Abstract
The number of distinct biomolecules that can be visualized within individual cells and tissue sections via fluorescence microscopy is limited by the spectral overlap of the fluorescent dye molecules that are coupled permanently to their targets. This issue prohibits characterization of important functional relationships between different molecular pathway components in cells. Yet, recent improved understandings of DNA strand displacement reactions now provides opportunities to create programmable labeling and detection approaches that operate through controlled transient interactions between different dynamic DNA complexes. We examined whether erasable molecular imaging probes could be created that harness this mechanism to couple and then remove fluorophore-bearing oligonucleotides to and from DNA-tagged protein markers within fixed cell samples. We show that the efficiency of marker erasing via strand displacement can be limited by non-toehold mediated stand exchange processes that lower the rates that fluorophore-bearing strands diffuse out of cells. Two probe constructions are described that avoid this problem and allow efficient fluorophore removal from their targets. With these modifications, we show one can at least double the number of proteins that can be visualized on the same cells via reiterative in situ labeling and erasing of markers on cells.
INTRODUCTION
Recent advances in the field of DNA nanotechnology have facilitated the creation of various dynamic DNA and RNA complexes that can function as programmable logic gates (1–3), chemical amplifiers (4,5), and reconfigurable molecular structures (6,7). A key feature of these complexes is that, instead of classical hybridization reactions, they can operate via a process called strand displacement—the exchange of oligonucleotides possessing partially or fully identical sequences between different thermodynamically stable multi-strand complexes (8) (examples are shown in Schemes 1 and 2). Using this mechanism, long nucleic acid complexes possessing many matched base pairs can be hybridized and dehybridized multiple times at room temperature. Moreover, since strand displacement reactions are sequence dependent and tend to be more sensitive to base mismatches than classical hybridization reactions (9), different dynamic complexes can be designed to operate independently of one another, or, alternatively, integrated into programmable reaction networks that can perform complex computations (10–12). Such capabilities now offer opportunities to create new classes of molecular probe technologies for molecular-cell analyses.
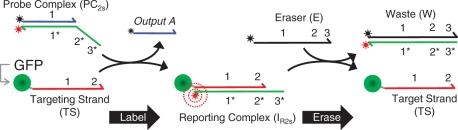
Scheme 1.
Labeling and erasing reactions for two-strand probe constructs.
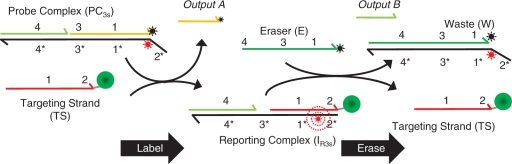
Scheme 2.
Labeling and erasing reactions for three-strand probe constructs.
The potential of dynamic nucleic acid complexes for various biological applications are beginning to be realized (13–15). Engineered RNA hairpin devices have been used as ‘smart’ therapeutic technologies that can selectively react with mutant RNA transcripts in vitro, and, in response, produce double stranded RNA polymers that trigger cell apoptosis (14). Multiplexed (5-color) in situ detection of mRNA transcripts in fixed zebrafish embryos has also been demonstrated using similar hairpin systems (15). Yet, despite these advances, the translation of dynamic oligonucleotide complexes towards such applications remains generically challenging. Most candidate probe constructions are first evaluated in a test tube where displacement reactions occur in homogeneously mixed solutions (4,9,16). The environment inside cells is much more complex and heterogeneous, and even if samples are fixed and permeabilized, issues surrounding the sample penetration and probe dispersion must now be addressed. Other environmental factors may also potentially interfere with the strand displacement process, and such effects could result in unwanted reverse or side reactions. For example, many dynamic DNA complexes are designed to consume a single-stranded input (a target) via toehold-mediated strand displacement. Here, a small single-stranded domain (∼6 bp), called a toehold, is used to partially hybridize to the target and accelerate the initiation rates of the strand displacement reaction. Once the composite reaction is completed, a new, fully duplexed complex is produced that is considered to be an ‘inert’ or unreactive byproduct since it no longer possesses a single stranded toehold domain. However, molecular crowding agents present in cells (e.g. proteins and other biomacromolecules) and enhanced concentration effects due to local confinement could potentially accelerate non-toehold-mediated strand exchange events between strands within duplexed DNA complexes and other unhybridized strands present in the sample (17). In turn, the products formed from a displacement reaction would no long be fully inert. Thus, addressing the unique challenges of detecting biomacromolecules within cells and tissues requires characterization of the strand exchange processes within these environments and the development of approaches to potentially circumvent these types of problems.
Our group is interested in developing dynamic DNA complexes that can function as reconfigurable (erasable) molecular imaging probes for in situ analyses of proteins (13). These probes harness the strand displacement mechanism to selectively couple fluorophore-labeled oligonucleotides to DNA-tagged protein markers on cells. After these markers are visualized via fluorescence microscopy, a second strand displacement reaction is then performed to strip the dye-bearing strands from the reporting complexes that are coupled to the protein targets. With this capability, the total number of proteins that can be visualized on an individual sample is no longer limited by the spectral overlap of fluorescent dyes since the removal of dyes now allows new sets of protein markers to be detected on the same cell or tissue section. For this application, the strand displacement reactions driving the labeling and erasing of markers must be selective and efficient (high labeling and dye removal yields) to facilitate quantitation of marker levels and to ensure residual signals that remain on a sample after an erasing step do not compromise subsequent image analyses of other proteins. As with other probe systems, the use of small diffusible probe complexes is likely important to ensure their even probe dispersion throughout a sample. Finally, since individual samples will be inspected multiple times, the time required to complete the entire reiterative marker imaging procedure should be kept to a minimum. Thus, short marker labeling and erasing reaction times are highly desirable.
