Abstract
A coiled-coil microtubule-bundling protein, p180, was originally identified as one of the ribosome receptor candidates on the rough endoplasmic reticulum (ER) and is highly expressed in secretory tissues. Recently, we reported that p180 plays crucial roles in upregulating collagen biosynthesis, mainly by facilitating ribosome association on the ER. Here, we provide evidence that p180 is required to form translationally active polysome/translocon complexes on the ER. Assembly of highly-developed polysomes on the ER was severely perturbed upon loss of p180. p180 associates with polysome/translocon complexes through multiple contact sites: it was coimmunoprecipitated with the translocon complex independently of ribosomes, while it can also bind to ribosomal large subunit specifically. The responsible domain of p180 for membrane polysome assembly was identified in the C-terminal coiled-coil region. The degree of ribosome occupation of collagen and fibronectin mRNAs was regulated in response to increased traffic demands. This effect appears to be exerted in a manner specific for a specified set of mRNAs. Collectively, our data suggest that p180 is required to form translationally active polysome/translocon complexes on the ER membrane, and plays a pivotal role in highly efficient biosynthesis on the ER membrane through facilitating polysome formation in professional secretory cells.
INTRODUCTION
Recently, broad functions for endoplasmic reticulum (ER)-bound ribosomes have been demonstrated. Genome-wide studies examining mRNA populations on cytosolic and ER-bound polysomes have revealed an unexpected overlap between the two mRNA pools in eukaryotic cells (1,2), and a significant fraction of cytosolic proteins undergo synthesis on ER-bound ribosomes (3).
While translation of mRNAs is potentially regulated at multiple levels, regulation at initiation has been most intensely studied as a key step (4,5). The 5′- and 3′-untranslated regions of mRNAs play crucial roles in various stages of translational regulation, including mRNA translational efficiency, stability and localization (6). The degree of polysome assembly can be postulated to be important aspect of translational control, possibly through a direct impact on translational efficiency linking with translational initiation. While recent advances in cryoelectron tomography have provided important insights into the organization of translating polysomes in cell lysates and intact cells (7,8), it still remains obscure whether ribosome occupation of mRNAs is solely dependent on the lengths of the mRNAs or is regulated by an unknown mechanism (9–11). For membrane and secretory proteins in particular, the situation is more complicated because of subsequent translocation across the membrane. Limited information has been available for how the ribosome and translocon machineries are structurally and functionally coupled (12). ER-associated ribosomes have been shown to mediate more efficient biosynthesis than free ribosomes (3), although it remains unknown whether membrane-associated ribosomes are structurally distinguishable from free cytosolic ribosomes. Moreover, on a single polysome, higher-order coordination should be essential between each unit of a ribosome/translocon complex to accomplish synchronized translation and subsequent translocation across the membrane. However, fundamentally nothing is known about the molecular basis for such coordination.
Collagens are one of the major components of the extracellular matrix in connective tissues such as skin, tendon and bone. They are synthesized on the ER membrane as precursor forms, i.e. procollagens, and secreted by professional secretory cells, including fibroblasts, chondroblasts and osteoblasts. These specialized cells for secretion have a highly developed network of rough ER to accommodate the high-rate synthesis, similar to other secretory cells such as pancreatic cells and plasma cells. Nevertheless, little is known about the mechanisms underlying the highly efficient activity of protein biosynthesis in professional secretory cells. Ascorbate is a popular and long-used stimulator of procollagen secretion during in vitro culture. It acts as a cofactor of prolylhydroxylase and promotes procollagen folding in the ER, thereby initiating its subsequent transport from the ER to the Golgi complex (13). If cells lack ascorbate, procollagens stay in the ER due to the immature folding. Therefore ascorbate treatment can activate de novo biosynthesis in response to resumption of ER-to-Golgi transport and subsequent increased traffic demands (14). However, it has remained unknown how the de novo biosynthesis is activated in the professional secretory cells (14,15).
p180 is an integral ER membrane protein and is highly expressed in secretory tissues (16). It was initially identified as one of the candidate ribosome receptors on the rough ER membrane (17). Its unique repeat domain was reported to have binding capacity for ribosomes and is composed of 54 tandem repeats of a dodecapeptide with a highly basic pI (18). However, it remains elusive whether p180 directly binds to ribosomes in animal cells. Recently, we reported that p180 plays a crucial role in upregulating collagen biosynthesis following ascorbate stimulation (19). Collagen biosynthesis appeared to be enhanced at the translational level by a novel activity of p180 that facilitated ribosome association with the ER (19). Recently, a crucial role for p180 in general protein biogenesis at the rough ER was also suggested (20,21). Therefore, we sought to examine the molecular mechanisms by which p180 facilitates the enhanced protein biosynthesis on the ER membrane. We investigated whether the enhancement of biosynthesis occurred by modulated organization of ribosome-translocon complexes or by more efficient polysome assembly. Here, we provide evidence that p180 is required to form translationally active polysome/translocon complexes on the ER. Furthermore, we show that the degree of ribosome occupation of collagen and fibronectin mRNAs is regulated in response to increased traffic demand. This effect appears to be exerted in a manner specific for certain mRNAs. In addition, the responsible domain for polysome assembly in the p180 sequence is identified.
MATERIALS AND METHODS
Cell culture and short interfering RNA oligonucleotide transfection
HEL fibroblasts were maintained in Dulbecco's modified Eagle's medium (DMEM) (Nissui) supplemented with 10% fetal bovine serum (FBS) (Intergen). l-Ascorbic acid phosphate magnesium salt n-hydrate (Wako) was added to a final concentration of 0.2 mM unless otherwise indicated. The protocols for cell culture and short interfering RNA (siRNA) transfection were carried out as described previously (22). siRNA duplexes against human p180 (DDBJ accession number: AB287347) corresponding to nucleotides 153–173 (p180-si) were used unless otherwise indicated. In some experiments, siRNA duplexes against human p180 corresponding to nucleotides 271–291 (p180-si2) were used. For control samples, control non-silencing siRNAs [either FITC-labeled siRNA (Qiagen) or scrambled siRNA (Stealth™ RNAi Negative Control Duplex; Invitrogen)] were transfected. To estimate the activity of de novo protein biosynthesis in membrane fractions, we used a non-radioisotope (RI) labeling system for newly synthesized proteins with azidohomoalanine (AHA) (23) as described previously (19). Samples were precipitated with trichloroacetic acid, and examined by immunoblotting analyses with an anti-biotin antibody. A stable HeLa cell line overexpressing p180 (HeLa/p180) was described previously (22).
Plasmids and transfection
Expression plasmids for wild-type and mutant forms of human p180 were used as described previously (24). HEL cells were transfected with 5 µg of the plasmids using an Amaxa Nucleofector transfection system (Lonza) according to the manufacturer’s protocols. At 2 days after transfection, cells were subjected to sequential detergent extractions of cytosolic and membrane fractions.
Antibodies, reagents and western blot analysis
Western blot analyses of p180 were performed under strong blotting conditions as described previously (25). The following antibodies were used: rabbit antibodies against GFP (Clontech), protein disulfide isomerase and calnexin (Stressgen), ribosomal protein L10 (Santa Cruz Biotechnology), ribosomal protein S6 (Cell Signaling), biotin (Bethyl Laboratories), Sec61 beta (Upstate), Sec61 alpha and TRAP alpha (generous gifts from C.V. Nicchitta, Duke University Medical Center), human p180 (N1) (24); goat antibody against ribophorin II (Santa Cruz Biotechnology); and mouse monoclonal antibody against CLIMP-63 (26) (a generous gift from H.P. Hauri, University of Basel), (His)5 tag (Qiagen). The samples for western blot analyses were normalized by the DNA amounts and equivalent amounts of samples were analyzed using a fixed exposure time unless otherwise indicated. For protease inhibitors, we used EDTA-free Complete Inhibitor Mixture (Roche Applied Science). Micrococcal S1 nuclease was purchased from New England Biolabs.
