Abstract
The DNA-dependent protein kinase (DNA-PK) was identified as an activity and as its three component polypeptides 25 and 15 years ago, respectively. It has been exhaustively characterized as being absolutely dependent on free double stranded DNA ends (to which it is directed by its regulatory subunit, Ku) for its activation as a robust nuclear serine/threonine protein kinase. Here, we report the unexpected finding of robust DNA-PKcs activation by N-terminal constraint, independent of either DNA or its regulatory subunit Ku. These data suggest that an N-terminal conformational change (likely induced by DNA binding) induces enzymatic activation.
INTRODUCTION
The DNA-dependent protein kinase (DNA-PK), composed of its regulatory subunit (the DNA end binding factor Ku) and the large catalytic subunit (DNA-PKcs) initiates the process of ‘classical’ non-homologous end joining (c-NHEJ) when Ku detects and binds free DNA ends to which DNA-PKcs then targets (1–3). Assembly of a Ku molecule and a DNA-PKcs molecule onto a single DNA end is termed DNA-PK. Synapses of two DNA-PK complexes provides the platform for repair which requires appropriate assembly of c-NHEJ factors (XLF, XRCC4, ligase IV, Artemis) accessory factors (pol µ, pol λ, PNK), and a growing list of ‘gluing’ factors and their activators (53BP1, H2AX, ATM, MRN) that likely stabilize synapsis of the two broken ends and facilitate repair (4,5). Recent reports also implicate XRCC4/XLF filaments in bridging and stabilizing synapsis of the DNA ends (S. N. Andres et al., submitted for publication and S. Roy et al., submitted for publication).
In physiologic salt conditions, DNA-PKcs has no (or very low) affinity for DNA ends. However, in low salt conditions, DNA-PKcs binds and is weakly activated by double stranded DNA ends (6,7). These data suggest a model whereby DNA binding by DNA-PKcs (facilitated by Ku) results in activation of DNA-PK's robust catalytic activity. Once activated, DNA-PK phosphorylates a plethora of targets including many of the other factors in the c-NHEJ pathway, including itself. Although most c-NHEJ factors are excellent in vitro and in vivo targets of DNA-PK's enzymatic activity, DNA-PKcs itself is the only c-NHEJ factor that has been shown (to date) to be a functionally relevant (for c-NHEJ) target of its own enzymatic activity (8–14). It has been well established that autophosphorylation on DNA-PK's catalytic subunit results in dissociation of DNA-PKcs from the Ku/DNA end complex (8,15), and considerable effort has been put forth to define phosphorylations that mediate kinase dissociation. Our previous studies have shown that two autophosphorylation site clusters (‘ABCDE’ and ‘PQR’, located roughly in the middle of the DNA-PKcs polypeptide) reciprocally regulate end access during c-NHEJ. Although phosphorylation within these two clusters regulates end processing, additional phosphorylations are required to facilitate dissociation of DNA-PKcs from Ku-bound DNA (9). An additional phosphorylation site within the putative activation loop in the C terminal kinase domain regulates kinase activity but not assembly (10).
Several recent studies ascribe an important role in kinase activation to the extreme N-terminus of DNA-PKcs. We have previously identified two regions within the N-terminus of DNA-PKcs as being critical for DNA binding: the leucine rich region (LRR) and the N-terminal 426 residues (10,16). Moreover, recent structural studies similarly suggest that the extreme N-terminus may be important for DNA-PKcs interactions with DNA (17,18). Finally, we have recently characterized four additional phosphorylation sites within the N-terminus of DNA-PKcs that negatively regulate c-NHEJ (19). Two of these sites are located at the extreme N-terminus (S56, S72); phospho-mimicking these sites severely impairs kinase activation, explaining the negative impact on c-NHEJ; DNA binding is not impaired by the phospho-mimicking mutations.
In 1998, Chu and colleagues (20) reported that DNA-PKcs mediates synapsis of DNA ends and that activation of DNA-PK's enzymatic activity was cooperative, leading these authors to propose a model whereby DNA bound by one DNA-PK complex activated the synapsed opposing complex in trans. We have previously demonstrated that autophosphorylation within the two major clusters (ABCDE and PQR) can occur in trans both in vitro and in living cells, although these experiments in no way preclude cis autophosphorylations (i.e. intra-molecular autophosphorylation) at these sites (or at other phosphorylation sites) occurring as well (21). Moreover, DNA-PKcs is phosphorylated in living cells at many additional sites [likely more than 40, (18)]; it seems quite likely that autophosphorylation of some sites may occur in cis whereas others occur in trans. In fact, recent studies from Lieber and colleagues (22) as well as Dynan and colleagues (23) using immobilized antibodies or biotin labeled DNA immobilized by streptavidin (respectively) are more consistent with a model whereby DNA-PK is activated (and autophosphorylated) in cis. Although in both studies, care was taken to make sure that DNA-PKcs was not saturating, with studies using antibodies, protein G or streptavidin to immobilize either DNA or DNA-PK, there is the caveat that multiple DNA-PK molecules may be co-localized because of the valence of antibodies, protein G or streptavidin. In sum, there is some discordance in the published literature as to whether DNA-PK activation results from an interaction of a single DNA-PK complex with the end of the DNA to which it is bound, or whether activation of one DNA-PK complex requires synaptic interaction with the DNA end bound by a second DNA-PK complex.
In an attempt to address questions of cis as opposed to trans activation, a variety of tagged DNA-PKcs expression plasmids were constructed. Although histidine tags affect the function of DNA-PKcs (whether placed at the C terminus, N-terminus or at a position internally), N-terminal GFP and FLAG tagged human DNA-PKcs fully complement the radiosensitivity of the DNA-PKcs deficient V3 CHO cell strain. Here we exploit the mono-valent nature of camelid antibodies (24), to immobilize DNA-PKcs onto agarose beads. We report the unexpected finding that N-terminal constraint of DNA-PKcs (in the absence of Ku or DNA) results in robust kinase activation. These data suggest a model whereby an N-terminal conformational change (likely induced by DNA binding) activates DNA-PK. As discussed earlier, we have recently shown that phospho-mimicking (by mutation) two extremely well conserved S/hydrophobic sites at the extreme N-terminus of the large DNA-PKcs polypeptide severely impedes kinase activation (19). Thus, we suggest that conformational changes at the extreme N-terminus may explain how DNA-PKcs ‘senses’ DNA and that phosphorylations within this region regulate kinase activation.