Herein, we evaluate the in situ marker labeling/erasing efficiencies and reaction kinetics of three different erasable dynamic DNA probes that contain different numbers of component strands and reactive domains. We also compare the labeling and erasing performance of probes that react with their target strands via three- and four-way strand displacement mechanisms. These analyses indicate that efficient in situ erasing of DNA-tagged protein targets requires probe constructions that minimize the ability of product complexes that are produced during erasure to react with and relabel free ssDNA targets within cells via strand exchange processes that are not mediated by a toehold. In particular, the avoidance of these processes appears to be important to facilitate the rapid diffusion of fluorophore-bearing strands out of the cells and into their surrounding medium during erasing reactions. Overall, we find that three-way strand displacement reactions where a single strand liberates two different strands from labeled targets support efficient erasing. Alternatively, fluorescent dyes can be removed efficiently using probe designs that exchange nucleotides via four-way strand displacement processes since these reactions produce much more ‘inert’ dsDNA products. While facilitating the optimization of probe designs for our application, such information should aid the development and implementation of other dynamic DNA systems that are designed for analogous biological applications.
MATERIALS AND METHODS
Materials
Oligonucleotides were purchased from Integrated DNA Technologies (IDT). The protein target, a recombinant green fluorescent protein, GFP-ZE, was produced using standard cloning and cell transfection procedures. The C-terminal leucine zipper (ZE) is used as an affinity tag for DNA labeling. This zipper associates strongly (KD ∼ 10−15 M) with a complementary basic zipper (ZR) that is incorporated into a DNA-conjugated artificial protein (ZR-ELS6-ssTS) as a gene fusion (18). These polymers were produced according to previously reported procedures (19). The GFP-ZE offers some advantages as a protein target for the present study since it can be outfitted with ssDNA stoichiometrically. The ZR-ELS6-ssDNA polymer is not depicted in Schemes 1 and 2 for simplicity.
DNA probe design
Probe sequences—probe complex (PC), eraser (E) and targeting (TS) strands—were designed using similar methods to those described in Ref. (13). All sequences, excluding those adopted from Zhang et al. (20), were selected using a custom MATLAB script that generates random domains of specified lengths having pre-determined GC% range, while excluding previously generated domains or other prohibitive sequences (i.e. G quadruplexes), and avoiding secondary structures (e.g. hairpins). The generated domains are ranked according to their normalized two-state hybridization energies with existing probe strands using mFold (21). The domains are then screened through the BLAST database to minimize probe sequence homology with the mRNA transcriptome. The final domain sequences are then selected manually from this list and concatenated with other domains to create full oligonucleotide sequences that will be incorporated into a probe complex. Other global criteria such as temperature, strand concentration, and salt concentration are specified prior to domain design. A table listing the oligonucleotide sequences for all complexes and calculations of their standard free energies can be found in the Supplementary Data (Tables S1 and S2).
Fluorophores (Cy3 or Cy5) and quencher molecules (Iowa Black FQ or RQ) were incorporated in opposing strands at positions that minimized their intermolecular distances in both the probe complexes and their waste products (Schemes 1 and 2). For the three-strand probes (PC3s), this requires that the dye molecule is positioned internally within the longer strand of the complex (Scheme 2).
Cell labeling and erasing procedures
CHO cells were grown on glass coverslips in F12 media supplemented with 10% FBS. After 24 h, the culture medium was replaced and the cells were transiently transfected with vector containing the GFP-ZE construct using Fugene (Roche) according to the manufacturer's protocol. Cells were cultured for an additional 12 h to allow for GFP-ZE production. The cells were then fixed using freshly prepared 4% paraformaldehyde for 30 min. Activated aldehydes resulting from the fixation procedure were quenched using 1 mg/ml NaBH4 for 5 min at room temperature. Afterwards, the cells were permeabilized using 0.2% Triton X-100, washed twice with PBS, and stored overnight a 4°C.
Prior to cell labeling experiments, the coverslips were rinsed twice in PBS, dried under an airstream, and then affixed to custom-fabricated micro-well chambers (10-round wells with 0.36 cm2 culture area and culture volume of 400 µl) using a precision-cut double sided adhesive film. The cells were re-hydrated with PBS prior to the labeling procedure. To minimize non-specific binding of the ZR-ELS6-ssTS, cells were first blocked for 2 h using a solution containing 1% BSA, 1 mg/ml Herring Sperm DNA and 0.5 µM polyT DNA in PBS. The cells were then incubated with a 400-nM solution of ZR-ELS6-ssTS for 2 h, and washed twice with PBS.
Cell labeling experiments were performed by incubating cells with solutions of 100 nM probe complexes in TAE buffer supplemented with 12.5 mM Mg2+. Probe deactivation/erasing reactions were performed using 1 µM of the eraser strands (Es) or complexes (Ec). All cell labeling/erasing reactions were carried out for 2 h at 30°C using a rotating incubator shaker (200 rpm), except for the kinetic experiments where the reactions were performed directly on the microscope at room temperature and without shaking.