Sequential detergent extractions of cytosolic and membrane fractions
Cytosolic and membrane fractions were obtained by sequential detergent extractions using digitonin as described previously (27) with minor modifications (19). Briefly, HEL cell monolayers in six-well plates were precultured with 200 µM cycloheximide for 15 min and washed with PBS. Extraction was performed in the presence of 200 µM cycloheximide, 80 U/ml RNase inhibitor (Toyobo), protease inhibitor cocktail and 1 mM dithiothreitol (DTT) throughout the procedure and the following analyses. The cells were incubated with 0.2 ml of permeabilization buffer [110 mM KOAc, 25 mM HEPES pH 7.5, 2.5 mM Mg(OAc)2, 1 mM EGTA, 0.015% digitonin) for 5 min on ice. After collection of the supernatant (cytosolic fraction), the cells were carefully washed twice with wash buffer [110 mM KOAc, 25 mM HEPES pH 7.5, 2.5 mM Mg(OAc)2, 1 mM EGTA, 0.004% digitonin]. The membrane fraction was then extracted with 0.2 ml of lysis buffer [25 mM HEPES pH 7.5, 400 mM KOAc, 15 mM Mg(OAc)2, 1 mM EGTA, 2% digitonin] for 30 min on ice. The solution was carefully collected and cleared by centrifugation at 7500g for 10 min at 4°C to remove insoluble debris. The resulting supernatant containing the lysate extracted with digitonin from the intracellular membranes was referred to as the membrane fraction in this study, and used for subsequent analyses. Detergent extractions of HeLa cells were performed using permeabilization buffer containing 0.05% digitonin for 10 min on ice, followed by the same procedure described above. Isolation of total RNA and quantification of mRNAs by real-time PCR were performed as described previously (19).
Preparation of ribosome-stripped membranes
Ribosome-stripping was performed by in situ EDTA treatment followed by membrane preparation. Briefly, after removal of the cytosolic fraction with permeabilization buffer as described above, the cells were treated with 50 mM EDTA at 4°C for 30 min in permeabilization buffer lacking digitonin. The dissociated ribosome subunits were removed by careful washing. Subsequently a ribosome-stripped membrane fraction was prepared with lysis buffer containing 2% digitonin as described in the above section. To thoroughly remove residual ribosomes in the ribosome-stripped membranes, sedimentation through a 0.5 M sucrose cushion was performed. rRNA analyses confirmed that about 10% of the total ribosomes were recovered in the pellet, while the supernatant (ribosome-free membrane fraction) contained no rRNA, indicating that 90% of the total ribosomes were released by EDTA under these conditions.
Polysome analysis by velocity sedimentation
The membrane fractions were loaded onto a linear 15–50% sucrose gradient in buffer containing 25 mM K-HEPES (pH 7.5), 400 mM KOAc, 15 mM Mg(OAc)2, 1 mM EGTA. The following analyses were performed in the presence of 200 µM cycloheximide, 80 U/ml RNase inhibitor (Toyobo), protease inhibitor cocktail and 1 mM dithiothreitol (DTT) throughout the procedure unless otherwise indicated. The gradients were centrifuged at 150 000g for 70 min in a TLS-55 rotor (Beckman) at 4°C. Fractions were collected manually. The rRNAs, mRNAs and proteins in the gradient fractions were analyzed. The positions of the 40S and 60S subunits and 80S monosome were defined by analyzing EDTA-treated and nuclease-treated samples. In some experiments, polysome-containing fractions (tube no. 19–29) were collected, and precipitated at 100 000g for 40 min. The resulting pellets were resuspended in lysis buffer lacking DTT and cycloheximide, and subjected to immunoprecipitation (IP) assays.
Isolation of 80S monosome, 60S and 40S subunits
The membrane fractions were treated with 1 mM puromycin in membrane buffer [25 mM HEPES pH 7.5, 400 mM KOAc, 15 mM Mg(OAc)2, 1 mM EGTA, 0.015% digitonin, 1 mM DTT]. Released ribosomes were sedimented by centrifugation at 100 000g for 40 min through a 0.5 M sucrose cushion, and washed five times. The ribosome pellets were subjected to 15–50% sucrose gradient to isolate monosomes. Collected monosome fractions were sedimented through a 0.5 M sucrose cushion at 100 000g for 40 min, and resuspended in membrane buffer lacking digitonin. For purification of 40S and 60S subunits, the 80S monosome was treated with 50 mM EDTA in membrane buffer lacking digitonin and DTT. After 15–25% sucrose density gradient centrifugation, 40S and 60S subunits were collected respectively, and washed five times with membrane buffer lacking Mg(OAc)2. The resulting 40S or 60S pellets were resuspended in membrane buffer lacking digitonin and DTT. Abs 260/280 of these ribosome preparations ranged between 1.65 and 1.77.
Immunoprecipitation assays
An anti-p180 antibody or control rabbit IgG was incubated with sheep anti-rabbit IgG-conjugated magnetic beads (Dynabeads-M280; Veritas), followed by washing according to the manufacture's instruction. Samples were then incubated with these beads at 4°C for 1 h in lysis buffer lacking DTT and cycloheximide. After extensive washing, the captured samples were analyzed by western blotting or agarose electrophoresis.
In vitro binding assay with ribosomes and p180-beads
Ribosome-stripping by EDTA was carried out as described above and the membrane fractions were prepared with lysis buffer containing 1% NP-40 instead of digitonin, and were further treated with 0.5% deoxycholate and 0.1% SDS at 4°C for 1 h. After centrifugation at 100 000g for 40 min through a 0.5 M sucrose cushion, the resulting supernatant was subjected to the IP assay with anti-p180 antibody to prepare p180-beads. The p180- or control-beads were incubated at 4°C for 1 h in membrane buffer with monosome, isolated 60S or 40S subunits, respectively.
Electron microscopy
For transmission Electron microscopy (EM) analysis, cells cultured in 2% FBS/DMEM were treated with medium containing 10 µM taxol for 3 min at 37°C, followed by a conventional fixation procedure for transmission EM as described previously (25). Ultrathin sections were cut parallel or perpendicular to the substrate and viewed under a Model 7650 EM (Hitachi) at 80 kV. Surface views of the rough ER membranes were frequently observed when ultrathin sections were cut parallel to the substrate.