METHODS
Cell culture, expression plasmids and transfectants
Methods to derive V3 transfectants have been described previously (25). Briefly, cells were cultured in α-MEM with 10% fetal calf serum, penicillin/streptomycin, and cipro (complete medium) and maintained at 37°C with 5% CO2. N-terminal FLAG tagged and GFP tagged DNA-PKcs (wild-type) expression plasmids were described previously (21,26).
Clonogenic survival assays
For zeocin sensitivity, V3 transfectants were cultured in the indicated dosages of zeocin. After 7 days, cell colonies were stained with 1% (w/v) crystal violet in ethanol, and colony numbers were assessed. Survival was plotted as percentage survival compared to untreated cells.
Immunoprecipitations
Cell pellets (from a confluent 150 mm2 dish) were lysed in 400 µl of the following IP buffer: [150 mM NaCl, 50 mM Tris, pH 7.5, 1% NP40, 1 mM NaFl, 1 mM Na3VO4, 5% glycerol with protease inhibitor tablet added (Pearce, Rockford, IL, USA)] on ice for 10 min, and subsequently spun in a microcentrifuge at maximum speed for 10 min. Resulting lysate was quantified and diluted 1:4 with IP dilution buffer: (150 mM NaCl, 50 mM Tris, pH 7.5) and ethidium bromide added to a final concentration of 50 µg/ml. Diluted lysate (1600 µl) was incubated with 50 µl anti-GFP agarose beads (GFP-trap, Allele Biotechnology, San Diego, CA, USA) rocking, for 1 h at 4°C. Agarose beads were washed three times in IP dilution buffer with protease inhibitors, and then three times in Buffer A: (25 mM HEPES, pH 7.5, 50 mM KCl, 10 mM MgCl2, 10% glycerol, 1 mM EDTA, 1 mM EGTA) with protease inhibitors. Alternatively (Figure 2B), immunoprecipitations were performed in 50 mM HEPES (pH 7.5), 150 mM NaCl, 0.1% Triton X-100, 5 mM MnCl2, 50 mM NaFl, 50 µg/ml ethidium bromide and protease inhibitors. This Triton-containing buffer resulted in more efficient pull-downs. Beads were washed three times in this buffer, and then three times in Buffer A (suitable for kinase reactions) as described earlier. Washed beads were divided into aliquots (generally nine) during the final wash for IP kinase assays.
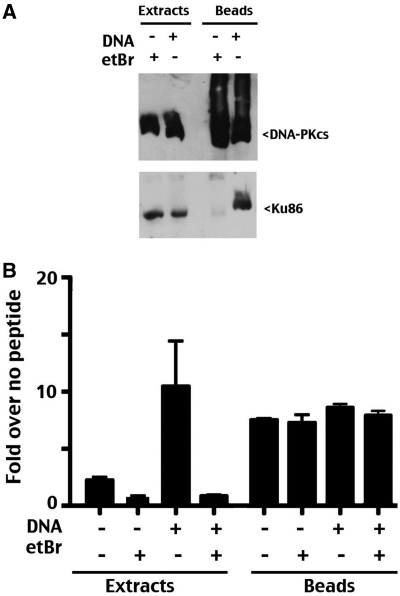
Figure 2.
DNA-independent activation of N-terminally constrained DNA-PKcs. (A) Whole cell extracts or anti-GFP immunoprecipitates in the presence or absence of DNA (20 µg/ml) or ethidium bromide (50 µg/ml) as indicated from V3 transfectants expressing GFP-tagged wild-type DNA-PKcs were analyzed by immunoblotting using anti-DNA-PKcs or anti-Ku antibodies as indicated. (B) DNA-PK enzymatic levels were determined in extracts or anti-GFP immunoprecipitates from V3 transfectants expressing GFP-tagged wild-type DNA-PKcs in the presence or absence of DNA (20 µg/ml), or ethidium bromide 50 µg/ml). Relative activity is depicted as fold-increase over no peptide as described in the ‘Methods’ section. Error bars depict SDs of triplicate assays. This assay is representative of no less than three independent assays.
In some experiments, anti-GFP agarose beads were produced by first synthesizing the cDNA encoding a published anti-GFP domain (27) by multiplex PCR using Phusion DNA polymerase and synthetic oligonucleotides (DNA sequences available upon request). The resulting PCR product was subcloned into pCRblunt to generate pCRblunt-GFPbinder, and then subcloned into pET28a (pET28a-GFP binder). Recombinant camel anti-GFP was expressed in E. coli and purified by nickel agarose affinity, and subsequently covalently coupled to NHS activated agarose (Thermo Scientific, Rockford, IL, USA) according to the manufacturers recommended protocol.
DNA-PK kinase assays
The DNA-PK enzymatic assay is an adaptation of the assay originally described by Finnie et al. (28), modified by Kienker et al. (29), utilizing a commercially available DNA-PK assay kit (Promega, Madison, WI, USA). After extensive washing, aliquots of immunoprecipitates were incubated in kinase reaction buffer [50 mM HEPES (pH 7.4), 100 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 0.2 mM EGTA, 1 mM DTT; 0.1 mM ATP, 80 µCi/ml γ−32P-ATP and with or without 20 µg/ml calf thymus DNA, 0.8 mM biotinylated peptide substrate, 50 µg/ml ethidium bromide as indicated]. For extract kinase reactions, (freeze/thaw) extracts prepared by the method described by Finnie et al. (28) were prepared in the following buffer (50 mM NaF, 20 mM HEPES pH 7.8, 450 mM NaCl, 25% glycerol, 0.2 mM EDTA); 100 µg extract was utilized/reaction. Kinase reactions (25 µl) were incubated for 1 h at room temperature and the reaction stopped by the addition of 10 µl 7.5 M guanidine hydrochloride. Ten microliter of each reaction was spotted onto SAM2 membrane (avidin impregnated membrane; Promega, Madison, WI, USA); washed extensively in 2 M NaCl, and then 2 M NaCl in 1% HCPO4. Dried membranes were subjected to scintillation counting. DNA-PK activity is expressed as fold increase over kinase reactions with no added peptide (determined for each extract or IP triplicate). All assays were performed in triplicate. Experiments presented are representative of no less than three independent experiments. To detect autophosphorylated DNA-PKcs, kinase reactions were stopped by the addition of SDS–PAGE loading buffer; kinase reactions were electrophoresed, gels dried and subjected to phosphorimaging. To detect phosphorylation of p53, 2 µg purified recombinant p53 (generous gift Bill Henry, Michigan State University) was added to kinase reactions. Kinase reactions were analyzed by SDS–PAGE as described earlier.
