Abstract
The eukaryotic ribosomal protein S26e (rpS26e) lacking eubacterial counterparts is a key component of the ribosomal binding site of mRNA region 5′ of the codon positioned at the exit site. Here, we determined the rpS26e oligopeptide neighboring mRNA on the human 80S ribosome using mRNA analogues bearing perfluorophenyl azide-derivatized nucleotides at designed locations. The protein was cross-linked to mRNA analogues in specific ribosomal complexes, in which the derivatized nucleotide was located at positions −3 to −9. Digestion of cross-linked rpS26e with various specific proteolytic agents followed by identification of the resulting modified oligopeptides made it possible to map the cross-links to fragment 60–71. This fragment contains the motif YxxPKxYxK conserved in eukaryotic but not in archaeal rpS26e. Analysis of X-ray structure of the Tetrahymena thermophila 40S subunit showed that this motif is not implicated in the intraribosomal interactions, implying its involvement in translation process in a eukaryote-specific manner. Comparison of the results obtained with data on positioning of ribosomal ligands on the 40S subunit lead us to suggest that this motif is involved in interaction with both the 5′-untranslated region of mRNA and the initiation factor eIF3 specific for eukaryotes, providing new insights into molecular mechanisms of translation in eukaryotes.
INTRODUCTION
Prokaryotic and eukaryotic ribosomes perform the same function of carrying out protein synthesis using incoming mRNA as template that bears genetic information presented as the sequence of trinucleotide-codons to be translated into the amino acid sequence of the polypeptide synthesized. Structures of the ribosomes from both kingdoms, that are now well known due to remarkable progress in X-ray crystallography (1–5) and cryo-electron microscopy (cryo-EM) (6–11), share significant similarity. Arrangements of prokaryotic ribosomal functional sites have been deciphered at the atomic level (2,12–15), while eukaryotic ribosomal sites remain much less studied. This concerns in particular the mRNA binding site since the mRNA has been visualized in the eukaruotic ribosomal complexes by neither cryo-EM nor X-ray crystallography. In particular, the resolution of cryo-EM, that has been extensively applied to study eukaryotic ribosome (9–11), is so far insufficient to directly visualize mRNA codons; two recently published X-ray crystallographic structures of the eukaryotic ribosome (4,5) do not contain mRNA. Data on rRNA nucleotides and ribosomal proteins neighboring the mRNA in the 80S ribosome have been obtained by site-directed cross-linking (16–18). This approach made it possible to reveal both the conserved rRNA ‘core’ of the ribosome, which is structurally similar in all kingdoms, and differences related to protein environment of the mRNA in eubacterial and eukaryotic ribosomes (17). The most striking difference concerns the arrangement of the binding site of the mRNA region 5′ of the codons interacting with tRNA anticodons in the A and P sites. Ribosomal protein S26 (rpS26e), which has no eubacterial counterparts, is the main component of this site. In early studies it was reported that this protein cross-links to derivatives of oligo(U) bearing an alkylating group at the 5′-termini in their complexes with rat liver (19,20) and human ribosomes (21). Later, with the use of a set of short mRNA analogues bearing a perfluorophenyl azide cross-linker at G or U at the designed locations, it was demonstrated that rpS26e is the main target for cross-linking from positions −4 to −9 with respect to the first nucleotide of the P site-bound codon on the human 80S ribosome (17). Finally, very similar results were obtained using longer synthetic mRNAs carrying 4-thiouridine, a ‘zero-length’ cross-linker, in rabbit 48S and 80S initiation complexes assembled with a set of appropriate initiation factors (22,23). The amino acids of rpS26e involved in the formation of the mRNA binding site remain unknown. Knowledge on the mutual positioning of rpS26e and the mRNA in the ribosome is of paramount importance to understand the functional significance of rpS26e as one of the key components of the 40S ribosomal mRNA binding site that has no homologs in eubacteria.
In the current study, to identify amino acid residues of rpS26e neighboring the mRNA in the ribosome, this protein was cross-linked to short labeled mRNA analogues with a perfluorophenyl azide cross-linker at the uridines in human 80S ribosomal complexes where derivatized U was in positions −3 to −9. Mapping of cross-linking sites on the rpS26e was carried out using an approach based on the application of various proteolytic agents for selective cleavage of the cross-linked protein with subsequent SDS-PAGE separation of the labeled modified peptides, their identification and mapping of the cross-links. This approach has been successfully used to identify cross-linking sites of mRNA analogues in rpS15e (24) and eRF1 (25,26) in human 80S ribosomal complexes where the nucleotide with the cross-linker was positioned at the decoding site. We show here that mRNA analogues with modified nucleotides in positions −3 to −9 in 80S ribosomal complexes cross-link to the same dodecapeptide in positions 60–71 in the central part of the protein. A comparative analysis of protein sequences of the rpS26e family revealed that motif YxxPKxYxK within the fragment mentioned is conserved in eukaryotes but not in archaea. Application of the cross-linking results to available models of the 40S subunits makes it possible to propose a role of motif YxxPKxYxK of rpS26e in the translation process in eukaryotes.