RESULTS
p180-dependent formation of highly developed polysomes on the ER membranes in collagen-secreting fibroblasts
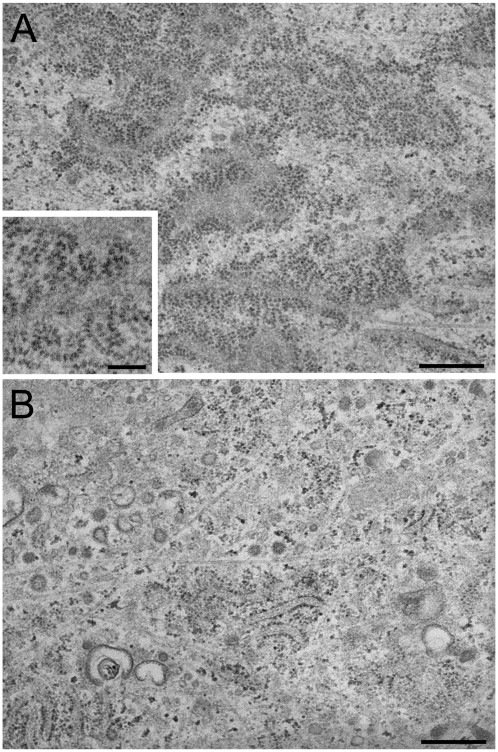
Recently, we reported that ascorbate treatment increases membrane-associated ribosomes without altering the levels of mRNAs (19). These findings suggest that ascorbate upregulates biosynthesis via a novel translational control mechanism in response to high traffic loads, either by altered ribosome-translocon complexes or enhanced polysome organization. To determine whether p180 affects polysome organization, we reexamined the ultrastructure of control and p180-depleted cells. Further inspection of the EM images revealed the presence of a large number of extremely long polysomes on the ER surface of horizontally cut sections from ascorbate-stimulated HEL fibroblasts (Figure 1A). A higher magnification image showed spiral arrays or double-row shaped polysomes consisting of more than 28 ribosomes (Figure 1A, inset), closely resembling those reported in rat dermal fibroblasts (28). In contrast, p180 depletion caused a drastic reduction in membrane polysomes despite ascorbate stimulation (Figure 1B).
Figure 1.
Transmission EM images of the rough ER surface in collagen-secreting HEL fibroblasts. Control (A) and p180 siRNA-transfected (B) HEL cells were grown in F12/DMEM containing 2% FBS in the presence of ascorbate. A large number of highly developed polysomes are clearly seen on the membrane surface in ascorbate-stimulated HEL fibroblasts, while p180 depletion results in dramatically reduced numbers of polysomes on the ER. Polysomes are frequently seen in the membrane surface view when the sections were cut parallel to the substrate. A higher magnification image of control cells (inset in A) shows spiral arrays of polysomes consisting of about 25–30 ribosomes. Bars: A and B, 500 nm: inset in A, 200 nm.
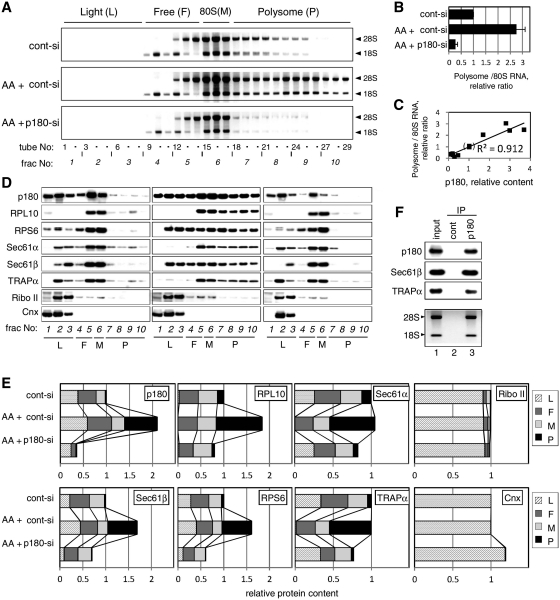
To examine the specific roles for p180 in polysome formation on the ER, we performed polysome gradient analyses under stringent conditions using a high-salt buffer. The membrane fractions of ascorbate-treated and/or p180-depleted cells were fractionated on linear 15–50% sucrose gradients in a buffer containing 0.4 M potassium acetate. To maintain the formation of ribosome-translocon complexes, a lysis buffer containing digitonin was used for membrane extraction (29). Western blot analyses confirmed a large increase in the endogenous p180 level by >2-fold upon ascorbate stimulation (19), which was decreased by p180 siRNA treatment to <16% of the control (Supplementary Figures S1A and S1B). Analyses of rRNAs showed that ascorbate stimulation greatly enhanced polysome formation in the membrane fractions (Figure 2A, middle) compared with those in non-stimulated cells (upper). Consistent with the TEM images mentioned above, heavy fractions were profoundly assembled in the ascorbate-treated cells. Densitometric scanning revealed that ascorbate-treated cells contained ∼2.8-fold higher levels of polysomal RNAs than non-stimulated cells (Figure 2B). Loss of p180 caused a drastic reduction of the membrane-associated polysomes (Figure 2A, bottom), and their RNA levels were decreased to <20% of the non-depleted cells (Figure 2B). The use of another siRNA sequence (p180-si2) resulted in the same effects (Supplementary Figure S2A). No significant changes were observed for the relative amounts of free 60S and 40S subunits per 80S monosome (data not shown).
Figure 2.
p180 facilitates polysome assembly on the ER membrane. HEL cells were cultured in the presence or absence of ascorbate (AA) and treated with a control scrambled (cont-si) or p180-specific (p180-si) siRNA. At post-transfection Day 4, sequential digitonin extractions were carried out to obtain the membrane fractions, followed by polysome analysis using a 15–50% sucrose gradient. Equal amounts of samples normalized by the total DNA contents were subjected to the polysome analysis. (A) The ribosomal RNAs in each fraction were analyzed by agarose electrophoresis. The positions of the polysome (P), monosome (M), free 40S and 60S subunits (F) and light (L) fractions are shown at the top. Top panel: cont-si-treated cells without ascorbate; middle panel: cont-si-treated cells with ascorbate; bottom panel: p180-si-treated cells with ascorbate. (B) The total RNA contents of the polysome fractions were estimated by densitometric scanning of the gels, and the relative ratios of polysomes to 80S monosome are shown (means ± SD, n = 4). (C) The relative amounts of p180 and polysomes in the membrane fractions were plotted to evaluate their correlation. The 12 samples used were obtained from five separate experiments under various conditions. The values for cont-si-treated cells without ascorbate stimulation were set as 1. (D) The distributions of p180 and other translocon-related proteins in the collected fractions were analyzed by western blotting. Non-stimulated cells with cont-si (left), ascorbate-stimulated cells with cont-si (middle), and ascorbate-stimulated p180-si treated cells (right) are shown. (E) Quantitative data for the protein markers in the polysome (P), monosome (M), free 40S and 60S subunits (F) and light (L) fractions are shown. The total amounts of each protein in the membrane fractions were estimated by densitometry using samples prior to sucrose density centrifugation, and their relative contents were assigned to the P, M, F and L fractions by densitometric scanning of the data shown in panel D. The total amounts for the membrane fractions from the cont-si-treated cells without ascorbate stimulation were set as 1. (F) Polysome fractions were collected from the cont-si treated cells after ascorbate stimulation and subjected to IP assays using an anti-p180 antibody or control rabbit IgG bound to magnetic beads conjugated with sheep anti-rabbit IgG. The samples captured by the beads were immunoblotted with antibodies against marker proteins or analyzed for rRNAs. Lane 1, input samples; lane 2, immunoprecipitates with control IgG; lane 3, immunoprecipitates with anti-p180 antibody. Sec61α was not visible because of co-existing immunoglobulin chains.
When p180 was further depleted in ascorbate-untreated cells that contained small amounts of p180 and membrane-associated polysomes, a significant decrease in polysomes was observed (Supplementary Figure S2A and S2B), suggesting that the levels of membrane-associated polysomes were correlated with the p180 levels. The correlation between p180 and polysomes in the membrane fractions was estimated by their relative amounts obtained in five separate experiments under various conditions. The plotted data indicated a positive correlation (R2 = 0.91) (Figure 2C). These findings suggest that p180 is required to enhance polysome formation on the ER in response to high traffic loads upon ascorbate stimulation, and that its protein levels appear to have an impact on the levels of membrane-associated polysomes.