Cells were imaged using an inverted Nikon microscope outfitted with a ×40 0.95 NA objective, electronic shutters, and a 14-bit depth EMCCD camera (LucaR; Andor). A mechanical translation stage and electronic focusing mechanism was used to collect 5–10 different image fields for each sample. Images were processed using Nikon (NIS-Elements) or ImageJ software, and are presented as heat maps since this rendering enhances the contrast of low-level, remnant fluorescence signals within the ‘erased’ images. Average cell intensities were determined using an algorithm in NIS image that marks the boundary of a selected cell and calculates the average pixel intensity within that region.
RESULTS
Dynamic DNA probe designs
Our previous report showed that a class of three-strand DNA complexes (PC3s) that have been previously integrated into catalytic networks (20) can also function effectively as erasable molecular imaging probes, providing linearly correlative labeling intensities and efficient >95% dye removal (13). Despite this success, we believe our application would benefit from the development of somewhat smaller, 2-component probe complexes that incorporate terminal (3′ or 5′) dye molecules instead of the internal dyes in our prior design. There was concern that the internal dye placement could potentially interfere with the strand-displacement process and restrict the types of dyes that can be incorporated into a probe. To address this issue, we created several DNA probe complexes composed of two partially complementary DNA oligonucleotides (PC2s; Scheme 1). The marker labeling and erasing reactions for the original three-strand complexes are illustrated in Scheme 2. In both cases, the probes react with their ssDNA targets (TS) via toehold-mediated strand displacement to produce a fluorescent reporting complex (IR) containing an unquenched fluorophore. As a result, molecular targets that are conjugated with TS can be visualized using fluorescence microscopy. Analogously, the fluorescent reporting complexes (IR2s and IR3s for the two- and three-strand probes, respectively) can be displaced using a single-stranded eraser oligonucleotide (E). In principle, the quenched waste product (W) of this reaction can then be washed off the sample. Of note, the strand displacement in the labeling and erasing reactions of each probe system proceed via a three-way branch migration process (22), where ssDNA components displace one another.
The present PC2s probes contain three distinct domains (Scheme 1): an 18-bp domain that is completely hybridized (domains 1/1*) and two 6-bp toehold domains that are positioned adjacent to one another at one end of the complex (domains 2* and 3*). Reporting complexes of the PC2s system are formed through a toehold-mediated exchange reaction that is initiated by binding of the probe complex to domain 2 of the TS strand. However, unlike the three-strand complexes whose reaction is completed by the release of an output strand from a second toehold domain (output A releases from domain 3 in Scheme 2), the PC2s probes were designed so that TS displaces the output strand (output A) within PC2s completely during the marker labeling reaction. Similarly, the IR2s reporters are disassembled/erased through a displacement reaction where an eraser strand (E) binds to toehold 3* on IR2s and then displaces the TS completely from the reporting complex. Since neither reaction require the dehybidization of a toehold domain, the forward reactions during marker labeling and erasing both results in the accumulation of six additional matched bps. This design was originally chosen so that each reaction would be energetically favorable. Additionally, since the release of outputs from toehold domains could potentially lower probe-target strand exchange rates, we generally expected that this feature would accelerate our labeling and erasing reactions.
Another important distinction between the PC2s and PC3s probes is that their erasing reactions produce different numbers of products. For the three-strand probes, the marker erasing involves a reaction where E binds to a 4-bp domain in IR3s and then displaces two strands (output B and TS) from the reporting complex. Thus, even though this reaction results in a net loss of two matched bps in the system, there is an increase in configurational entropy as the reaction proceeds forward (20). As discussed below, an important consequence of this design for our application is that reverse (relabeling) reactions can only occur if both B and TS bind to W simultaneously.
Finally, in an attempt to drive each reaction to completion, our protein labeling and erasing reactions all use relatively high concentrations of E (1 µM) relative to TS (∼2 nM in the total reaction volume). Thus, so long as the reactions of probes with their targets mimic the behavior one would expect in a homogenously mixed solution at these concentrations, each probe reaction should be able to reach an equilibrium distribution where the vast majority of the TS strands (>>95%) were either incorporated into an IR complex, or erased (see Supplementary Data). In principle, such behavior should support efficient labeling and erasing.
Selective in situ labeling and erasing of DNA-conjugated proteins
To evaluate the labeling and erasing performance of the PC2s and PC3s probes, we performed two sets of in situ cell imaging experiments where expressed GFP proteins within fixed and permeabilized CHO cells were first outfitted with TS strands using an ssDNA-artificial protein conjugate and then reacted with either two- or three-strand probe complexes that incorporate Cy5 fluorophores (Figures 1 and 2). In each case, labeling intensities were evaluated after the probes were allowed to react for a period of 2 h. For practical purposes, reactions will likely need to be completed within a short time frame to implement our reiterative marker imaging technique.