Immunoblotting
Cell extracts or immunoprecipitated fractions were analyzed after electrophoresis on 4.5% SDS polyacrylamide gels and transferred to PVDF membranes. To detect total DNA-PKcs, a monoclonal antibody raised against DNA-PKcs (42-27, a generous gift of Dr Tim Carter, St Johns University, New York, NY, USA) was used as the primary antibody. Phospho-specific antibodies that recognize phosphorylated S2056 (‘R’ site) or T2609 (‘A’ site), (Abcam; Cambridge, MA, USA) were utilized to detect specific phosphorylations.
RESULTS
DNA-independent activation of N-terminally constrained DNA-PKcs
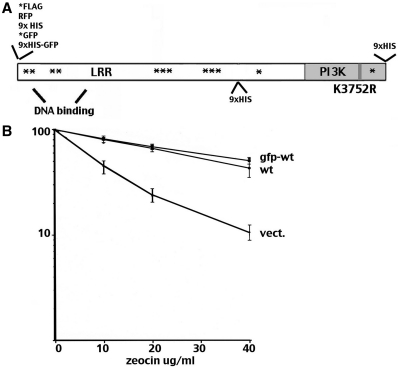
Several strategies were tested for tagging full-length human DNA-PKcs (Figure 1A) including N-terminal GFP, FLAG, RFP and HIS as well as C-terminal and internal HIS tags. Of the seven strategies employed, only N-terminal GFP and FLAG tagged constructs were able to completely reverse the radiosensitive phenotype of the DNA-PKcs deficient cell strain V3 as tested by zeocin sensitivity (a bleomycin analogue that induces double strand breaks) resistance (Figure 1B); those constructs were chosen for further study.
Figure 1.
N-terminally GFP-tagged DNA-PKcs reverses V3's radiosensitive phenotype. (A) Diagram of DNA-PKcs coding sequence showing position of HIS, GFP, FLAG and RFP tags positioned in this study. Asterisks denote positions of previously studied phosphorylation sites. (B) V3 transfectants expressing either wild-type DNA-PKcs, or the GFP-tagged wild-type DNA-PKcs, or no DNA-PKcs were plated into complete media with increasing doses of zeocin as indicated at cloning densities. Colonies were stained after 7 days later and percent survival was calculated. Error bars represent SEM.
A commercially available (Allele Biotech, San Diego, CA, USA) agarose immobilized camelid antibody was utilized in immunoprecipitation assays. GFP pull-downs are remarkably efficient using this reagent. In some experiments, immobilized anti-GFP agarose was produced by direct coupling of recombinant camel anti-GFP to NHS agarose as described in the ‘Methods’ section. Immunoprecipitations were performed either in the presence of DNA (20 µg/ml calf thymus DNA) or ethidium bromide (50 µg/ml) to enhance or block interaction of DNA-PKcs with Ku bound DNA as has been reported previously (3,30). As can be seen (Figure 2A), GFP-tagged wild-type DNA-PKcs is efficiently immunoprecipitated in both conditions. Consistent with previous studies, DNA-PKcs only co-immunoprecipitates with Ku in the presence of DNA (3,30).
Next, DNA-dependent phosphorylation of a biotinylated p53 peptide spanning its well studied N-terminal SQ DNA-PK target site (S15) was assessed in whole V3 cell extracts or in immunoprecipitates performed in the presence of 50 µg/ml ethidium bromide (immunoprecipitates were washed extensively to remove ethidium bromide prior to enzymatic assays). The kinase assay utilized is a modification of the DNA cellulose ‘pull-down’ assay described previously (29). In this assay, extracts or immunoprecipitates are incubated in the presence or absence of a biotinylated p53 peptide spanning the S15 phosphorylation site. The phosphorylated peptide is then captured onto a biotin-coated membrane; after extensive washing, bound phosphorylated peptide is assessed by scintillation counting. Enzymatic activity is expressed as fold increase over no peptide control reactions (performed for each extract or immunoprecipitate). As expected, phosphorylation of the p53 peptide when measured in extracts is highly dependent on DNA. In the extract kinase assays, addition of ethidium bromide (which disrupts Ku-DNA interaction) completely ablates phosphorylation in the presence of DNA. This demonstrates that the concentration of ethidium bromide utilized here and in all subsequent experiments is sufficient to completely abrogate kinase activation. Further, this level of ethidium bromide also suppresses the minimal phosphorylation observed in extracts without additional DNA added (this phosphorylation is presumably the result of DNA contamination in whole cell extracts). The GFP immunoprecipitates efficiently phosphorylated the p53 peptide, but to our surprise, the addition of DNA, ethidium bromide or recombinant Ku plus DNA (not shown) had no affect on the robust peptide phosphorylation observed in the anti-GFP pull-down fractions.