Ternary complex formation of polysome, translocon and p180
Ascorbate stimulation enhances biosynthetic activity, especially in the membrane fraction, if a high level of p180 is expressed on the ER (19), suggesting that the organization of ribosome-translocon complexes may change in a p180-dependent manner. To characterize organization of the polysomes in more detail, the distributions of p180 and translocon proteins were analyzed using collected fractions of the sucrose gradients (Figure 2D). In non-stimulated cells, the polysome fractions (Fractions 7–10) contained small amounts of p180, Sec61α, Sec61β and TPAPα together with ribosomal markers RPL10 and RPS6 (Figure 2D, left). Upon ascorbate treatment, the levels of all six proteins were elevated especially in the polysome fractions, while that of ribophorin II or calnexin was not (Figure 2D, middle, Fractions 7–10). Although Sec61α and TRAPα contents in the total membrane fractions remained unchanged after ascorbate stimulation (Figure 2E, Supplementary Figure S1A), their shift into polysomes occurred. Upon p180 depletion, reduced levels of the translocon-related proteins or ribosomal markers were observed in the polysome fractions as expected (Figure 2D, right), with their reduced contents in the total membrane fractions (Figure 2E). In contrast, little change was observed in the distributions and contents of other rough ER marker proteins, including ribophorin II and calnexin (Figure 2D and E). When analyzed on an RNA basis, the distributions of the translocon-related proteins among Fractions 6–10 did not vary significantly (data not shown), suggesting that heavily assembling polysomes have similar components to light polysomes except for higher loadings. To ensure that p180/polysome/translocon complexes were formed in these cells, co-IP assays were performed using magnetic beads conjugated with an anti-p180 antibody or control IgG. When the collected polysome fractions (tube nos. 18–29) were subjected to the co-IP assays, the anti-p180 antibody specifically captured Sec61β, TRAPα and ribosomes (Figure 2F), indicating that ribosome/translocon and p180 complexes were certainly present in these fractions.
To determine whether p180 can associate with the translocon in the absence of ribosomes, the membranes of permeabilized cells were either left untreated or stripped of ribosomes by in situ EDTA treatment followed by membrane fraction preparation. The ribosome-stripped membrane fractions or control (non-stripped) membrane fractions were analyzed on a 5–20% sucrose gradient. It was found that p180 comigrated with translocon complexes in the vicinity of the bottom in both samples regardless of ribosome stripping (Figure 3A). The possible association of p180 with translocon complexes in the absence of ribosomes was further assessed by IP experiments (Figure 3B). The residual ribosomes in the stripped membranes were thoroughly removed by sedimentation, and the resulting ribosome-free membrane fractions were used for IP experiments. An anti-p180 antibody efficiently captured Sec61β and TRAPα, but not ribophorin II, from the ribosome-free membranes (lane 3), while control IgG did not (lane 2). These findings were further confirmed by a reciprocal test using an anti-Sec61β antibody (lane 4). These data indicate that p180 has the ability to associate with translocon complexes independently of ribosomes.
Figure 3.
Independent association capacity of p180 with the translocon and ribosomes. (A) After removal of the cytosolic fractions with the permeabilization and wash buffers, the membrane fractions of ascorbate-treated cells were either left untreated or stripped of ribosomes by in situ EDTA treatment. Subsequently, membrane fractions were prepared with lysis buffer containing digitonin. The control (untreated) or ribosome-stripped membrane fractions were analyzed on a 5–20% sucrose gradient at 0.4 M KOAc. Marker proteins on western immunoblots and rRNAs in agarose gels analyzed for control membranes (left panel) and ribosome-stripped membranes (right panel) are shown. Flow schema for preparing the ribosome-stripped and untreated membranes are shown in Supplementary Figure S4. (B) Ribosome-stripped membrane fractions were prepared as described for (A) and subsequently centrifuged through a 0.5 M sucrose cushion to remove residual ribosomes. The resulting ribosome-free membrane fractions were subjected to IP analysis with control IgG (lane 2), an anti-p180 antibody (lane 3) and an anti-Sec61β antibody (lane 4) in the presence of 0.4 M KOAc. In the lanes containing the IP samples 10 times higher amounts were loaded compared with the input samples (lane 1). Flow schema for preparing the ribosome-free membranes are shown in Supplementary Figure S4. (C) Ribosome-stripped membrane fractions prepared with 1% NP-40 lysis buffer were further treated with 0.5% deoxycholate and 0.1% SDS at 4°C for 1 h to dissociate possible complexes in the presence of 0.4 M KOAc. After sedimentation at 100 000g for 40 min through a 0.5 M sucrose cushion, the supernatants were subjected to the IP assay with an anti-p180 antibody to prepare p180-beads. Marker proteins were analyzed for the input sample (lane 1), p180-beads (lane 2) and control-beads (lane 3) to confirm the loss of the translocon-related proteins. (D) The p180-beads or control-beads were incubated with isolated monosomes or the subunits at 4°C for 1 h at 0.4 M KOAc. After careful washing, the tested beads were recovered by a magnet and analyzed for rRNA by agarose electrophoresis and for p180 by western blotting. The monosome, 60S and 40S preparations contained no detectable levels of Sec61β, ribophorin II, TRAPα or p180 (data not shown). Lanes 1, 5 and 9: input p180-beads; lane 2: input 80S ribosomes; lanes 6 and 10: input ribosome subunits; lanes 3, 4, 7, 8, 11 and 12: recovered bead fractions after incubation.
Since p180 has been reported to have ribosome-binding domain (18,30), we have addressed whether p180 associates with ribosomes independently of translocon. When the ribosome-stripped membranes were further treated with deoxycholate and SDS, neither Sec61β nor TRAPα was no-longer co-precipitated by the p180 antibody (Figure 3C). In vitro binding experiments using the resulting p180 beads were performed to determine if p180 is able to bind isolated monosomes or the subunits. It is shown that p180 can associate with ribosomes through the 60S subunit, but not with 40S subunit in high salt buffer (Figure 3D). Based on these data, it is likely that 180 associates with polysomes/translocon complexes through multiple sites.
Overexpression of p180 leads to enhanced formation of membrane-associated polysomes
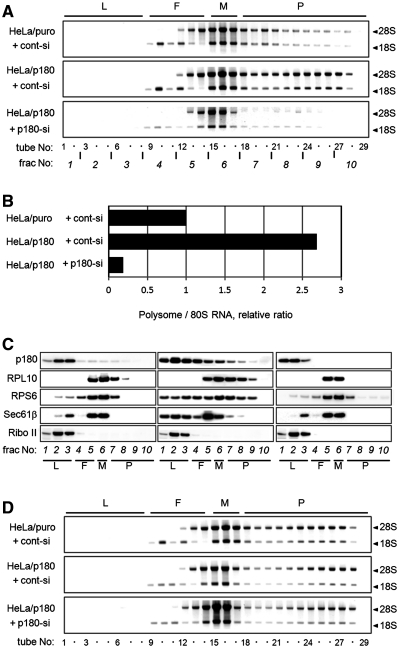
To further address the specific roles of p180 in the formation of membrane-associated polysomes, we examined the polysome organization in HeLa transfectants overexpressing p180 (HeLa/p180 cells). p180 overexpression led to a considerable increase in membrane-associated polysomes in a p180-dependent manner (Figure 4A). The relative RNA levels compared with HeLa/puro cells were increased by ∼2.5-fold (Figure 4B). On the contrary, the polysome patterns in the cytosolic fractions were not significantly affected by the p180 manipulation (Figure 4D). Similar to the data in Figure 2D, co-migration of p180 with polysomes/translocon was observed (Figure 4C), while levels of Sec61 comigration with polysomes appeared to be somewhat low.
Figure 4.