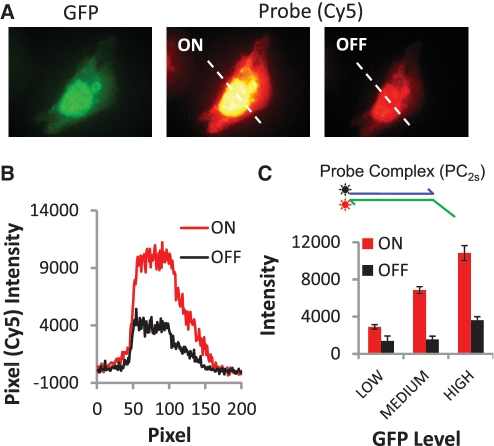
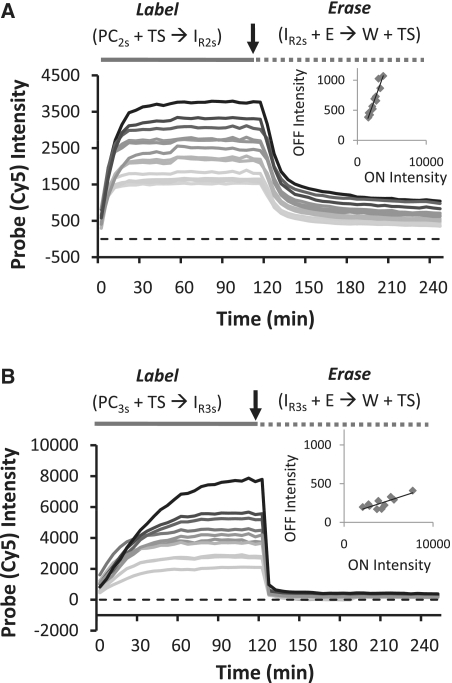
Figure 1.
Labeling and removal of Cy5 fluorophores from protein markers via strand displacement reactions of a two-strand probe complex (PC2s). (A) Selective labeling of expressed GFP proteins in CHO cells. The images display a strong correspondence between the GFP and two-strand probe (Cy5) signals; pixel intensities of the GFP and Cy5 signals are linearly correlated (r2 = 0.94). However, OFF signal intensities indicate ∼20% of active Cy5 dye remains on the cells after the erasing reaction. (B) Pixel intensities for cross section indicated in the probe images in A for both the ON and OFF states of the cells. (C) Histogram of the average Cy5 signal intensities for the ON and OFF states of 20 cells. Cells are grouped based on their GFP whole-cell fluorescence intensities (low, medium and high levels correspond to 2000–5500, 5500–10 000 and 10 000–15 800 intensity units, respectively).
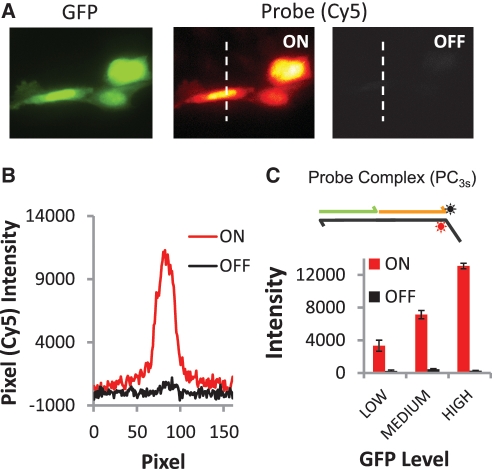
Figure 2.
Labeling and removal of Cy5 fluorophores from protein markers via strand displacement reactions of a three-strand probe complex (PC3s). (A) Selective labeling of expressed GFP proteins in CHO cells. The OFF reactions are now efficient, and yield a signal (cell intensity) to background (slide surface intensity) ratio of 1.08. The ratio of labeled/erased probe intensities, or ON/OFF ratio, is 28.6. This result is reflected in (B) pixel intensities for cross sections as well as (C) histogram of average, whole-cell Cy5 signal intensities for 20 cells in their ON and OFF states.
Comparisons of GFP and Cy5 signals produced after a labeling reaction show that both types of probes can selectively couple fluorophores to DNA-conjugated proteins on cells. In each case, intracellular distributions of the GFP molecules (e.g. nuclear–cytoplasm signal ratios) are reproduced, while the cells that were not transfected in the sample are not labeled. Yet, despite their similar protein labeling performance, the erasing reaction of the PC2s probe system (Figure 1) is found to be much less efficient than the PC3s system (Figure 2). Here, the erasing reaction that disassembles the two-strand IR2s reporter results in residual Cy5 signals that range between 20–30% of the signal amplitudes produced by the prior marker labeling reaction, yielding an average ON/OFF ratio of 3.14. Furthermore, these unwanted ‘OFF-state’ signals are positively correlated with ‘ON-state’ Cy5 intensities (Figure 1C, inset). The erasing performance of the three-strand complex is much better (Figure 2). As in our prior work (13), ‘OFF-state’ signals with the three-strand complexes can barely be detected over background autofluorescence of the unlabeled cells (‘OFF’-signals are only one to three times the RMS noise of the slide background), and average ON/OFF ratios are much higher (26.8). Thus, of the two different probes systems evaluated above, only the three-strand systems can function effectively as an erasable molecular imaging probe.