MATERIALS AND METHODS
Ribosomes, tRNA and mRNA analogues
tRNAPhe (1300 pmol/A260 unit) was a kind gift from Dr V. Katunin (B.P. Konstantinov's St Petersburg Institute of Nuclear Physics RAS, Gatchina, Russia). Isolation of the 40S and 60S ribosomal subunits from unfrozen human placenta and their association to produce 80S ribosomes were performed as described (27). The activity of the ribosomes in the poly(U)-directed binding of [14C]Phe-tRNAPhe was ∼80% (maximum binding level was about 1.6 mol of Phe-tRNAPhe per mol of the 80S ribosomes). Synthesis and purification of derivatives of oligoribonucleotides bearing an 4-azido-2,3,5,6-tetrafluorobenzoyl group attached via an ethylene diamine spacer to the C5 of the uridines was carried out as described (28). Before use in cross-linking experiments, the derivatives were 5′-32P-labeled and purified according to (29).
Ribosomal complexes and cross-linking procedures
Complexes of 80S ribosomes (1.6 × 10−6 M) with tRNAPhe (1.5 × 10−5 M) and mRNA analogues (1.3 × 10−5 M) were obtained by incubating these components for 40 min in buffer A (20 mM Tris–HCl, pH 7.5, 120 mM KCl, 13 mM MgCl2 and 0.6 mM EDTA) at room temperature. The reaction mixtures for the cross-linking experiments typically contained 15–30 pmol of 80S ribosomes. To obtain cross-links of mRNA analogues to the ribosomes, the complexes were irradiated with mild UV light (λ > 290 nm) as described (17) and then supplied with 1/30 v of 5% 2-mercaptoethanol. Cross-linked ribosomal components were separated by SDS–PAGE on a 15% gel as described (17).
Isolation of cross-linked rpS26e
Total protein was extracted from the irradiated complexes with acetic acid according to ref. (30) and lyophilized. The preparations were dissolved in 10 µl of 2 M urea, treated with 0.1 µg of RNase A at 37°C for 2 h and acetone precipitated. Cross-linked rpS26e was isolated from these protein preparations by 15% SDS–PAGE as described (17). The gels were dried and autoradiographed, and a major radioactive band in the lower part of the gels corresponding to cross-linked rpS26e (17) was excised.
Mapping of cross-linking site on the S26e protein
Cleavage of the cross-linked S26e protein with endoproteases GluC or ArgC (Roche), or with pepsin (Sigma) was carried out as described (24). The reaction mixtures of a total volume of 50 or 100 µl contained GluC, ArgC or pepsin in amounts providing exhaustive cleavage of the cross-linked protein. These amounts were selected for each enzyme preparation in parallel experiments based on cleavage of cross-linked rpS26e with increasing enzyme concentrations.
Cleavage of the cross-linked rpS26e with 2-nitro-5-thiocyanobenzoic acid (NTCB) was carried out as follows. The gel slice containing cross-linked rpS26e was incubated in 50 µl of buffer B (0.2 M Tris–HCl, pH 8.0 and 6 M guanidine chloride) containing 5 mM DTT at 37°C for 6 h. Then the same volume of 0.2 M NTCB in buffer B was added together with 25 µl of 1 M NaOH and the mixture was incubated at 37°C for 12 h. The reaction was stopped with 1/5 v of 85% formic acid, diluted with ½ v of water, after which 1 ml of bovine serum albumin (10 g/l) was added as a carrier and the polypeptides were precipitated with 6 v of cold acetone (incubation at −20°C for 2 h).
Treatment of the cross-linked oligopeptides with endoprotease AspN (Roche) was carried out in 100 µl of 10 mM Tris–HCl, pH 7.5, by incubating of the mixture overnight at 37°C.
The fragments resulting from the hydrolysis of cross-linked rpS26e were analyzed by 16.5% Tris–tricine SDS–PAGE in parallel with polypeptide size markers obtained as a result of selective digestion of a specially designed recombinant protein S26C (see below). The gels were stained with Coomassie Brilliant Blue G-250 to develop the positions of the markers, dried and radioautographed.
Preparation of molecular mass markers
To obtain size markers to evaluate the molecular masses of modified oligopeptides resulting from the cleavage of cross-linked rpS26e, a special 66 amino acid-long recombinant protein S26C was constructed whose sequence is:
MGSSHHHHHH SSGLVPRGSH MLHYCVSCAI HSKVVRNRSR EARKDRTPPP RFRPAGAAPR PPPKPM.
It consists of His-tag (marked in bold) followed by a sequence corresponding to the C-terminal 71–115 fragment of human rpS26e. This 7.4 kDa protein bears the only site for cleavage by endoprotease GluC (after E41) and two sites for pepsin (before L14 and L22) that makes it possible to obtain peptides of known molecular masses. Fragments obtained as a result of S26C cleavage with the endoproteases mentioned are more suitable size markers for the cross-linked rpS26e fragments than standard commercially available size markers since ribosomal proteins (and especially rpS26e) have higher electrophoretic mobilities in SDS–PAGE than those expected from their molecular masses.