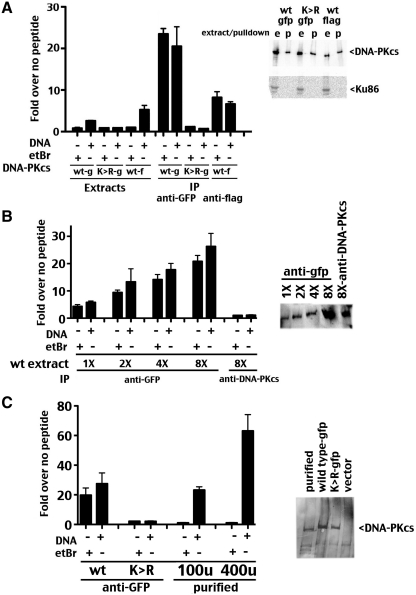
Since DNA-independent enzymatic activity of DNA-PK has not been reported in physiologic kinase conditions, we considered that a contaminating kinase with activity towards the p53 peptide was responsible for the observed activity. Similar paired extract and pull-down kinase assays were performed with extracts derived from cells expressing GFP wild-type, GFP-kinase inactive and FLAG wild-type DNA-PKcs constructs (Figure 3A). We have described previously a Lys to Arg mutant at amino acid 3752 (K3752R) that was designed to disrupt ATP-binding (29,31), although the precise ATP-binding site has not been determined in DNA-PKcs. As expected, phosphorylation of the p53 peptide when measured in extracts containing either FLAG or GFP-tagged wild-type DNA-PKcs is highly dependent on DNA; further, no DNA-dependent phosphorylation was observed in extracts containing catalytically inactive GFP-tagged DNA-PKcs. However, phosphorylation of the p53 target peptide was observed in immunoprecipitated FLAG or GFP-tagged DNA-PKcs extracts. This activity was not affected by the addition of either DNA or ethidium bromide, and the activity was not observed in immunoprecipitates of GFP-tagged kinase inactive DNA-PKcs. Immunoblotting revealed similar levels of immunoprecipitated DNA-PKcs from each extract as well as the absence of Ku. Several additional IP kinase experiment were performed using the GFP reagent as well as a well-studied anti-DNA-PKcs monoclonal antibody (42-27, immobilized by protein G sepharose) that recognizes a determinant in the C-terminal 150 kD of the DNA-PKcs polypeptide. Since retrieval of DNA-PKcs with the anti-DNA-PKcs was not as efficient as with the anti-GFP reagent, a titration of different extract concentrations was immobilized with the anti-GFP reagent (Figure 3B). As can be seen, more DNA-PK activity is retrieved in anti-GFP pull-downs as extract concentration is increased. No significant DNA-PK activity was observed in pull down fractions using the anti-DNA-PKcs antibody, although retrieval of DNA-PKcs was similar to that retrieved using the anti-GFP reagent and approximately one-half as much extract. These data suggest that immobilizing DNA-PKcs via its N-terminus (by antibodies to either FLAG or GFP N-terminal tags) results in robust phosphorylation of the S15 p53 peptide target.
Figure 3.
DNA-independent activation of N-terminally constrained DNA-PKcs. (A, left panel) DNA-PK enzymatic levels were determined in extracts, anti-GFP immunoprecipitates or anti-FLAG immunoprecipitates from V3 transfectants expressing GFP-tagged wild-type DNA-PKcs or FLAG-tagged wild-type DNA-PKcs, or GFP-tagged catalytically inactive mutant DNA-PKcs, K3752R (as indicated) in the presence or absence of DNA (20 µg/ml), or ethidium bromide 50 µg/ml). Relative activity is depicted as fold-increase over no peptide as described in the ‘Methods’ section. Error bars depict SDs of triplicate assays. This assay is representative of no less than three independent assays. (A, right panel) Whole cell extracts (e), anti-GFP or anti-FLAG immunoprecipitates (p) from V3 transfectants expressing GFP-tagged or FLAG-tagged wild-type DNA-PKcs or GFP-tagged catalytically inactive mutant K3752R were analyzed by immunoblotting using anti-DNA-PKcs or anti-Ku antibodies as indicated. (B, left panel) DNA-PK enzymatic levels were determined in extracts, anti-GFP immunoprecipitates (1X = 2.5 mg extract. 2X = 5 mg extract. 4X = 10 mg extract. 8X = 20 mg extract) or anti-DNA-PKcs immunoprecipitates (20 mg extract, using mAB 42-27) from V3 transfectants expressing GFP-tagged wild-type DNA-PKcs in the presence or absence of DNA (20 µg/ml), or ethidium bromide 50 µg/ml). Relative activity is depicted as fold-increase over no peptide as described in the ‘Methods’ section. Error bars depict SDs of triplicate assays. This assay is representative of no less than three independent assays. (B, right panel) Immunoblotting of IP fractions probed with anti-DNA-PKcs antibody. (C, left panel) DNA-PK enzymatic levels were determined from anti-GFP immunoprecipitates, GFP-tagged catalytically inactive mutant DNA-PKcs, K3752R (as indicated), or 100 or 400 U purified DNA-PK in the presence or absence of DNA (20 µg/ml), or ethidium bromide 50 µg/ml). Relative activity is depicted as fold-increase over no peptide as described in the ‘Methods’ section. Error bars depict SDs of triplicate assays. This assay is representative of no less than three independent assays. (C, right panel) Anti-GFP immunoprecipitates from V3 transfectants expressing GFP-tagged wild-type DNA-PKcs, GFP-tagged catalytically inactive mutant K3752R, or vector alone were analyzed by silver staining and compared to 100 U purified DNA-PK.
In the titration experiment, (at higher concentrations of extract) DNA-PK activity did not double when extract concentration was doubled, and we considered that the peptide assay might be saturated. To test this possibility, relative DNA-PK activity of GFP-tagged DNA-PKcs immobilized on anti-GFP agarose was compared to increasing levels of a commercial preparation of purified human DNA-PKcs (Promega, Madison, WI, USA). As can be seen (Figure 3C), the amount of activity retrieved by anti-GFP IP of 20 mg of V3 transfectant extract is similar to the activity of 100 U of the commercial preparation, whereas substantially higher activity (∼3-fold more) is observed in assays utilizing 400 U.
Ku independent activation of N-terminally constrained DNA-PKcs
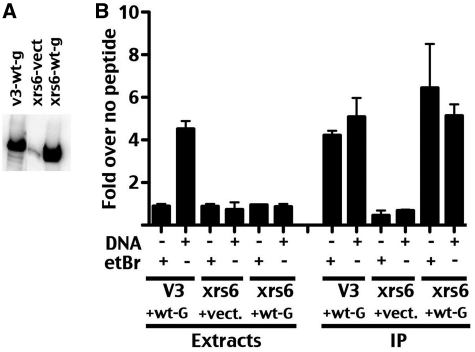
To further confirm that the observed enzymatic activity represented DNA and Ku independent phosphorylation by DNA-PKcs, GFP-tagged DNA-PKcs was expressed in xrs6 cells that lack Ku (Figure 4). As can be seen, stable transfection of the GFP-tagged wild-type DNA-PKcs expression vector in xrs6 cells markedly increases DNA-PKcs expression compared to the level of endogenous hamster DNA-PKcs (Figure 4A). Although DNA-dependent p53 phosphorylation is readily detected in V3 extracts over-expressing GFP-tagged human DNA-PKcs (as in Figure 2), no activity is detected in xrs6 extracts from cells over-expressing GFP-tagged human DNA-PKcs (since these cells lack Ku86). However, robust, DNA-independent activity is detected in GFP immunoprecipitates of both V3 and xrs6 cells expressing GFP-DNA-PKcs but not vector controls. These data are consistent with DNA and Ku independent activation of DNA-PKcs tethered by its N-terminus.