In situ kinetics of strand displacement reactions
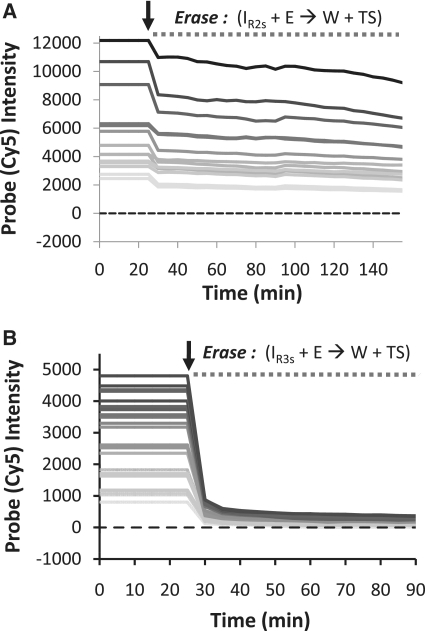
We next characterized the in situ marker labeling and erasing kinetics of the PC2s and PC3s systems by monitoring the rates that probe (Cy5) signals colocalized to, and then are removed from, expressed GFP molecules that are labeled with single TS strands via a DNA-conjugated protein polymer (Figure 3). For these analyses, cells were imaged every 5 min during the marker labeling/erasing procedures. Analyses of probe intensities show that TS-tagged GFP proteins are labeled rapidly by the two-strand probes, and that probe signals saturate within 20 min, even for the cells possessing the highest GFP expression levels. The PC3s labeling reactions were somewhat slower than the PC2s reactions, indicating that the release of the output strand from domain 3* in IR3s and the accumulation of 2 instead of 6 bp during this forward reaction affects the rates that markers are labeled. Nevertheless, Cy5 intensities reached their plateau levels in <1 h, except for the very brightest cells within the sample, and are linearly correlated with the GFP intensities on a pixel-by-pixel basis, as was the case in our prior report (13). Thus, so long as the PC3s probes are allowed to react for a sufficient period of time, both probe complexes can support efficient (relatively fast and proportionally correlative) marker labeling. Yet, Figure 3 also shows there are significant differences in the erasing kinetics of these systems. Although Cy5 fluorescence intensities decrease rapidly after the initial addition of eraser strands (E) in both cases, the PC2s system erases more slowly than the PC3s system. In addition, and, more importantly, the PC2s erasing reaction slows appreciably after a period of ∼20 min, and significant Cy5 signals remain on the sample after the full 2-h incubation period. In contrast, fluorescence intensities drop rapidly to a value that is <5–10% of their ON-state values within minutes during the reaction of E with three-strand IR3s complex.
Figure 3.
Kinetics of DNA strand displacement reactions on fixed cells. (A) Labeling and erasing reactions of a two-strand probe complex (PC2s). Each curve represent the average intensity of an individual cell within the sample. The erasing reactions are inefficient and significant signals remain on the sample even after a 2-h incubation period. (B) Labeling and erasing reactions of a three-strand probe complex (PC3s) showing rapid and efficient erasing. The arrows in each plot indicate the time point where the labeling reactions were stopped and the erasing reactions were initiated.
To gain further insight into the difference between the erasing behaviors of the PC2s and PC3s probes, we repeated the in situ kinetics experiments using eraser strands that do not contain quencher molecules (Figure 4A and B). With this modification, duplexed IR, W complexes, and other unidentified intermediates should all produce fluorescence signals if they remain bound to their targets or trapped within the cells during the erasing reaction. As shown in Figure 4B, the kinetic curves measured for unquenched three-strand erasing reaction (IR3s + E → W + TS + Output B) are very similar to the original plots in Figure 3B. We therefore conclude that W is able to release from the 2* toehold domain within TS during the erasing reaction. The lack of persistent fluorescence in this setting rules out the possibility that the dye molecule remains tethered to the GFP target but is quenched by an interaction with E (as depicted in a complex in Supplementary Figure S1). The rapid drop of Cy5 signals during the erasing reaction in the three-strand reporter complex setting suggests that the released W complex can freely diffuse out of fixed cells. In contrast, removing the quencher from reactions of two-strand reporter complexes IR2s with E results in appreciable remnant Cy5 signals on GFP transfected cells due to incomplete erasing (Figure 4A). In fact, the residual Cy5 signal remaining on the sample during the erasing reaction (‘OFF-state’) is nearly 75% of that seen for in the ‘ON-state’.
Figure 4.
Erasing kinetics using eraser strands (E) that do not incorporate quencher molecules. (A) Erasing reactions of a two-strand complex (PC2s) showing appreciable (∼75%) residual signals. (B) Erasing reactions of a three-strand complex PC3s.
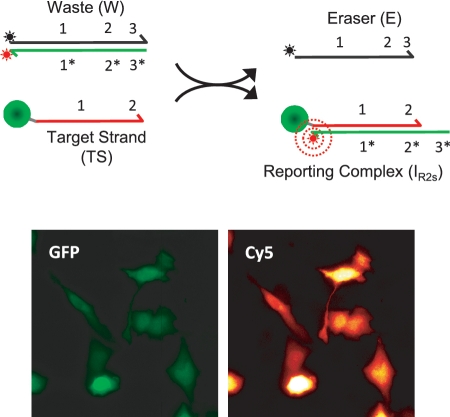
The in situ kinetic analyses of the PC2sC erasing reactions with and without quencher in Figures 3A and 4 A indicate that, while significant numbers of IR2s reporters remain on the cells after 2-h incubations, fluorescently active W complexes are also somehow trapped within the cells. Because TS does not release from the waste complex via toehold dehybridization in this reaction (TS is displaced completely from the complex), we attribute that majority of the signal intensities present in Figure 3A to fluorescently active W complexes as opposed to an unidentified, metastable intermediate-state complex. Importantly, we also find that TS-labeled GFP proteins can be selectively labeled using in situ reactions where the cells are simply incubated with a quenched W complex (Figure 5). This would suggest that the W complex can directly bind to the TS strand and become fluorescently activated through a non-toehold mediated exchange of oligonucleotides. Thus, it appears that in the two-strand reporter setting (but not in the three-strand reporter setting), there may be some reverse (relabeling) reactions occurring during the erasing reaction resulting in a decreased diffusion rate of the W complex from the cell.