A fragment of human rpS26e cDNA coding for the C-terminal 45 amino acid-long fragment of the protein was obtained by PCR using a pair of primers bearing on their 5′-termini sequences necessary for the presence of restriction sites in the PCR product. The sequences of the forward and reverse primers were 5′-CGGGAATTCCATATGCTACATTACTGTGTGAGTTG-3′ and 5′-GGGAATTCGGATCCTTACATGGGCTTTGGTG-3′, respectively. Full size cDNA of human rpS26e was used as a template (31). The PCR product was digested with NdeI and BamHI and cloned into plasmid pET-15b (Novagen) at the corresponding sites. The integrity of the insert was validated by sequencing. The resulting plasmid was expressed in Escherichia coli BL-21 (DE-3) strain. The recombinant protein was synthesized and purified as in ref. (31) and refolded by dialysis against buffer containing 20 mM Tris–HCl, pH 7.5 and 100 KCl. It was then acetone precipitated (12 h), resuspended in 2 M urea and stored at −20°C as 20 µl aliquots (each used once).
Digestion of S26C with endoprotease GluC and with pepsin was carried out as described in the previous section; each reaction mixture contained 10 µg of S26C. Pepsin and GluC were added in amounts providing exhaustive hydrolysis of the protein (the exact amounts of the enzymes were selected in each set of experiments separately). The resulting protein fragments were resolved by Tris–tricine SDS–PAGE together with the cross-linked rpS26e fragments; the gels were stained with Coomassie Brilliant Blue G-250 and dried. It should be noted that in the lanes corresponding to products of S26C digestion only the larger fragments are clearly seen, while the shorter ones (<25 amino acids long) are poorly stained with the dye and therefore are hardly detectable.
RESULTS
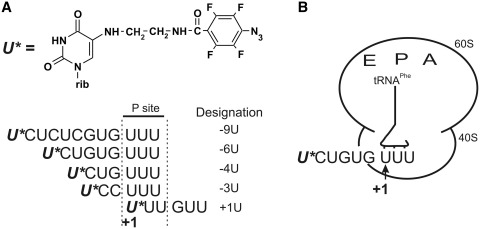
The mRNA analogues used here are presented in Figure 1A. They contain the triplet UUU coding for Phe at the 3′-terminus and a perfluorophenyl azide-modified U 5′ of the triplet or in the first position of the triplet. Protein rpS26e was cross-linked to labeled mRNA analogues in specific human 80S ribosomal complexes obtained in the presence of tRNAPhe that directed triplet UUU to the ribosomal P site and positioned the modified mRNA nucleotides in designed locations (Figure 1B).
Figure 1.
mRNA analogues (A) and schematic representation of ribosomal complex type (B) used here. Designations of mRNA analogues reflect positions of the modified uridines (marked with asterisks in their sequences) with respect to the first nucleotide of UUU codon targeted to the P site.
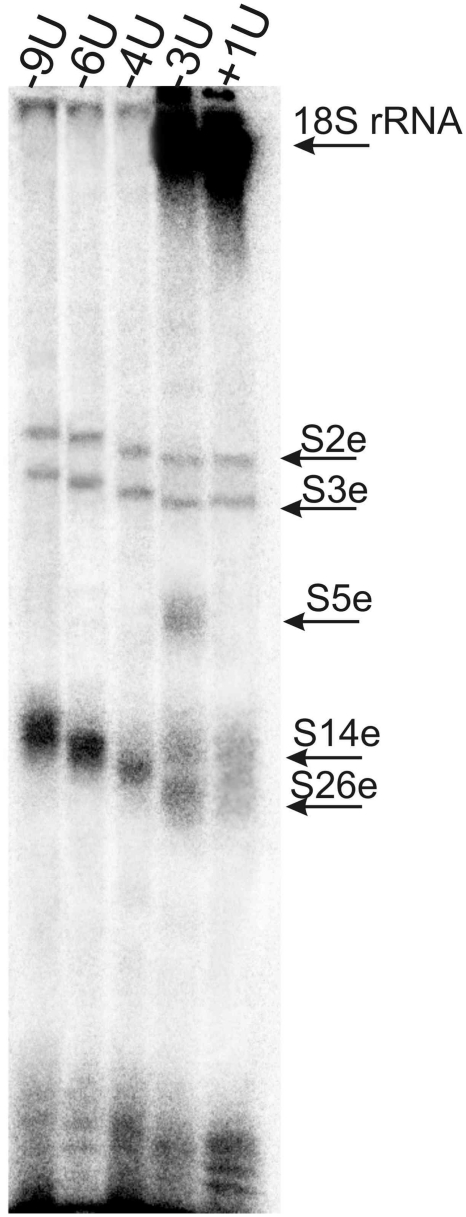
The 5′-32P-labeled mRNA analogues were cross-linked to human 80S ribosomes by mild UV-irradiation, and the labeled ribosomal components were resolved by SDS–PAGE (Figure 2). Weak protein bands in the upper parts of the gels correspond to cross-linked ribosomal proteins S2e and S3e, and the major (with all mRNA analogues with except for +1U) protein bands in the lower parts relate to cross-linked rpS26e. This assignment is based on our earlier studies in which these proteins cross-linked to mRNA analogues were identified (17,32,33). One can see that the level of the cross-linking to rpS26e strongly depends on the position of modified nucleotide in the mRNA analogue. In particular, the level of cross-linking is maximum with mRNA analogues -6U and -9U, somewhat less with analogue -4U (with these three analogues cross-link with rpS26e is dominating), much less with analogue -3U and almost negligible with mRNA analogue +1U [the latter two analogues cross-linked mainly to the 18S rRNA, which is in agreement with our earlier report (32)]. Cross-linking of mRNA analogues to rpS26e is completely specific since it does not take place in binary complexes of the analogues with 80S ribosomes. In contrast, the extent of weak cross-linking to rpS2e and rpS3e barely depends on the position of the derivatized nucleotide with respect to the first nucleotide of the P site codon and on the presence of tRNAPhe. Lanes corresponding