Manipulation of the p180 level affects membrane-associated polysomes. The amounts of ER-associated polysomes were analyzed using stable HeLa transfectants overexpressing p180 (HeLa/p180) and a control cell line (HeLa/puro). (A) The membrane fractions were obtained by sequential digitonin extractions from cont-si-treated HeLa/puro cells, cont-si-treated HeLa/p180 cells and p180-si-treated HeLa/p180 cells, and subjected to polysome analyses using a 15–50% sucrose gradient. (B) The RNA contents of the polysome fractions were estimated by densitometric scanning of the gels, and the relative ratios of polysome:80S are shown. (C) The distributions of p180 and other translocon-related proteins in the collected fractions were analyzed by western blotting. Data for cont-si-treated HeLa/puro cells (left), cont-si-treated HeLa/p180 cells (middle) and p180-si-treated HeLa/p180 cells (right) are presented. (D) The cytosolic fractions were subjected to polysome analyses on a 15–50% sucrose gradient, and the rRNAs were analyzed.
Enhancement of biosynthetic activity in the ER-associated polysomes
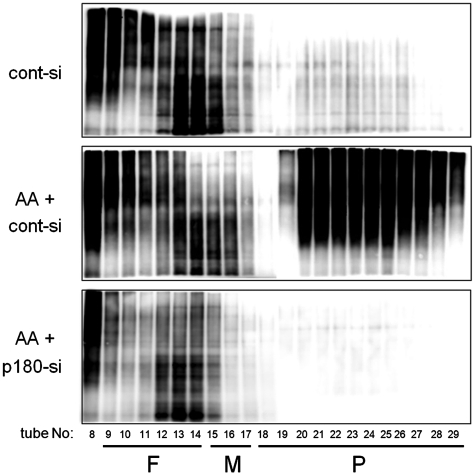
Membrane fractions with abundant p180 exhibit high biosynthetic activity in a p180-dependent manner (19). To determine whether the activated biosynthesis is accompanied by changes in the polysome organization, we estimated the activity of de novo biosynthesis using a non-RI labeling system with AHA, which is specifically incorporated into newly synthesized proteins as a surrogate for methionine (23). Cells were treated with ascorbate and/or depleted with a p180 siRNA, followed by labeling with AHA. Density gradient analysis of the membrane fractions revealed extremely high levels of activation of protein biosynthesis in the polysome fractions after ascorbate stimulation (Figure 5, top and middle). Again, loss of p180 led to almost complete blockade of biosynthesis in the corresponding fractions (Figure 5, bottom). These findings directly indicate that the translational activity is correlated with the occupation levels of polysomes, and that this upregulation seems to be mainly driven by augmented p180/translocon complexes on the ER membrane.
Figure 5.
p180 depletion perturbs ascorbate-stimulated de novo biosynthesis in polysome fractions on the ER membrane. The de novo biosynthesis activity was estimated using a non-RI labeling system with AHA in HEL cells. The cells were cultured in the presence or absence of ascorbate and treated with a control or p180-specific siRNA. After incubation with AHA, the membrane fractions were fractionated on a 15–50% sucrose gradient. The AHA-incorporating proteins were labeled with biotin-alkyne by click chemistry and immunoblotted with an anti-biotin antibody. Top panel: non-stimulated cells with cont-si; middle panel: ascorbate-stimulated cells with cont-si; bottom panel: ascorbate-stimulated cells with p180-si.
Ribosome occupation of mRNAs for specific proteins is accelerated upon ascorbate stimulation in a p180-dependent manner
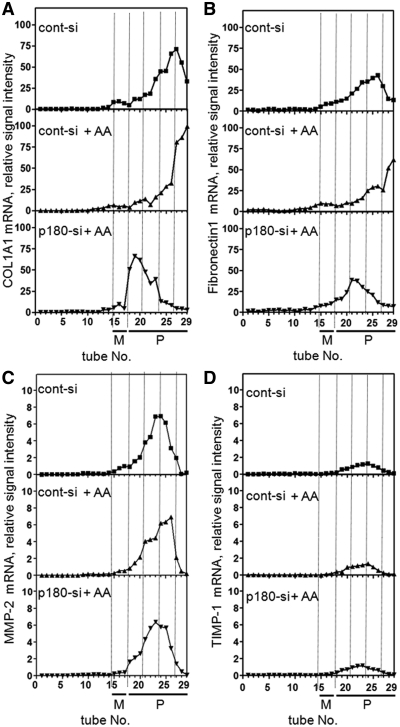
In our previous study p180 knockdown resulted in preferential blockade of the secretion of large extracellular matrix proteins including procollagens and fibronectin (19). To further characterize the molecular basis for these observations, the patterns of procollagen and fibronectin mRNAs in the sucrose gradient were compared with those of non-responding proteins (Figure 6). Prior to ascorbate addition in control cells, the procollagen and fibronectin mRNAs peaked at tubes 26–27 (Figure 6A and B, top). Ascorbate stimulation caused profound shifts of these mRNAs into extremely-heavy polysomes (tubes 28 and 29, middle). On the contrary, loss of p180 caused their dramatic shifts into lighter fractions that peaked at tubes 19 and 20 (bottom), although they were located in polysome fractions. In contrast, the mRNA patterns for MMP-2 and TIMP-1 remained unchanged at around tubes 23 and 24 regardless of p180 manipulation (Figure 6C and D), which is consistent with their constant secretion levels (19).
Figure 6.
Ribosome occupation of mRNAs for specific proteins is accelerated upon ascorbate treatment in a p180-dependent manner. HEL cells were cultured in the presence or absence of ascorbate and treated with a control siRNA or p180-specific siRNA. The membrane fractions obtained by sequential digitonin extractions were subjected to polysome analyses using a 15–50% sucrose gradient. cDNAs were synthesized from RNA samples extracted from individual tubes. The distributions of mRNAs from non-stimulated cells with cont-si, ascorbate-treated cells with cont-si and ascorbate-treated cells with p180-si are plotted. Relative signal intensity of mRNAs for procollagen type 1 alpha chain (A), fibronectin (B), MMP-2 (C) and TIMP-1 (D) are shown. M, monosome fraction; P, polysome fraction.
Expression of the coiled-coil domain of p180 in the cytosol perturbs the membrane association of polysomes
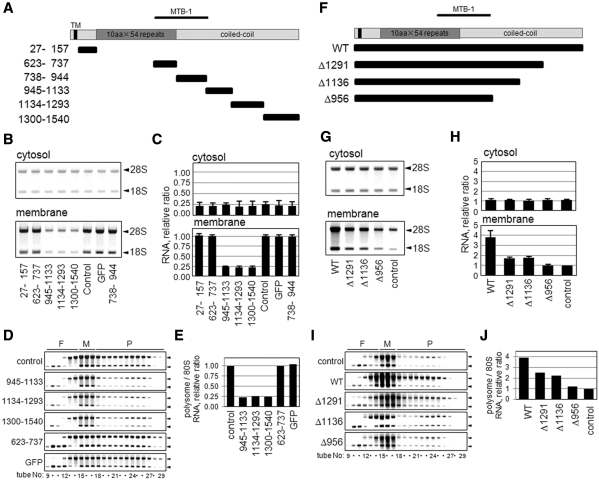
Based on its deduced primary sequence, human p180 is predicted to have a highly basic N-terminal region that includes a reported ribosome-binding repeat domain, an MTB-1 microtubule-bundling domain and an acidic C-terminal region consisting of a predicted coiled-coil domain (18,24). To determine the responsible region for the membrane-associated polysome formation, a series of GFP-tagged polypeptides containing different regions of human p180 were expressed in HEL cells (Figure 7A), and their effects on ribosome/polysomes in the membrane fractions were examined. Western blotting analyses of the chimeras confirmed that the tested proteins were expressed at the correct sizes in the cytosolic fraction (Supplementary Figure S3A). Of the six chimeric proteins analyzed, three C-terminal polypeptides in the coiled-coil domain (amino acids 945–1133, 1134–1293 and 1300–1540) unexpectedly led to dramatic decreases in membrane-bound ribosomes, while cytosolic ribosomes remained unaffected (Figure 7B). Their inhibitory effects were >75% (Figure 7C) and elicited by preventing polysome assembly on the membrane (Figure 7D and E), indicating that these coiled-coil polypeptides expressed in the cytosol exhibit dominant-negative effects. Other chimeras including the repeat domain and MTB-1 had no significant effects (Figure 7B and C). Similar results were found in analyses of HeLa/p180 transfectants (data not shown).