Figure 5.
Labeling of DNA-tagged GFP proteins through non-toehold mediated exchange via a reaction of TS with a quenched W complex as a probe. The concentration of the W complex was 200 nM, and the reaction was performed without adding E to the reaction mixture.
Four-way branch migration reactions facilitate efficient signal erasing
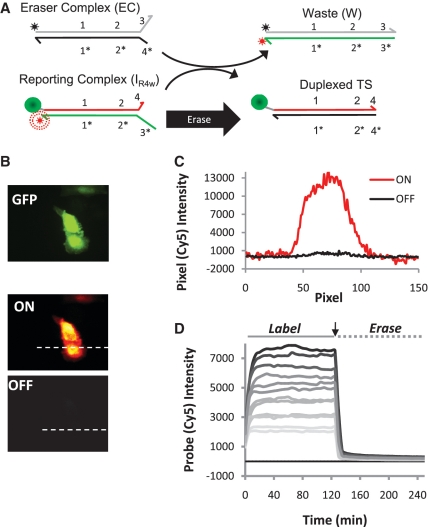
With the hypothesis that non-toehold-mediated exchange of oligonucleotides between W and TS complexes reduces the erasing performance of the PC2s probes systems, we next tested whether a fully erasable probe composed of only two different strands could be created that exchange nucleotides via four-way (23) as opposed to three-way strand displacement mechanisms. To do so, the original two-strand complexes were modified such that they still bind the same TS sequence, but leave a second 2-bp toehold unhybridized within the IR complex (domain 4 in Figure 6A). As a result, while the labeling reaction proceeds near-identically to that of the original PC2s systems, the four-way branch migration processes of the erasing reaction now produces two reaction products that are fully duplexed (i.e. TS is incorporated into a duplexed complex after the reaction). In this case, probe erasing rates and efficiencies are quite similar to those of the three-strand complexes (Figure 6D). Measured ‘OFF-state’ intensities and reaction rates are now very close to those of the PC3s system (Figure 6B and C). Thus, four-way branch migration reactions can be used as an alternative to the multi-strand release in the PC3s systems in order to create more efficient erasable probes.
Figure 6.
Labeling and removal of Cy5 fluorophores from protein markers using the two-strand probe complex that exchanges via a four-way branch migration process. (A) A scheme depicting the modified erasing reaction. (B) Selective labeling and erasing of expressed GFP proteins in CHO cells. (C) Pixel intensities for cross sections indicated in (B). (D) Kinetics of DNA strand displacement reactions on fixed cells showing that four-way branch migrations facilitate efficient marker erasing.
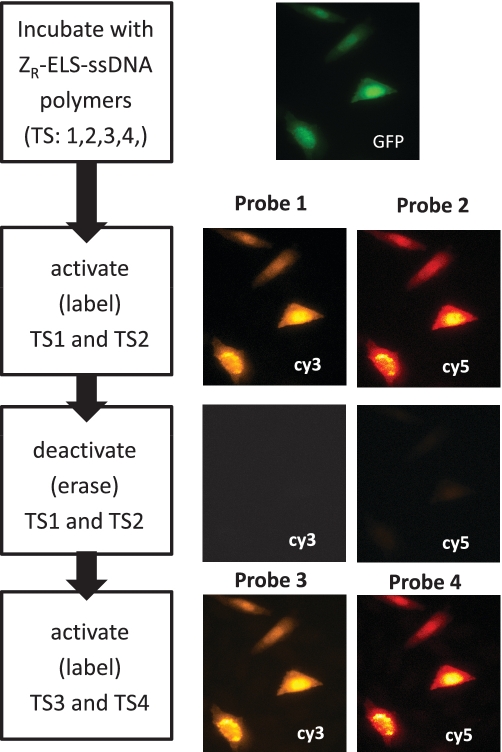
To further evaluate the utility of the four-way branch migrations for marker erasing, we also labeled expressed GFP-ZE with mixture of four different DNA conjugates in a single incubation step at approximately equal molar concentrations (Figure 7). The cells were then reacted simultaneously with two probes that couple Cy3 and Cy5 dye molecules to the GFP targets bearing the TS1 and TS2 strands. In a single incubation step, these two dyes can be removed from the sample using the four-way displacement mechanism, yielding average ON/OFF ratios that are >10:1. Subsequently, two new complexes can be used to label GFP molecules coupled to the TS3 and TS4 targets in a second round of marker labeling. After this step, the probe Cy3 and Cy5 signals reappear at levels that are similar to those produced in the first labeling step. The second labeling produced Cy3 and Cy5 signals were 60% and 120% of their corresponding ON-state values (note: the dyes are now coupled to different TS strands on the sample; some of this variability can come from differences in the concentrations of the conjugated polymers used to couple the four different TS strands to the GFP target). Importantly, ‘OFF’-state signals of the cells only constitute 2–6% of the measured ‘ON’-state signals produced by the second labeling reaction, and, hence, contribute little to the signals generated after the second set of TS markers were labeled. These experiments therefore indicate the four-way displacement mechanism can be used to create multiple erasable probes that can be used to label multiple sets of markers with the same color dyes, or even to perform replicate measurements using different probes for standardization purposes.
Figure 7.