to cross-linking experiments without tRNA are not shown here [the respective results with mRNA analogues used in this study were reported earlier (17,32)]. It should be noted that tRNA-dependent cross-linking to rpS26e and tRNA-independent labeling of rpS2e and rpS3e were also observed with mRNA analogues similarly derivatized at guanines (17). Thus, it is reasonably to suggest that cross-linking to rpS2e and rpS3e takes place in fractions of binary complexes of the ribosomes with mRNA analogues that remain in the reaction mixtures because of incomplete occupation of the P site by tRNAPhe. Proteins S2e and S3e are homologous prokaryotic rpS5p and rpS3p, respectively that are involved in the formation of the mRNA entry site on the 30S subunit (12). Proteins S2e and S3e are believed to take part in organization of the mRNA entry site on the 40S subunit (4). Apparently, without tRNA, mRNA analogues independently of their sequences bind at the ribosomal entry site in a labile transient manner and cross-link to rpS2e and rpS3e located nearby.
Figure 2.
SDS–PAGE analysis of ribosomal components cross-linked to 5′-32P-labeled mRNA analogues in their complexes with 80S ribosomes and tRNAPhe. Autoradiogram. Bands of the cross-linked 18S rRNA and cross-linked ribosomal proteins are marked according to their positions observed earlier (17,32,33), cross-linked proteins were identified in the mentioned studies by one-dimensional SDS–PAGE and 2D PAGE in various electrophoretic systems; the identity of cross-linked rpS26e was confirmed by immunoblotting (17). Mobilities of the proteins cross-linked to mRNA analogues decrease from mRNA +1U to mRNA -9U according to the increase of the lengths of the mRNA analogues (Figure 1A).
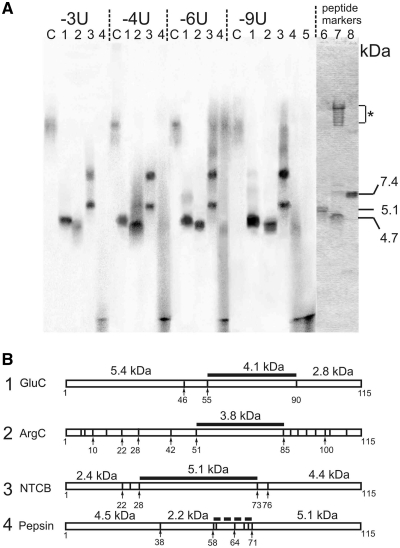
Identification of rpS26e peptides cross-linked to mRNA analogues was carried out with all analogues with except for mRNA +1U whose cross-linking yield was too low for the analysis. For this purpose, total protein was isolated from the irradiated ribosomal complexes and cross-linked RNA moieties of mRNA analogues were hydrolyzed with RNase A to reduce their effects on electrophoretic mobilities of the cross-linked peptides. RNase treatment did not lead to loss of the label cross-linked to proteins since in all mRNA analogues the cross-linker was at the very 5′-terminal U that was coupled with the 32P labeled phosphate. After hydrolysis with RNase A, mono- or dinucleotide fragments (whose masses were about 0.5 or 0.8 kDa, respectively) of mRNA analogues remained cross-linked to the protein. Modified labeled rpS26e was isolated by SDS–PAGE and subjected to exhaustive hydrolysis with specific proteolytic agents that cleave the polypeptide chain at selective sites. We applied 2-nitro-5-thiocyanobenzoate (NTCB) that cleaves peptide bonds before Cys, endoprotease GluC that hydrolyses the polypeptide chain after Glu, endoprotease ArgC that cleaves after Arg, and pepsin. The cleavages were carried out in parallel experiments with subsequent analysis of the resulting labeled products by Tris–tricine SDS–PAGE. To evaluate the sizes of the modified labeled oligopeptides based on their electrophoretic mobilities, we used peptide size markers derived from the especially designed recombinant protein S26C consisting of a sequence corresponding to C-terminal sequence 71–115 of the human rpS26e preceded by a 21 amino acid-long His-tag sequence (see ‘Materials and Methods’ section). This protein has the only cleavage site for endoprotease GluC and two sites for pepsin, and its treatment with these enzymes makes it possible to obtain unambiguously identifiable products, that are easily detected in Coomassie-stained gels. Comparing the positions of the bands of labeled oligopeptides with the bands of size markers, we took into account that all cross-linked oligopeptides contained residues of mRNA analogues, which remained after treatment of the cross-linked rpS26e with RNase A (see above) that somewhat reduced their electrophoretic mobilities.