Figure 4.
Ku independent activation of N-terminally constrained DNA-PKcs. (A) Whole cell extracts from V3 or xrs-6 cells expressing wt-GFP tagged DNA-PKcs or vector only were analyzed by immunoblotting using anti-DNA-PKcs antibody, 42-27. (B) DNA-PK enzymatic levels were determined in extracts or anti-GFP immunoprecipitates from V3 transfectants expressing GFP-tagged wild-type DNA-PKcs or xrs6 transfectants expressing wilt type GFP-tagged wild-type DNA-PKcs or vector alone xrs6 transfectants (as indicated) in the presence or absence of DNA (20 µg/ml), or ethidium bromide 50 µg/ml). Relative activity is depicted as fold-increase over no peptide as described in the ‘Methods’ section. Error bars depict SDs of triplicate assays. This assay is representative of no less than three independent assays.
DNA-PKcs can be activated by N-terminal constraint in physiologic salt
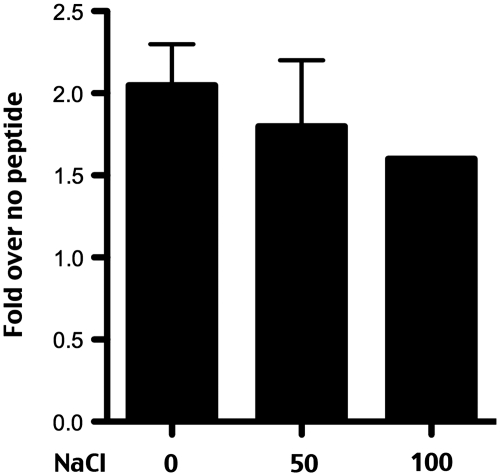
As discussed earlier, DNA-PKcs has no (or very low) affinity for DNA ends in physiologic salt conditions (6,7). However, in low salt conditions (less than ∼50 mM), DNA-PKcs binds and is weakly activated by double stranded DNA ends. Although immunoprecipitations are performed in buffer containing 150 mM NaCl, the buffer is exchanged to a standard kinase buffer during washing. The kinase buffer utilized includes 100 mM KCl, 50 mM HEPES (pH 7.5), 10 mM MgCl2, but lacks NaCl. We next determined whether DNA-PK activated by N-terminal constraint was salt labile. As can be seen (Figure 5), there is no significant reduction of kinase activity by the addition of 50 mM or 100 mM NaCl. Hammarstan and Chu (6) reported virtually no DNA-PKcs activity (without Ku) in 100 mM NaCl, and West et al. (7) reported a 3-fold reduction in activity in 100 mM compared to 50 mM NaCl. The modest reduction in enzymatic activity observed in Figure 5 does not approach the level of reduction observed by others for DNA-dependent activation of isolated DNA-PKcs (i.e. without Ku), and we conclude that the activity described here resembles authentic DNA-PK activity (i.e. that ascribed to DNA-PKcs + Ku) with regards to its salt lability.
Figure 5.
DNA-PKcs can be activated by N-terminal constraint in physiologic salt. DNA-PK enzymatic levels were determined in anti-GFP immunoprecipitates from V3 transfectants expressing GFP-tagged wild-type DNA-PKcs in the presence of DNA (20 µg/ml) and no salt, 50 mM NaCl, or 100 mM NaCl as indicated. Relative activity is depicted as fold-increase over no peptide as described in the ‘Methods’ section. Error bars depict SDs of triplicate assays. This assay is representative of no less than three independent assays.
DNA-independent autophosphorylation of N-terminally constrained DNA-PKcs
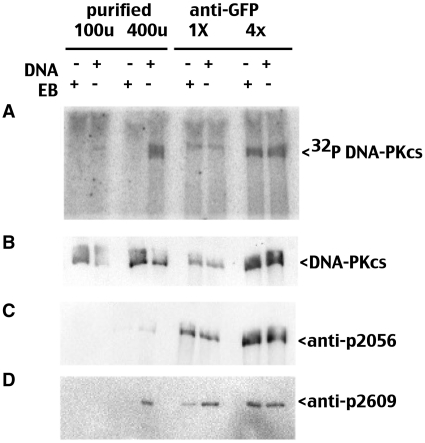
We next determined whether DNA-PKcs autophosphorylation occurs under these activation conditions. In an effort to match DNA-PKcs concentrations, in vitro kinase reactions were performed with different amounts of immobilized GFP-DNA-PKcs and different amount of the purified enzyme preparation; kinase assays were electrophoresed on 4.5% SDS–PAGE gels and 32P incorporation assessed by phosphorimager. As can be seen (Figure 6), autophosphorylation of DNA-PKcs is DNA dependent in assays utilizing purified enzyme but DNA independent in assays using immobilized GFP-DNA-PKcs. Autophosphorylation was also assessed by immunoblotting with a phospho-specific antibodies that recognizes phosphorylated S2056 (‘R’ site in the PQR cluster) and T2609 (‘A’ site in ABCDE cluster). S2056 phosphorylation was more robust in the pull-down assays, even though P-32 incorporation is similar in both pull-down assays and assays using purified enzyme. We have previously shown that both S2056 and T2609 phosphorylations can occur in trans both in vitro and in living cells. However detection of autophosphorylation of DNA-PKcs that is immobilized by a monovalent reagent (both by P-32 incorporation and by phospho-specific antibodies) suggests a single DNA-PKcs molecule can autophosphorylate itself.
Figure 6.
DNA-independent autophosphorylation of N-terminally constrained DNA-PKcs. Phosphorimager analysis of kinase reactions electrophoresed in 4.5% SDS–PAGE anti-GFP immunoprecipitates or purified DNA-PK (as indicated) in the presence or absence of DNA (20 µg/ml), or ethidium bromide 50 µg/ml). (A) phosphorimager analysis; (B) immunoblot with anti-DNA-PKcs antibody 42-27; (C) immunoblot analysis with pS2056; (D) immunoblot analysis with pT2609.