Multiplexed (multi-color) and reiterative (sequential) labeling of four different TS strands coupled to the GFP-ZE targets at equimolar ratios. The scheme at the left depicts the labeling steps for each set of probe images.
DISCUSSION
The ability to control the exchange of nucleotides between different dynamic DNA complexes offers opportunities to create new classes of molecular-cell detection and imaging probes that are reconfigurable, adaptive, and that can perform complex logic functions. However, realizing this potential requires more detailed characterization of strand displacement reactions within cellular environments. Herein, we compared the efficiencies and rates that different dynamic DNA complexes can exchange strands within fixed cell samples as they undergo chemical reactions that either selectively couple (label) or remove (erase) oligonucleotides bearing fluorescent dye molecules to and from DNA-tagged proteins. One of the goals of this study is to define simple probe designs containing a minimal number of component strands that can function effectively as erasable molecular imaging probes. Since this capability allows fluorophores to be removed from a sample and then the same color dyes to be reused to label new proteins, this functionality should facilitate more comprehensive biomarker analyses where multiple sets of proteins are visualized on the same cell or tissue samples. Due to the spectral overlap of fluorescent dye molecules, the number of markers that can be visualized using fluorescence microscopy and permanently stained samples is typically limited to three to five proteins. However, this number can be at least doubled using our reiterative labeling and erasing procedure, and hence, one could detect at least 6–10 proteins using standard, three to five color fluorescence microscopy procedures, while avoiding the use of temperature, bleaching, or caustic chemicals to remove dyes from sample. Moreover, several spectral deconvolution microscopy approaches have been developed that offer opportunities to increase the number of markers that are detected during a single round of fluorescence microscopy. Combining this capability with our reiterative labeling techniques could therefore facilitate even more extensive in situ analyses of protein markers.
In the course of optimizing our probe designs, we found that dyes cannot be removed efficiently from their targets using three-way displacement reactions between relatively simple two-strand duplexes and ssDNA targets. The mechanistic basis for this poor response is revealed in the kinetic analyses of our probes’ erasing reaction. Erasing reactions that incorporate quenchers in E show that significant numbers of reporters complexes (IR2s) remain on the cells after a 2-h reaction; it is our experience that >12-h incubations are necessary to achieve significantly lower fluorescent levels. Further experiments where the quencher is removed from E suggests that the W complexes are kinetically trapped within the cells’ volume and do not diffuse beyond the cell boundary within this time period. This behavior is somewhat surprising given the high E concentrations (1 µM) and reaction volume (100 µl) used for the erasing experiments. With such an excess of E relative to the total number of IR2s reporting complexes on the cells, there should be a strong driving force to push the erasing reaction forward. However, given the compartmentalization of the GFP targets within the cells, the local concentration of TS and W can be quite high: 1400–15 000 GFP/µm3 or 25–250 µM according to GFP intensity analyses. Thus, high local target concentrations could serve to drive the reverse (relabeling) reaction (W + TS → IR2s + E). Of note, the analogous reverse reaction (IR2s + output A → TS + PC2s) could be affecting the marker labeling step. Yet, the probe labeling intensities appear to saturate rapidly (<30 min) and still yield marker intensities that are linearly correlated with GFP levels.
Interestingly, the failure of the two-strand probes to erase efficiently occurs even though the W complex of the two-strand probes (PC2s) do not contain a toehold, implying the reverse (relabeling) reactions occur via non-toehold mediated exchange process. Indeed, experiments involving reactions of quenched W also produce fluorescent signals that colocalize with the TS-tagged GFP molecules, confirming non-toehold-mediated exchange can occur. Furthermore, reactions between ssDNA and duplexed complexes have been shown to be enhanced when molecular crowding agents are present in solution (17). Thus, it is possible that, in addition to the local confinement of targets, proteins and other macromolecules present within the cells (e.g. blocking reagents) act similarly as crowding agents that further accelerate the rates of non-toehold-mediated strand exchange.
An important consequence of non-toehold-mediated exchange reactions is that they can influence the effective diffusive mobilities of oligonucleotides and DNA complexes within cell samples. Through this reaction, strands containing the fluorophores will interact transiently with multiple immobilized TS strands as they diffuse towards the cell boundary. These interactions can therefore lower the rates at which the fluorophore containing strands are released from the sample. While this effect limits removal of dyes from cell samples during our marker erasing procedure, analogous reverse and side reactions could potentially influence abilities to integrate the reactions between different dynamic DNA complexes for other biological detection applications. Nevertheless, the analyses of the three-strand probe complexes (PC3s) indicate these issues can be mitigated using probe designs where the reaction of a probe with its DNA target is entropically favorable and two output strands are produced by the reaction. In the case of the three-strand probe complexes, both outputs must bind to the W complex simultaneously in order to produce a fluorescently active complex that is stably bound to TS. As a result, W complexes are less likely to reassociate with TS and can more readily diffuse out of the cells. Alternatively, efficient marker erasing can be achieved using probes that react via four-way branch migration processes. This mechanism produces two fully duplexed waste products instead of only one, and hence, both products are much less likely to exchange their strands, but also, should be less reactive towards other oligonucleotide complexes present within cells. While such control has allowed our group to create a series of erasable imaging probes that facilitate the reiterative labeling of cell samples, overall, we anticipate these adaptations will be generically important to the development of other probe technologies that harness the unique functionalities of dynamic DNA complexes.
SUPPLEMENTARY DATA
Supplementary Data including are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figure 1 and Supplementary References [24–26].