The data presented in Figure 3A show that the patterns of rpS26e cleavage with each proteolytic agent are very similar with all mRNA analogues, indicating a common cross-linking site. With endoproteases GluC and ArgC only one labeled oligopeptide is formed, its mass is about 4 kDa, the fragment resulting from ArgC treatment being a little bit shorter than the one obtained with GluC. These results are consistent with the only candidate pair of labeled fragments, namely, amino acids 55–90 with GluC and amino acids 51–85 with ArgC (Figure 3B). With NTCB, two bands are visible; the upper bands evidently relate to the undercleaved cross-linked rpS26e since they correspond to a product whose mobility is drastically less than that of any product that could result from the complete cleavage of rpS26e with NTCB (compare data of Figure 3A and B). The lower bands fit well the modified fragment 28–73 that overlaps with the mentioned above labeled fragments found with GluC and ArgC. Finally, treatment of cross-linked rpS26e with pepsin leads to the formation of very short labeled fragments similar to those produced with unspecific proteinase K that digests proteins to amino acids, di- and tripeptides (bands at the bottom, compare lanes 4 and 5 in Figure 3A). Such fragments can be formed only if the cross-link is located within positions 59–71 of rpS26e (Figure 3B); such a position of the cross-link agrees with the results obtained with NTCB and the endoproteases ArgC and GluC.
Figure 3.
Identification of mRNA analogues cross-linking sites on the rpS26e. (A) Autoradiograms of an SDS–PAGE analysis of labeled peptides resulting from digestion of the cross-linked protein with endoprotease GluC (lanes 1), endoprotease ArgC (lanes 2), NTCB (lanes 3) or pepsin (lanes 4). Cross-linked mRNA analogues were hydrolysed with RNase A prior to the analysis. Control lanes C correspond to the cross-linked rpS26e untreated with proteolytic agents, lane 5 represents a pattern of hydrolysis of rpS26e cross-linked to mRNA -6U with proteinase K. Approximately equal amounts of labeled rpS26e were applied to each lane. The right panel represents a part of the Coomassie stained gel showing bands of peptide size markers obtained by digestion of an especially designed recombinant protein S26C (lane 8) with endoprotease GluC (lane 7) or pepsin (lane 6) (for details, see ‘Materials and Methods’ section). The group of bands in the upper part of the lane 7 marked with an asterisk corresponds to the GluC protein (these bands are observed as well when the enzyme is incubated without rpS26C; data not shown). (B) Schematic representations (diagrams) of human rpS26e cleavages with endoproteases GluC and ArgC, NTCB and pepsin. Thick black lines correspond to the fragments, which were found to be cross-linked; their calculated molecular masses are shown.
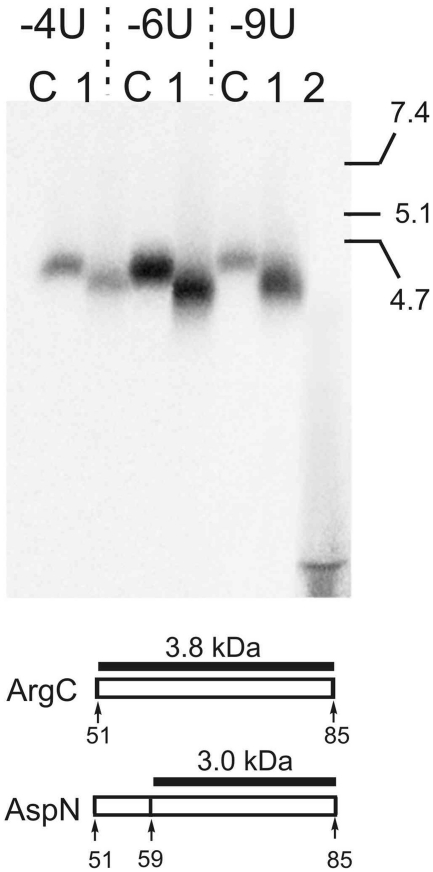
To map cross-link more precisely, labeled fragment obtained by digestion of the cross-linked rpS26e with ArgC (amino acids 51–85) was cut off the gels and treated with endoprotease AspN that cleaves before Asp, which is present in the only position 60 in this fragment. The results presented in Figure 4 show that treatment with AspN leads to modest shortening of the labeled fragment with all the mRNA analogues resulting in formation of a labeled product whose electrophoretic mobility fits well the expected mobility of the cross-linked 60–85 fragment and is incompatible with that of the cross-linked 51–59 fragment. Therefore, the cross-link is located within positions 60–85 and not within positions 51–59. The combination of the results presented in Figures 3 and 4 allows us to map the cross-link to rpS26e in positions 60–71 with all mRNA analogues examined.