Figure 7.
Cytoplasmic expression of the coiled-coil domain perturbs polysome assembly on the membranes. (A) Schematic representations of full-length and truncated constructs of various regions of p180. Human p180 has a predicted transmembrane domain (TM) close to the N-terminus, a highly basic tandem repeat domain (dark gray box), a microtubule binding and bundling domain (MTB-1) and a C-terminal acidic coiled-coil domain (gray boxes). The numbers on the left of each truncated mutant denote the amino acid residues of human p180 (DDBJ accession number: AB287347). (B) A series of GFP-tagged polypeptides containing different regions of human p180 were expressed in HEL cells, and the rRNAs (indicated by 28S and 18S, respectively) in the cytosolic and membrane fractions are shown. (C) The RNA contents in the cytosolic and membrane-bound fractions were estimated by densitometric scanning, and the relative amounts are shown. Data represent means ± SD (n = 3). (D) Polysome analyses were performed using the membrane fractions of cells overexpressing peptides containing amino acid residues 945–1133, 1134–1293 and 1300–1540 compared with those of control cells or cells overexpressing control peptides. Arrowheads indicate 28S (upper) and 18S (lower) rRNAs. (E) The relative ratios of polysomal RNA to 80S RNA are shown. (F) Schematic representations of wild-type and various C-terminal deletion mutants of p180. (G) Expression plasmids encoding wild-type p180 and p180 mutants were transfected into HeLa cells. The rRNAs in the cytosolic and membrane fractions were analyzed. (H) The relative RNA contents in the cytosolic and membrane-bound fractions are shown. Data represent means ± SD (n = 3). (I) Polysome analyses were performed using the membrane fractions of these cells. Arrowheads indicate 28S (upper) and 18S (lower) rRNAs. (J) The relative ratios of polysomal RNA to 80S RNA are shown.
Next, the role of the coiled-coil polypeptide was further examined in parental HeLa cells, which contained very low levels of endogenous p180. Similar to the stable transfectants (Figure 4), transient overexpression of wild-type p180 resulted in increased ribosomes and polysomes on the membrane while truncation mutants lacking C-terminal domains (Δ956, Δ1136 and Δ1291) failed to enhance the assembly (Figure 7F–J, Supplementary Figure S3B). Therefore, exogenously expressed p180 is competent for the assembly of ER-associated polysomes in a manner dependent on the C-terminal domain.
DISCUSSION
A recent genome-wide study of mRNA translation profiles revealed the unexpected correlation that ribosome density decreases with increasing ORF length (9). However, little is known about the molecular mechanism of polysome assembly, especially on membranes. Here, we have demonstrated that p180 plays a pivotal role in the formation of actively translating heavy polysomes on the ER membrane. Tightly regulated cooperation of the ribosome and translocon on the mRNA seems to occur in a p180-dependent manner in collagen-secreting fibroblasts. In response to resumption of ER-to-Golgi transport and subsequent increased traffic demands, rough ER with high levels of p180 may provide an efficient platform for active protein biosynthesis through facilitation of heavy polysome formation.
The unique repeat domain in p180 was reported to have proliferating activity for the rough ER membrane itself (18,30). However, it remains elusive whether p180 directly binds to ribosomes in animal cells. Based on our data, p180 most likely accelerates the membrane association of polysomes and thereby leads to densely studded ribosomes/polysomes on the ER. This novel feature of p180 may account for its reported proliferative activity toward rough ER membranes (18,30), although the effect was minimal in our study as well as in previous studies (20,22). The polysome assembly activity of p180 is consistent with its enrichment in professional secretory cells undergoing highly active protein biosynthesis, such as pancreas and plasma B cells (16,31), and also fits well with a recent report showing polysome-dependent localization of p180 in rough ER sheets (20).
The formation of a ribosome/translocon/p180 ternary complex was demonstrated in a recent proteome study (21) and by the biochemical analyses of membrane-associated polysomes presented here. The importance of the complex formation for active protein biosynthesis is fully consistent with our previous finding of disturbed protein biosynthesis upon p180 depletion (19). Moreover, we unexpectedly found that the C-terminal coiled-coil domain, but not the tandem repeat domain, plays a key role in the enhanced polysome assembly on the ER by both dominant-negative expression and gain-of-function assays. These findings strongly suggest the involvement of other factor(s) in the interaction between p180 and the polysome/translocon complex. In addition, p180 associates with translocon complexes in the absence of ribosomes. On the other hand, the in vitro binding assay revealed that p180 can bind the 60S subunit, but not the 40S subunit, under high salt conditions independently of the translocon, similar to reported findings of the close disposition of p180 with ribosomes (17,32). Although the binding of p180 to the 60S subunit in preference to the 40S subunit indicates that it is not based on a charge-dependent nonnon-specific interaction, further studies with recombinant proteins are needed to define whether it is a direct interaction. Thus p180 is likely to have multiple contact sites with translocon/polysome complexes and thereby play a role in the complicated translational machinery. Based on these findings, the ability of p180 to enhance the polysome assembly may be exerted primarily by the coiled-coil domain via indirect interactions with the translocon complex through unknown factor(s). The molecular basis for the polysome enhancing mechanism remains to be elucidated.
Thus far, several molecules have been reported to bind to the C-terminal coiled-coil domain of p180 using in vitro assays. One of the candidates is a kinesin motor (33), although little is known about the involvement of microtubule motor proteins in the translation step on the ER. Interestingly the C-terminal domain of p180 shares conserved motifs with kinectin, a potent kinesin-binding partner on the ER (34) which binds to an elongation factor (35). In addition, a key regulator of polysome assembly for the fibronectin gene was identified as a subunit of microtubule-associated protein 1 (36), which is reminiscent of the microtubule-bundling domain in p180. Consistent with previous in vitro data showing polysome targeting to microtubules (37), a role for microtubule-binding protein and/or kinesin motor might be suggested in assembling polysomes for secretory proteins. Alternatively, p180 may drive more efficient docking of polysomes onto the translocon in a manner dependent on conformational change upon translocon/ribosome interaction (12). A further study is now being undertaken to explore the novel mechanism underlying the formation of the polysome/translocon/p180 complex.
In the present study, we found that the degree of polysome occupation on ER-associated mRNAs was regulated, rather than formed at random, in intact cells. Several proteins have been reported to be specifically associated with polysomes, although none of them are expected to function preferentially on the ER (38–40). Recently, a shift of certain mRNAs into polysome fractions from non-polysomal ones was reported in response to specific stimuli (41,42). In these studies, the type of regulatory mode was considered to be ‘turning on/off’ and was different from that observed in the present study. In polysome analyses of ascorbate-untreated cells, the procollagen and fibronectin mRNAs were mainly located in light polysome fractions, which probably correspond to biosynthesis at a basal level (19). Without altered levels of these mRNAs on the ER, greater loading of ribosomes and their shift into very heavy polysome fractions were accelerated in a p180-dependent manner upon ascorbate treatment, conditions in which biosynthetic activity was pronouncedly enhanced. As expected from the preferential secretion loss (19), this shift was seen for the mRNAs of procollagens and fibronectin, but not for the mRNAs of TIMP-1 and MMP-2. p180 may facilitate augmented ribosome occupation of certain mRNAs probably by coordination with other factors. Regarding the site of activation by ascorbate, it remains to be clarified what kinds of signal transducers drive the upregulation of p180 and translocon components after resumption of procollagen transport out of the ER.