DNA-PKcs activated by N-terminal constraint phosphorylates full length p53, albeit less efficiently than DNA activated DNA-PK
To determine whether the observed enzymatic activity was active towards other well-studied DNA-PK targets, kinase assays were done in the presence of recombinant p53. In these experiments, we compared phosphorylation of recombinant p53 by either immunoprecipitated GFP-DNA-PKcs or purified DNA-PK (Promega, Madison, WI, USA). Phosphorylation of p53 (Figure 7) by purified DNA-PK is robust and DNA dependent as expected. In contrast, p53 is phosphorylated by immunoprecipitated DNA-PKcs in the presence or absence of DNA or ethidium bromide. The enzymatic activity towards recombinant full length p53 by immobilized DNA-PKcs is considerably less than the activity of DNA-PK in solution; however, in the same reactions, autophosphorylation of immobilized GFP-DNA-PKcs is more robust than that of 100 U purified DNA-PK. Peptide assays were performed concurrently; enzymatic activity of GFP-DNA-PKcs was similar to 100 U purified enzyme; whereas activity from 400 U was ∼5-fold higher. We conclude that DNA-PKcs tethered by its N-terminus is constitutively active towards a variety of known DNA-PK targets, although its autophosphorylation and peptide phosphorylations are more efficient when the enzyme is immobilized, whereas phosphorylation of full length p53 is less efficient when the enzyme is immobilized.
Figure 7.
DNA-PKcs activated by N-terminal constraint phosphorylates p53, albeit slightly less efficiently than DNA activated DNA-PK. Phosphorimager analysis of kinase reactions electrophoresed in 7% SDS–PAGE using purified DNA-PK or anti-GFP immunoprecipitates (as indicated) to phosphorylate recombinant p53 in the presence or absence of DNA (20 µg/ml), or ethidium bromide (50 µg/ml).
N-terminal tethering cannot rescue enzymatic activity in N-terminal phospho-mimics at sites that regulate kinase activity
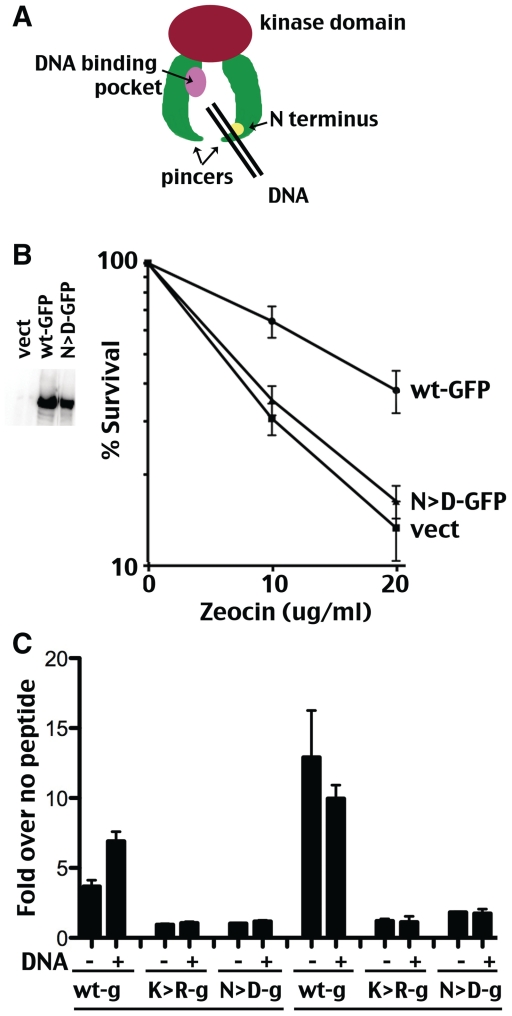
Recently we have reported mutational studies of several additional phosphorylation sites within DNA-PKcs (19). Briefly, earlier work from our laboratory demonstrated that DNA-PKcs with 13 S/T to A substitutions of previously studied autophosphorylation sites still undergoes autophosphorylation induced kinase dissociation at rates similar to the wild-type kinase (10). Since, we had also shown that the extreme N-terminus (N-terminal 426 amino acids) and the Leucine Rich Region contribute to the stability of the DNA-PK complex (10,16), we reasoned that phosphorylation within regions important for complex assembly might mediate kinase dissociation. Examination of these regions for potential target sites revealed two highly conserved potential DNA-PKcs autophosphorylation sites, S-hydrophobic 56–57 (‘N1’) and S-hydrophobic 72–73 (‘N2’). N1 (S56) is present in every DNA-PKcs sequence available; the N2 target (S72) is also present in every species although the adjacent hydrophobic site is not conserved in slime mold. Previously, these sites were studied as combined S/T to D or S/T to A mutants (19). Although, the combined S/T to A mutant completely restores radioresistance to V3 cells, the S/T to D mutant completely fails to complement V3’s radiosensitive phenotype. This can be attributed to markedly reduced catalytic activity of the S/T to D phospho-mimic, which can only be demonstrated using DNA-cellulose pull-down assays. The S/T to D phospho-mimic associates with Ku-bound DNA and with chromosomal DSBs in living cells as evidenced by its ability to robustly inhibit homologous recombination mediated repair of chromosomal DSBs. This is the first mutation in DNA-PK outside of the PI3 kinase domain or extreme C terminus that affects enzymatic activity. The extreme phenotype observed by phospho-mimicking these extremely well conserved ‘N1 and N2’ sites are consistent with a model whereby N1 and N2 sites phosphorylation interferes with the interaction of DNA-PKcs with DNA, abrogating kinase activation. A proposed model of DNA-PKcs is depicted in Figure 8A. It is thought that the N-terminal ‘pincers’ (the green arms correspond to the N-terminal HEAT repeat regions) may tighten when DNA threads between the two pincers into a putative DNA binding pocket (shown in pink). The position of the extreme N-terminus is shown in yellow. Phosphorylation of sites in this area may interfere with how DNA interacts with the pincer domains. Similar, immobilization by tags attached to the N-termini might also affect how the pincer domains can interact with DNA.
Figure 8.