Figure 4.
Mapping of the cross-linking site within the 51–85 region of rpS26e. SDS–PAGE analysis of oligopeptides resulting from treatment of the labeled 51–85 fragment (obtained by digestion of the cross-linked rpS26e with ArgC; lanes C) with endoprotease AspN (lanes 1). mRNA analogues used for cross-linking are shown above the lanes. Lane 2 on the right represents a pattern of hydrolysis of rpS26e cross-linked to mRNA -6U with proteinase K. Schematic representation of the 51–85 fragment of rpS26e and its cleavage with endoprotease AspN is shown under the autoradiograms. Labeled oligopeptides and their expected molecular masses are indicated (without regard for the mass of the cross-linked mRNA residues). Positions of peptide size markers (for details, see the legend to Figure 3) are shown on the right of the electrophoregram.
DISCUSSION
The results of the current study show that modified uridines of mRNA analogues in positions −3 to −9 cross-link to the same dodecapeptide 60–71 of rpS26e. This dodecapeptide seems to be the only protein fragment neighboring mRNA nucleotides in these positions because rpS26e is practically the only target for cross-linking of the mRNA analogues, and dodecapeptide 60–71 is the only cross-linking site in the protein. We suggest that this fragment of rpS26e directly interacts with mRNA nucleotides in positions −6 to −9 because (i) the yield of cross-linking of mRNA analogues to rpS26e is maximum when nucleotides with the cross-linker are in these positions (Figure 2), and (ii) ‘zero-length’ cross-links of 4-thiouridines to rpS26e have been observed from positions −6 to −10 but not from positions −3 and −4 (23). Decrease of cross-linking yield of perfluorophenyl azide-modified uridines of the mRNA analogues to rpS26e when the derivatized uridine is shifted downstream of position −6 (Figure 2) clearly reflects its moving away from the 60–71 fragment of rpS26e. The more downstream from position −6 is the modified uridine, the less likely is its cross-link to the protein. Apparently, cross-linking of derivatized uridines in positions −3 and −4 to rpS26e is detectable here because the perfluorophenyl azide group used is linked to uridines via a flexible linker enabling this group to reach the target, that is located close to the mRNA but does not make direct contacts with it.
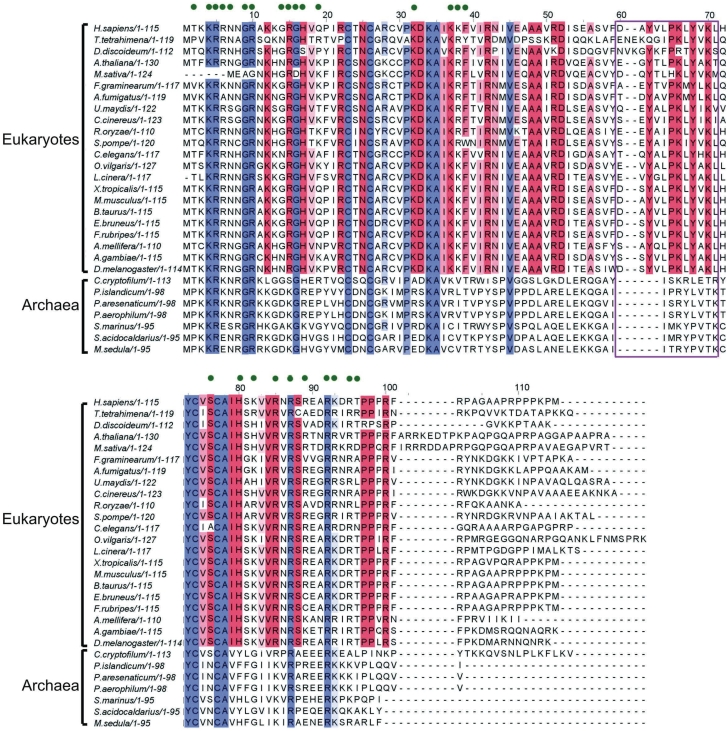
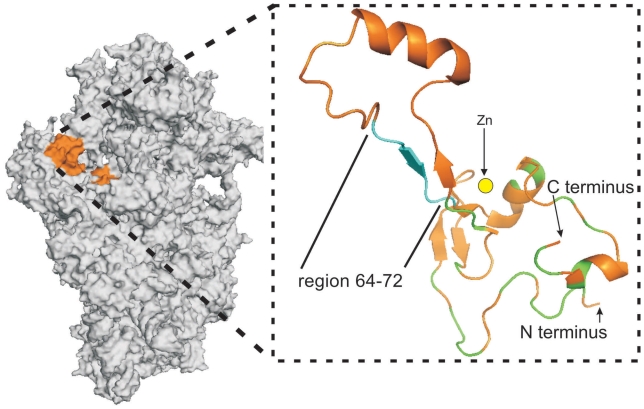
There are two currently available atomic models for the small subunit of the eukaryotic ribosome where rpS26e is localized; one is derived from cryo-EM data (11) and the other from X-ray crystallography (5). In both models rpS26e is located on the solvent side of the subunit at the level of the platform, but only a minor part of rpS26e is positioned similarly on both models, namely, on the platform side facing the head near the mRNA exit site. As for the remaining larger part of the protein, it is exposed on the top of platform surface in the cryo-EM structure (11), whereas in the X-ray structure (5) the respective site is occupied by another ribosomal protein, S1e (S3a). In the cryo-EM study the rpS26e structure has been tentatively modeled ab initio based on the prediction of the secondary structure of this protein, and therefore this structure has lower degree of accuracy than protein structures modeled based on the available X-ray and NMR structures (11). This is why we compared our cross-linking results with an X-ray structure of the 40S subunit of the lower eukaryote Tetrahymena thermophila (PDB accession number 2xzn, hereinafter we term the corresponding structure 2xzn model) (5). This