During the preferential control of ribosome loading, factors that bind to a specific sequence on a subset of mRNAs may be involved. One of the candidate factors is an hnRNP, a class of RNA-binding proteins that function in both transcription and translation steps (43,44). Previously, important roles of hnRNP E1 in the stabilization of procollagen mRNAs and in translational control were reported (45,46). To explore a role for hnRNP E1 in our system, we examined its expression levels following p180 knockdown. However hnRNP E1 was not affected by depletion of p180 (unpublished observations). This observation implies that the mRNAs for procollagens may be mainly stabilized by hnRNP E1 regardless of p180, while the efficiency of their translation can be modulated by p180 through pronounced assembly of polysomes. A key molecule that functions in such a specific regulation remains to be identified.
Based on the present biochemical data, extremely heavy polysomes are one of the hallmarks of collagen-secreting fibroblasts, in which the majority of the mRNAs for procollagens and fibronectin are located. They probably correspond to the highly assembled double-row or spiral-shaped polysomes (10) in our EM images, which bear a close resemblance to previously reported ones in active collagen-secreting cells (28). To accomplish efficient translation coupled with translocation, higher-order coordination is necessary between each single unit of the ribosome/translocon complex. To this end, the degree of ribosome occupation should be tightly controlled in accordance with the translocation process, which is especially true for extremely high-rate translation of heavy polysomes for the huge molecules. One possible hypothesis is that synchronized translation/translocation on a single mRNA molecule is controlled by a factor that is associated with the polysome/translocon and in turn prevents interference between the individual units of the complex. p180 might be involved in such a regulation process through the association of its C-terminal domain with other factors. For the sake of high-rate translation of secretory proteins, continuous supply of aminoacyl-tRNA and other factors is another important aspect. To achieve sufficient supply of these factors engaged in translation, the aminoacyl-tRNA channeling hypothesis is an attractive model which states that aminoacyl-tRNA is funneled through the translational machinery without intermittent diffusion (47). Indeed aminoacyl-tRNA synthetases and initiation and elongation factors were localized with the ER (48,49), consistent with the kinetic advantage of protein biosynthesis on the ER (3). Given that kinectin associates with elongation factor eEF1Bδ and thereby anchor the eEF1B complex on the ER (50), the highly conserved C-terminal domain among p180 and kinectin may play a role in providing a suitable platform for high-rate protein biosynthesis. Further studies on the complex formation of the ribosome/translocon machinery with p180 will provide important insights into the basic mechanism for orchestrating the high-rate biosynthesis achieved by professional secretory cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4.
FUNDING
Funding for open access charge: Nippi Research Institute of Biomatrix.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Reid Gilmore for helpful discussions and also Christopher Nicchitta and Hans-Peter Hauri for providing reagents. The authors are also grateful to Yuki Taga, Yuko Ushiki-Kaku and other members of the Nippi laboratory for helpful discussions.
REFERENCES
- 1.Diehn M, Bhattacharya R, Botstein D, Brown PO. Genome-scale identification of membrane-associated human mRNAs. PLoS Genet. 2006;2:39–50. doi: 10.1371/journal.pgen.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diehn M, Eisen MB, Botstein D, Brown PO. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat. Genet. 2000;25:58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
- 3.Stephens SB, Nicchitta CV. Divergent regulation of protein synthesis in the cytosol and endoplasmic reticulum compartments of mammalian cells. Mol. Biol. Cell. 2008;19:623–632. doi: 10.1091/mbc.E07-07-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell. Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abaza I, Gebauer F. Trading translation with RNA-binding proteins. RNA. 2008;14:404–409. doi: 10.1261/rna.848208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandt F, Carlson LA, Hartl FU, Baumeister W, Grunewald K. The three-dimensional organization of polyribosomes in intact human cells. Mol. Cell. 2010;39:560–569. doi: 10.1016/j.molcel.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Brandt F, Etchells SA, Ortiz JO, Elcock AH, Hartl FU, Baumeister W. The native 3D organization of bacterial polysomes. Cell. 2009;136:261–271. doi: 10.1016/j.cell.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kopeina GS, Afonina ZA, Gromova KV, Shirokov VA, Vasiliev VD, Spirin AS. Step-wise formation of eukaryotic double-row polyribosomes and circular translation of polysomal mRNA. Nucleic Acids Res. 2008;36:2476–2488. doi: 10.1093/nar/gkm1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson EM, Winkler MM. Regulation of mRNA entry into polysomes. Parameters affecting polysome size and the fraction of mRNA in polysomes. J. Biol. Chem. 1987;262:11501–11506. [PubMed] [Google Scholar]
- 12.Pool MR. A trans-membrane segment inside the ribosome exit tunnel triggers RAMP4 recruitment to the Sec61p translocase. J. Cell Biol. 2009;185:889–902. doi: 10.1083/jcb.200807066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levene CI, Bates CJ. Ascorbic acid and collagen synthesis in cultured fibroblasts. Ann. NY Acad. Sci. 1975;258:288–306. doi: 10.1111/j.1749-6632.1975.tb29289.x. [DOI] [PubMed] [Google Scholar]
- 14.Chan D, Lamande SR, Cole WG, Bateman JF. Regulation of procollagen synthesis and processing during ascorbate-induced extracellular matrix accumulation in vitro. Biochem. J. 1990;269:175–181. doi: 10.1042/bj2690175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sullivan TA, Uschmann B, Hough R, Leboy PS. Ascorbate modulation of chondrocyte gene expression is independent of its role in collagen secretion. J. Biol. Chem. 1994;269:22500–22506. [PubMed] [Google Scholar]
- 16.Langley R, Leung E, Morris C, Berg R, McDonald M, Weaver A, Parry DA, Ni J, Su J, Gentz R, et al. Identification of multiple forms of 180-kDa ribosome receptor in human cells. DNA Cell Biol. 1998;17:449–460. doi: 10.1089/dna.1998.17.449. [DOI] [PubMed] [Google Scholar]
- 17.Savitz AJ, Meyer DI. Identification of a ribosome receptor in the rough endoplasmic reticulum. Nature. 1990;346:540–544. doi: 10.1038/346540a0. [DOI] [PubMed] [Google Scholar]
- 18.Wanker EE, Sun Y, Savitz AJ, Meyer DI. Functional characterization of the 180-kD ribosome receptor in vivo. J. Cell Biol. 1995;130:29–39. doi: 10.1083/jcb.130.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueno T, Tanaka K, Kaneko K, Taga Y, Sata T, Irie S, Hattori S, Ogawa-Goto K. Enhancement of procollagen biosynthesis by p180 through augmented ribosome association on the endoplasmic reticulum in response to stimulated secretion. J. Biol. Chem. 2010;285:29941–29950. doi: 10.1074/jbc.M109.094607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143:774–788. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dejgaard K, Theberge JF, Heath-Engel H, Chevet E, Tremblay ML, Thomas DY. Organization of the Sec61 translocon, studied by high resolution native electrophoresis. J. Proteome Res. 2010;9:1763–1771. doi: 10.1021/pr900900x. [DOI] [PubMed] [Google Scholar]