FUNDING
National Institute of Health (Grant 1R21CA147912) and Nanobiology Interdisciplinary Graduate Training Program of the W. M. Keck Center for Interdisciplinary Bioscience Training of the Gulf Coast Consortia (NIH Grant No. T32 EB009379 to R.M.S.). Funding for open access charge: NIJ NCI.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Paul Rothemund and David Zhang for helpful discussions.
REFERENCES
- 1.Stojanovic MN, Mitchell TE, Stefanovic D. Deoxyribozyme-based logic gates. J. Am. Chem. Soc. 2002;124:3555–3561. doi: 10.1021/ja016756v. [DOI] [PubMed] [Google Scholar]
- 2.Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-free nucleic acid logic circuits. Science. 2006;314:1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 3.Hagiya M, Yaegashi S, Takahashi K. Computing with hairpins and secondary structures of DNA. Nanotechnol. Science Comput. 2006:293–308. [Google Scholar]
- 4.Dirks RM, Pierce NA. Triggered amplification by hybridization chain reaction. Proc. Natl Acad. Sci. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lubrich D, Green SJ, Turberfield AJ. Kinetically controlled self-assembly of DNA oligomers. J. Am. Chem. Soc. 2009;131:2422–2423. doi: 10.1021/ja807765v. [DOI] [PubMed] [Google Scholar]
- 6.Goodman RP, Heilemann M, Doose S, Erben CM, Kapanidis AN, Turberfield AJ. Roconfigurable, braced, three-dimensional DNA nanostructures. Nat. Nanotechnol. 2008;3:93–96. doi: 10.1038/nnano.2008.3. [DOI] [PubMed] [Google Scholar]
- 7.Zhang DY, Seelig G. Dynamic DNA nanotechnology using strand displacement reactions. Nat. Chem. 2011;3:103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 8.Zhang DY. Cooperative hybridization of oligonucleotides. J. Am. Chem. Soc. 2011;133:1077–1086. doi: 10.1021/ja109089q. [DOI] [PubMed] [Google Scholar]
- 9.Zhang DY, Winfree E. Robustness and modularity properties of a non-covalent DNA catalytic reaction. Nucleic Acids Res. 2010;38:4182–4197. doi: 10.1093/nar/gkq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian L, Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 11.Yurke B, Mills AP, Cheng SL. DNA implementation of addition in which the input strands are separate from the operator strands. Biosystems. 1999;52:165–174. doi: 10.1016/s0303-2647(99)00043-x. [DOI] [PubMed] [Google Scholar]
- 12.Benenson Y, Gil B, Ben-Dor U, Adar R, Shapiro E. An autonomous molecular computer for logical control of gene expression. Nature. 2004;429:423–429. doi: 10.1038/nature02551. [DOI] [PubMed] [Google Scholar]
- 13.Duose DY, Schweller RM, Hittelman WN, Diehl MR. Multiplexed and reiterative fluorescence labeling via DNA circuitry. Bioconjug. Chem. 2010;21:2327–2331. doi: 10.1021/bc100348q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkataraman S, Dirks RM, Ueda CT, Pierce NA. Selective cell death mediated by small conditional RNAs. Proc. Natl Acad. Sci. 2010;107:16777–16782. doi: 10.1073/pnas.1006377107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Choi HMT, Chang JY, Trinh LA, Padilla JE, Fraser SE, Pierce NA. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 2010;28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang DY, Winfree E. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 2009;131:17303–17314. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]
- 17.Feng B, Frykholm K, Norde B, Westerlund F. DNA strand exchange catalyzed by molecular crowding in PEG solutions. Chem. Commun. 2010;46:8231–8233. doi: 10.1039/c0cc03117h. [DOI] [PubMed] [Google Scholar]
- 18.Moll JR, Ruvinov SB, Pastan I, Vinson C. Designed heterodimerizing leucine zippers with a range of pIs and stabilities up to 10(-15) M. Protein Sci. 2001;10:649–655. doi: 10.1110/ps.39401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schweller RM, Constantinou PE, Frankel NW, Narayan P, Diehl MR. Design of DNA-conjugated polypeptide-based capture probes for the anchoring of proteins to DNA matrices. Bioconjug. Chem. 2008;19:2304–2307. doi: 10.1021/bc8003606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang DY, Turberfield AJ, Yurke B, Winfree E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science. 2007;318:1121–1125. doi: 10.1126/science.1148532. [DOI] [PubMed] [Google Scholar]
- 21.Rouillard JM, Zuker M, Gulari E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003;31:3057–3062. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lilley DMJ. Structures of helical junctions in nucleic acids. Q. Rev. Biophys. 2000;33:109–159. doi: 10.1017/s0033583500003590. [DOI] [PubMed] [Google Scholar]
- 23.Karymov M, Douglas D, Sankey OF, Lyubchenko YL. Holliday junction dynamics and branch migration: single-molecule analysis. Proc. Natl Acad. Sci. 2005;102:8186–8191. doi: 10.1073/pnas.0407210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 25.Dirks RM, Bois JS, Schaeffer JM, Winfree E, Pierce NA. Thermodynamic analysis of interacting nucleic acid strands. SIAM Rev. 2007;49:65–88. [Google Scholar]
- 26.Friedman LJ, Chung J, Gelles J. Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence. Biophys. J. 2006;91:1023–1031. doi: 10.1529/biophysj.106.084004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.