model makes it possible to reveal essential features of the rpS26e structure, in particular, its folding, the location of the Zinc finger, and the particular amino acids interacting with the 18S rRNA. The sequences of the human and the Tetrahymena thermophila rpS26e share significant similarity (Figure 5). The sequences have the same numbering up to amino acid 59; after this position, and up to aa 110 near the C-terminus, the Tetrahymena numbering is two amino acid residues ahead of the human numbering, so that human fragment 60–71 corresponds to the Tetrahymena sequence 62–73. Figure 6 shows the location of this sequence on the structure of rpS26e in the 40S subunit. The sequence 62–73 does not overlap with the Zinc finger, and does not contain fragments involved in the interactions of rpS26e with the 18S rRNA (Figure 5). Moreover, analysis of the 2xzn structure also shows that fragment 62–73 does not interact with any ribosomal protein. Thus, it is reasonable to suggest that fragment 60–71 of rpS26e in the human 40S subunit is not involved in any intraribosomal interaction too. This implies conformational flexibility of the fragment, which could be essential for specific functions of the fragment related to the binding of the mRNA region upstream of the E site codon and of other, eukaryote-specific, ligands.
Figure 5.
Alignment of amino acid sequences of eukaryotic (from lower fungal to mammalian) and archaeal proteins from the rpS26e family. The numbering corresponds to human rpS26e. Conserved protein fragments are marked; more intense color corresponds to the higher extent of conservation. Fragments conserved in all kingdoms are blue; fragments conserved only in eukaryotes are red. Protein residues contacting the 18S rRNA in the Tetrahymena thermophila 40S subunit (PDB accession number 2xzn) (5) are marked by green points above the sequences. Dodecapeptide in positions 60–71 is boxed. The protein sequences were taken from three databases, GenBank (codes ACR, ABP, AEB, AAC, AAL, AAN, AAG), the NCBI Reference Sequence (codes NP, XP, YP) and from the ribosomal protein gene database (http://ribosome.miyazaki-med.ac.jp): H. sapiens, Homo sapiens NP_001020.2; T. tetrahymena, Tetrahymena thermophila XP_001018846.1; M. musculus, Mus musculus NP_038793.2; A. gambiae, Anopheles gambiae AAG15374.1; A. mellifera, Apis mellifera XP_001122510.2; C. elegans, Caenorhabditis elegans ribosomal protein gene database; F. graminearum, Fusarium graminearum ribosomal protein gene database; S. pompe, Schizosaccharomyces pombe ribosomal protein gene database; C. cinera, Coprinopsis cinerea XP_001839003.2; U. maydis, Ustilago maydis XP_761090; R. oryzae, Rhizopus oryzae ribosomal protein gene database; D. discoideum, Dictyostelium discoideum XP_640573.1; F. rubripes, Fugu rubripes ribosomal protein gene database; D. melanogaster, Drosophila melanogaster AAN11004.1; A. thaliana, Arabidopsis thaliana NP_191193.1; A. fumigatus, Aspergillus fumigatus EAL88388.1; B. taurus, Bos taurus NP_001015561.1; L. cinera, lepidochitona cinerea ACR24972.1; O. vilgaris, Octopus vulgaris CAB57819.1; X. tropicalis, Xenopus tropicalis NP_001086904.1; E. bruneus, Epinephelus bruneus AEB31268.1; M. sativa, Medicago sativa AAC77928.1; P. aerophilum, Pyrobaculum aerophilum AAL64000.1; S. acidocaldarius, Sulfolobus acidocaldarius YP_256160.1; M. sedula, Metallosphaera sedula ABP96062.1; P. marinus, Prochlorococcus marinus XP_002769921; C. Korarchaeum, Candidatus Korarchaeum cryptofilum YP_001737625.1; P. aresenaticum, Pyrobaculum arsenaticum YP_001153040.1; P. islandicum, Pyrobaculum islandicum YP_930339.1.
Figure 6.