- 22.Ueno T, Kaneko K, Katano H, Sato Y, Mazitschek R, Tanaka K, Hattori S, Irie S, Sata T, Ogawa-Goto K. Expansion of the trans-Golgi network following activated collagen secretion is supported by a coiled-coil microtubule-bundling protein, p180, on the ER. Exp. Cell Res. 2010;316:329–340. doi: 10.1016/j.yexcr.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 23.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc. Natl Acad. Sci. USA. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogawa-Goto K, Tanaka K, Ueno T, Kurata T, Sata T, Irie S. p180 is involved in the interaction between the endoplasmic reticulum and microtubules through a novel microtubule-binding and bundling domain. Mol. Biol. Cell. 2007;18:3741–3751. doi: 10.1091/mbc.E06-12-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogawa-Goto K, Irie S, Omori A, Miura Y, Katano H, Hasegawa H, Kurata T, Sata T, Arao Y. An endoplasmic reticulum protein, p180, is highly expressed in human cytomegalovirus-permissive cells and interacts with the tegument protein encoded by UL48. J. Virol. 2002;76:2350–2362. doi: 10.1128/jvi.76.5.2350-2362.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klopfenstein DR, Kappeler F, Hauri HP. A novel direct interaction of endoplasmic reticulum with microtubules. EMBO J. 1998;17:6168–6177. doi: 10.1093/emboj/17.21.6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lerner RS, Seiser RM, Zheng T, Lager PJ, Reedy MC, Keene JD, Nicchitta CV. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christensen AK, Bourne CM. Shape of large bound polysomes in cultured fibroblasts and thyroid epithelial cells. Anat. Rec. 1999;255:116–129. doi: 10.1002/(SICI)1097-0185(19990601)255:2<116::AID-AR2>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 29.Potter MD, Nicchitta CV. Ribosome-independent regulation of translocon composition and Sec61alpha conformation. J. Biol. Chem. 2000;275:2037–2045. doi: 10.1074/jbc.275.3.2037. [DOI] [PubMed] [Google Scholar]
- 30.Becker F, Block-Alper L, Nakamura G, Harada J, Wittrup KD, Meyer DI. Expression of the 180-kD ribosome receptor induces membrane proliferation and increased secretory activity in yeast. J. Cell Biol. 1999;146:273–284. doi: 10.1083/jcb.146.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luckey CJ, Bhattacharya D, Goldrath AW, Weissman IL, Benoist C, Mathis D. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc. Natl Acad. Sci. USA. 2006;103:3304–3309. doi: 10.1073/pnas.0511137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collins PG, Gilmore R. Ribosome binding to the endoplasmic reticulum: a 180-kD protein identified by crosslinking to membrane-bound ribosomes is not required for ribosome binding activity. J. Cell Biol. 1991;114:639–649. doi: 10.1083/jcb.114.4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diefenbach RJ, Diefenbach E, Douglas MW, Cunningham AL. The ribosome receptor, p180, interacts with kinesin heavy chain, KIF5B. Biochem. Biophys. Res. Commun. 2004;319:987–992. doi: 10.1016/j.bbrc.2004.05.069. [DOI] [PubMed] [Google Scholar]
- 34.Toyoshima I, Yu H, Steuer ER, Sheetz MP. Kinectin, a major kinesin-binding protein on ER. J. Cell Biol. 1992;118:1121–1131. doi: 10.1083/jcb.118.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ong LL, Er CP, Ho A, Aung MT, Yu H. Kinectin anchors the translation elongation factor-1 delta to the endoplasmic reticulum. J. Biol. Chem. 2003;278:32115–32123. doi: 10.1074/jbc.M210917200. [DOI] [PubMed] [Google Scholar]
- 36.Zhou B, Rabinovitch M. Microtubule involvement in translational regulation of fibronectin expression by light chain 3 of microtubule-associated protein 1 in vascular smooth muscle cells. Circ. Res. 1998;83:481–489. doi: 10.1161/01.res.83.5.481. [DOI] [PubMed] [Google Scholar]
- 37.Hamill D, Davis J, Drawbridge J, Suprenant KA. Polyribosome targeting to microtubules: enrichment of specific mRNAs in a reconstituted microtubule preparation from sea urchin embryos. J. Cell Biol. 1994;127:973–984. doi: 10.1083/jcb.127.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 39.Skabkin MA, Kiselyova OI, Chernov KG, Sorokin AV, Dubrovin EV, Yaminsky IV, Vasiliev VD, Ovchinnikov LP. Structural organization of mRNA complexes with major core mRNP protein YB-1. Nucleic Acids Res. 2004;32:5621–5635. doi: 10.1093/nar/gkh889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lang BD, Fridovich-Keil JL. Scp160p, a multiple KH-domain protein, is a component of mRNP complexes in yeast. Nucleic Acids Res. 2000;28:1576–1584. doi: 10.1093/nar/28.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perlewitz A, Nafz B, Skalweit A, Fahling M, Persson PB, Thiele BJ. Aldosterone and vasopressin affect {alpha}- and {gamma}-ENaC mRNA translation. Nucleic Acids Res. 2010;38:5746–5760. doi: 10.1093/nar/gkq267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhamija S, Doerrie A, Winzen R, Dittrich-Breiholz O, Taghipour A, Kuehne N, Kracht M, Holtmann H. IL-1-induced post-transcriptional mechanisms target overlapping translational silencing and destabilizing elements in IkappaBzeta mRNA. J. Biol. Chem. 2010;285:29165–29178. doi: 10.1074/jbc.M110.146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaudhury A, Chander P, Howe PH. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: Focus on hnRNP E1's multifunctional regulatory roles. RNA. 2010;16:1449–1462. doi: 10.1261/rna.2254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 45.Thiele BJ, Doller A, Kahne T, Pregla R, Hetzer R, Regitz-Zagrosek V. RNA-binding proteins heterogeneous nuclear ribonucleoprotein A1, E1, and K are involved in post-transcriptional control of collagen I and III synthesis. Circ. Res. 2004;95:1058–1066. doi: 10.1161/01.RES.0000149166.33833.08. [DOI] [PubMed] [Google Scholar]
- 46.Lindquist JN, Kauschke SG, Stefanovic B, Burchardt ER, Brenner DA. Characterization of the interaction between alphaCP(2) and the 3′-untranslated region of collagen alpha1(I) mRNA. Nucleic Acids Res. 2000;28:4306–4316. doi: 10.1093/nar/28.21.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Negrutskii BS, Deutscher MP. Channeling of aminoacyl-tRNA for protein synthesis in vivo. Proc. Natl Acad. Sci. USA. 1991;88:4991–4995. doi: 10.1073/pnas.88.11.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gavrilova LP, Rutkevitch NM, Gelfand VI, Motuz LP, Stahl J, Bommer UA, Bielka H. Immunofluorescent localization of protein synthesis components in mouse embryo fibroblasts. Cell Biol. Int. Rep. 1987;11:745–753. doi: 10.1016/0309-1651(87)90134-2. [DOI] [PubMed] [Google Scholar]
- 49.Dang CV, Yang DC, Pollard TD. Association of methionyl-tRNA synthetase with detergent-insoluble components of the rough endoplasmic reticulum. J. Cell Biol. 1983;96:1138–1147. doi: 10.1083/jcb.96.4.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ong LL, Lin PC, Zhang X, Chia SM, Yu H. Kinectin-dependent assembly of translation elongation factor-1 complex on endoplasmic reticulum regulates protein synthesis. J. Biol. Chem. 2006;281:33621–33634. doi: 10.1074/jbc.M607555200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.