x. [DOI] [PubMed] [Google Scholar]
- 19.Betney R, de Silva E, Krishnan J, Stansfield I. Autoregulatory systems controlling translation factor expression: thermostat-like control of translational accuracy. RNA. 2010;16:655–663. doi: 10.1261/rna.1796210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 21.Hayden CA, Jorgensen RA. Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes. BMC Biol. 2007;5:32. doi: 10.1186/1741-7007-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29:5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajkowitsch L, Vilela C, Berthelot K, Ramirez CV, McCarthy JE. Reinitiation and recycling are distinct processes occurring downstream of translation termination in yeast. J. Mol. Biol. 2004;335:71–85. doi: 10.1016/j.jmb.2003.10.049. [DOI] [PubMed] [Google Scholar]
- 24.Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 25.Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol. Cell. Biol. 1989;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peabody DS. Translation initiation at non-AUG triplets in mammalian cells. J. Biol. Chem. 1989;264:5031–5035. [PubMed] [Google Scholar]
- 27.Lorsch JR, Dever TE. Molecular view of 43 S complex formation and start site selection in eukaryotic translation initiation. J. Biol. Chem. 2010;285:21203–21207. doi: 10.1074/jbc.R110.119743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinnebusch AG. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 2011;75:434–467. doi: 10.1128/MMBR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hood HM, Neafsey DE, Galagan J, Sachs MS. Evolutionary roles of upstream open reading frames in mediating gene regulation in fungi. Annu. Rev. Microbiol. 2009;63:385–409. doi: 10.1146/annurev.micro.62.081307.162835. [DOI] [PubMed] [Google Scholar]
- 30.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell. Mol. Life Sci. 2009;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hann SR, Sloan-Brown K, Spotts GD. Translational activation of the non-AUG-initiated c-myc 1 protein at high cell densities due to methionine deprivation. Genes Dev. 1992;6:1229–1240. doi: 10.1101/gad.6.7.1229. [DOI] [PubMed] [Google Scholar]
- 33.Ivanov IP, Loughran G, Atkins JF. uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc. Natl Acad. Sci. USA. 2008;105:10079–10084. doi: 10.1073/pnas.0801590105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl Acad. Sci. USA. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iacono M, Mignone F, Pesole G. uAUG and uORFs in human and rodent 5'untranslated mRNAs. Gene. 2005;349:97–105. doi: 10.1016/j.gene.2004.11.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.