Two highly conserved S/hydrophobic sites at the extreme N-terminus regulate kinase activation. (A) Cartoon depiction of DNA-PKcs. N-terminal pincer domains are shown in green. The extreme N-terminus is shown in yellow, presumable position of ‘N’ phosphorylation sites. The C-terminal ‘head’ domain that includes the catalytic domain is shown in red. A putative DNA binding pocket has been proposed and is shown in pink. (B) V3 transfectants expressing either GFP-tagged wild-type DNA-PKcs, GFP-tagged combined S/T to D mutant DNA-PKcs (N > D), or no DNA-PKcs were plated into complete media with increasing doses of zeocin as indicated at cloning densities. Colonies were stained after 7 days later and percent survival was calculated. Error bars represent SEM. Left panel depicts immunoblot analyses of DNA-PKcs expression in extracts from V3 cells expressing GFP-tagged wild-type DNA-PKcs, GFP-tagged combined S/T to D mutant DNA-PKcs, or no DNA-PKcs. (C) DNA-PK enzymatic levels were determined in extracts or anti-GFP immunoprecipitates from V3 transfectants expressing GFP-tagged wild-type (wt-g), GFP-tagged K3752R mutant (K > R − g) or GFP-tagged combined S/T to D mutant DNA-PKcs (N > D − g) in the presence or absence of DNA (20 µg/ml). Relative activity is depicted as fold-increase over no peptide as described in the ‘Methods’ section. Error bars depict SDs of triplicate assays. This assay is representative of no less than three independent assays. To illustrate minimal activity present in GFP-N > D mutant IP kinase reactions, grey line shows level for no enzymatic activity (1 = fold over no peptide control).
To ascertain whether N-terminal tethering can relieve negative impact of phospho-mimicking the N sites, a GFP-tagged construct with the above-mentioned combined S/T to D mutations was generated and expressed in V3 cells. Although expressed at similar levels as GFP-tagged wild-type DNA-PKcs, the GFP-tagged combined S/T to D mutant does not complement V3’s radiosensitive phenotype (Figure 8B) consistent with lack of complementation with the un-tagged mutant (19). Paired IP and extract kinase assays were done with extracts from V3 cells expressing wt-GFP tagged, K3752R-GFP tagged and combined S/T to D GFP tagged DNA-PKcs. As can be seen (Figure 8C), only minimal enzymatic activity is demonstrable in pull-downs of the combined S/T to D GFP mutant [consistent with DNA cellulose pull-downs in our previous study (19)]. However, the activity is DNA independent. We conclude that N-terminal tethering cannot relieve the negative impact of phospho-mimicking of S52/S72. These data are consistent with a model whereby DNA binding results in a conformational change at the extreme N-terminus that allows DNA-PKcs to ‘sense’ DNA; moreover, phosphorylations within this region abate kinase activation.
DISCUSSION
Experiments presented here were designed to address cis versus trans activation. Although activation in this artificial system occurs in cis, this N-terminal constraint likely mimics DNA binding induced conformation change. Thus, our work does not clarify the discordant published conclusions of whether DNA-PK is activated by DNA bound in cis versus trans. However, these data do provide additional insight into whether autophosphorylation occurs in cis or trans.
We have previously demonstrated (by using site ablated mutants to phosphorylate a kinase inactive molecule) that phosphorylation of the ABCDE and PQR sites can occur in trans both in vitro and in living cells. The experiments presented here showing autophosphorylation of DNA-PKcs molecules immobilized by a monovalent reagent on an 80 micron agarose bead suggest that cis autophosphorylation can also occur, a conclusion also reached by Lieber and colleagues working with multivalent immobilization techniques.
The DNA dependence of DNA-PK has been studied for decades. It has been proposed that Ku undergoes a conformational change when bound to DNA that allows DNA-PKcs to interact with both Ku and the DNA end (7). The interaction of DNA-PKcs with the DNA end (bound by Ku) or with the DNA alone (in low salt conditions), results in enzymatic activation. Here, we report the unexpected finding of robust DNA-PKcs activation by N-terminal constraint, independent of either DNA or its regulatory subunit Ku. These data suggest that immobilization of the extreme N-terminus mimics DNA binding; thus, we suggest that an N-terminal conformational change (likely induced by DNA binding) actually induces enzymatic activation. Consistent with this model ascribing a critical role for the N-terminus in kinase activation, recent studies from our laboratories demonstrate that phosphorylation at two conserved sites at the extreme N-terminus of DNA-PKcs affects the ability of the kinase to be activated (19). Thus, phosphorylation at the extreme N-terminus (at S56/S72) may preclude the N-terminal conformational change required to activate DNA-PK.
The low-resolution structure of DNA-PKcs in complex with the CTR of Ku80 reveals a horseshoe or pincer shaped molecule in which the large N-terminal domain forms two arms that surround a central open cavity that is suggested to bind dsDNA, while the FAT-kinase-FAT-C domain of DNA-PKcs resides at the apex of the structure (Figure 8A) (32). The region at the top of the arms is predicted to undergo conformational flexibility, which may account for autophosphorylation induced conformational changes in DNA-PKcs (18). Interestingly, the two arms do not meet at the base of the molecule, creating a gap, and we have suggested that conformational changes for example in the flexible regions at the apex of the arms may cause opening and closing of the gap and thus regulate interaction of the central cavity with DNA. Thus, DNA-PKcs is predicted to be a highly dynamic molecule that undergoes considerable conformational plasticity that is regulated at least in part by phosphorylation (18). Moreover, the N-terminus of DNA-PKcs is predicted to be located close to the gap at the base of the structure (32), thus phosphorylation at N-terminal regions of DNA-PKcs (for example at S56 and S72) might also induce conformational changes, causing opening and closing of the gap and regulating interactions with DNA.
In sum, our data suggest that conformational changes in the N-terminus of DNA-PKcs (likely induced by DNA binding) enable DNA-PK to ‘sense’ DNA binding which is then communicated to the kinase domain resulting in enzyme activation.
FUNDING
Public Health Service (Grant AI048758 to K.M.), Association for International Cancer Research (Grant 09-0633 to M.M.) and Association pour la Recherche contre le Cancer (Grant A09/2/5075 to M.M.). Funding for open access charge: Public Health Service (grant AI048758).
Conflict of interest statement. None declared.
REFERENCES
- 1.Walker AI, Hunt T, Jackson RJ, Anderson CW. Double-stranded DNA induces the phosphorylation of several proteins including the 90 000 mol. wt. heat-shock protein in animal cell extracts. EMBO J. 1985;4:139–145. doi: 10.1002/j.1460-2075.1985.tb02328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartley KO, Gell D, Smith GC, Zhang H, Divecha N, Connelly MA, Admon A, Lees-Miller SP, Anderson CW, Jackson SP. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 3.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 4.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammarsten O, Chu G. DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc. Natl. Acad. Sci. USA. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.West RB, Yaneva M, Lieber MR. Productive and nonproductive complexes of Ku and DNA-dependent protein kinase at DNA termini. Mol. Cell. Biol. 1998;18:5908–5920. doi: 10.1128/mcb.18.10.5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Block WD, Yu Y, Merkle D, Gifford JL, Ding Q, Meek K, Lees-Miller SP. Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic Acids Res. 2004;32:4351–4357. doi: 10.1093/nar/gkh761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui X, Yu Y, Gupta S, Cho YM, Lees-Miller SP, Meek K. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol. Cell. Biol. 2005;25:10842–10852. doi: 10.1128/MCB.25.24.10842-10852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douglas P, Cui X, Block WD, Yu Y, Gupta S, Ding Q, Ye R, Morrice N, Lees-Miller SP, Meek K. The DNA-dependent protein kinase catalytic subunit is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Mol. Cell. Biol. 2007;27:1581–1591. doi: 10.1128/MCB.01962-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douglas P, Gupta S, Morrice N, Meek K, Lees-Miller SP. DNA-PK-dependent phosphorylation of Ku70/80 is not required for non-homologous end joining. DNA Repair. 2005;4:1006–1018. doi: 10.1016/j.dnarep.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, et al. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Y, Mahaney BL, Yano K, Ye R, Fang S, Douglas P, Chen DJ, Lees-Miller SP. DNA-PK and ATM phosphorylation sites in XLF/Cernunnos are not required for repair of DNA double strand breaks. DNA Repair. 2008;7:1680–1692. doi: 10.1016/j.dnarep.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Y, Wang W, Ding Q, Ye R, Chen D, Merkle D, Schriemer D, Meek K, Lees-Miller SP. DNA-PK phosphorylation sites in XRCC4 are not required for survival after radiation or for V(D)J recombination. DNA Repair. 2003;2:1239–1252. doi: 10.1016/s1568-7864(03)00143-5. [DOI] [PubMed] [Google Scholar]
- 15.Chan DW, Lees-Miller SP. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J. Biol. Chem. 1996;271:8936–8941. doi: 10.1074/jbc.271.15.8936. [DOI] [PubMed] [Google Scholar]
- 16.Gupta S, Meek K. The leucine rich region of DNA-PKcs contributes to its innate DNA affinity. Nucleic Acids Res. 2005;33:6972–6981. doi: 10.1093/nar/gki990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, So S, et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J. Biol. Chem. 2010;285:1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobbs TA, Tainer JA, Lees-Miller SP. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair. 2010;9:1307–1314. doi: 10.1016/j.dnarep.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neal JA, Dang V, Douglas P, Wold MS, Lees-Miller SP, Meek K. Inhibition of HR by DNA-PK requires kinase activity, is titratable, and is modulated by autophosphorylation. Mol. Cell. Biol. 2011;31:1719–1733. doi: 10.1128/MCB.01298-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeFazio LG, Stansel RM, Griffith JD, Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meek K, Douglas P, Cui X, Ding Q, Lees-Miller SP. trans Autophosphorylation at DNA-dependent protein kinase’s two major autophosphorylation site clusters facilitates end processing but not end joining. Mol. Cell. Biol. 2007;27:3881–3890. doi: 10.1128/MCB.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu H, Shimazaki N, Raval P, Gu J, Watanabe G, Schwarz K, Swanson PC, Lieber MR. A biochemically defined system for coding joint formation in V(D)J recombination. Mol. Cell. 2008;31:485–497. doi: 10.1016/j.molcel.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jovanovic M, Dynan WS. Terminal DNA structure and ATP influence binding parameters of the DNA-dependent protein kinase at an early step prior to DNA synapsis. Nucleic Acids Res. 2006;34:1112–1120. doi: 10.1093/nar/gkj504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muyldermans S. Single domain camel antibodies: current status. J. Biotechnol. 2001;74:277–302. doi: 10.1016/s1389-0352(01)00021-6. [DOI] [PubMed] [Google Scholar]
- 25.Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, Lees-Miller SP, Meek K. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol. Cell. Biol. 2003;23:5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Convery E, Shin EK, Ding Q, Wang W, Douglas P, Davis LS, Nickoloff JA, Lees-Miller SP, Meek K. Inhibition of homologous recombination by variants of the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs) Proc. Natl Acad. Sci. USA. 2005;102:1345–1350. doi: 10.1073/pnas.0406466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas-Delucchi CS, Cardoso MC, et al. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17:133–138. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- 28.Finnie NJ, Gottlieb TM, Blunt T, Jeggo PA, Jackson SP. DNA-dependent protein kinase activity is absent in xrs-6 cells: implications for site-specific recombination and DNA double-strand break repair. Proc. Natl Acad. Sci. USA. 1995;92:320–324. doi: 10.1073/pnas.92.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kienker LJ, Shin EK, Meek K. Both V(D)J recombination and radioresistance require DNA-PK kinase activity, though minimal levels suffice for V(D)J recombination. Nucleic Acids Res. 2000;28:2752–2761. doi: 10.1093/nar/28.14.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan DW, Mody CH, Ting NS, Lees-Miller SP. Purification and characterization of the double-stranded DNA-activated protein kinase, DNA-PK, from human placenta. Biochem. Cell. Biol. 1996;74:67–73. doi: 10.1139/o96-007. [DOI] [PubMed] [Google Scholar]
- 31.Kurimasa A, Kumano S, Boubnov NV, Story MD, Tung CS, Peterson SR, Chen DJ. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol. Cell. Biol. 1999;19:3877–3884. doi: 10.1128/mcb.19.5.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010;463:118–121. doi: 10.1038/nature08648. [DOI] [PMC free article] [PubMed] [Google Scholar]