Positioning of rpS26e on the T. thermophila 40S subunit (PDB accession number 2xzn) (5). General view (on the left) and zoomed structure of rpS26e extracted from the model. Sequence 64–72 corresponding to the conserved motif 62-YxxPKxYxK-70 in human rpS26e is light blue, protein fragments interacting with the 18S rRNA are marked as green, location of the Zinc atom (presented as yellow circle) is highlighted.
Fragment 60–71 (human numbering) of rpS26e contains the motif 62-YxxPKxYxK-70 that is conserved in eukaryotic proteins of the S26e family but not in the corresponding archaeal proteins that contain an archaeal-specific motif in the respective region (Figure 5). This points to a significant role of motif 62-YxxPKxYxK-70 in the functioning of the ribosome in a manner specific for eukaryotes, while the respective archaeal-specific motif could be involved in functioning of the archaeal translating machinery in another manner. However, the latter possibility is not discussed here because of the lack of structural data on archaeal small ribosomal subunits. Analysis of the nature of protein–protein and protein–rRNA interactions in the 2xzn model shows that contacts of proteins with the 18S rRNA are mediated preferentially by lysines and/or arginines and rarely by tyrosines; the latter much more often participate in protein–protein interactions. Therefore, it is reasonable to assume that lysines K66 and/or K70 of the 62-YxxPKxYxK-70 motif interact with mRNA phosphates, while tyrosines Y62 and/or Y68 participate in binding of a protein involved in translation in a manner specific for eukaryotes. Of course, these do not rule out a possibility that Y62 and/or Y68 of rpS26e could be also involved in interactions with mRNA, e.g. to guide/stabilize mRNA conformation via stacking interactions. Proline in position 65 probably provides bending of the polypeptide chain, which can be important to maintain functionally correct conformation of the fragment. One candidate protein that could interact with the 62-YxxPKxYxK-70 motif is the eukaryotic initiation factor eIF3, that is involved in recruitment of the mRNA to the 40S subunit and has no counterparts in bacteria and archaea (34). A comparison of the 2xzn model of the 40S subunit discussed above (5) with available cryo-EM images of the 40S subunit complexed with eIF3 (35) shows that the location of rpS26e on the 40S subunit overlaps with the eIF3 binding site. This agrees with the data on the ability of mRNA nucleotides in positions -8 to -10 to cross-link to rpS26e as well as to eIF3d in the 48S initiation complex (23). Thus, one can suggest that motif 62-YxxPKxYxK-70 of rpS26e is involved in binding of both the 5′-untranslated region of the mRNA and eIF3, and that this interaction underlies a mechanism of mRNA recruitment by eIF3. However, the role of the rpS26e motif mentioned could extend beyond the initiation step, and contribute to the maintenance of the mRNA path from the region of codon-anticodon interactions to the exit site during translation. The 62-YxxPKxYxK-70 motif is partly accessible at the surface of the 40S subunit solvent side (5); it could therefore be a potential target for the action of some kind of regulatory factors modulating ribosomal function during translation.
Remarkably, recently it was reported that K66 from the 62-YxxPKxYxK-70 motif of rpS26e (together with K113) is acetylated in human cells (36). Acetylation is a reversible post-translational modification of proteins playing a key role in regulating gene expression; lysine acetylation preferentially regulates the functioning of large macromolecular complexes involved in diverse cell processes, such as modulation of chromatin architecture, cell cycle, splicing, nuclear transport, etc. [e.g. see (36) and refs therein]. However, rpS26e isolated from rat (37) and human (38) 40S ribosomal subunit lacks any posttranslational modifications. Thus, it is reasonable to suggest that rpS26e is deacetylated before or during ribosome assembly. Since acetylated lysines lack positive charges and are therefore unable to interact with RNA phosphates, acetylation of rpS26e at K66 can deprive the protein of its ability to interact with mRNA. Therefore, deacetylation of the protein involved in ribosome assembly could be of major importance to ensure interaction of its 62-YxxPKxYxK-70 motif with mRNA.
RpS26e lacks eubacterial counterparts. However, in the 40S subunit this protein occupies a site homologous to that for rpS18p in the 30S subunit (5,11) and both proteins neighbor the mRNA region 5′ of the E-site codon in the small ribosomal subunit. This suggests that rpS18p and rpS26e could function similarly in the small ribosomal subunit. Recently, it was found that the conserved structural motifs of rpS18p that contact the rRNA in the eubacterial ribosome, are present in rpS26e, and it was predicted that amino acids K83, R86, R92, K93 and R96 of human rpS26e contacts the 18S rRNA in the 40S subunit (39). Analysis of the 2xzn structure of the 40S subunit (5) shows that all these residues do contact the 18S rRNA together with 15 other amino acid residues that belong to the N-terminal part of rpS26e (positions 4–7, 9–10, 14–17, 19, 32 and 37–39, see Figures 5 and 6). One could expect that positioning of the mRNA with respect to rpS18p in the 30S subunit and to rpS26e in the 40S subunit share common features. However, cross-linking results argue against such similarities. In the 30S subunit, mRNA contacts only the very N-terminal fragment of rpS18p (40) that has nothing in common with the dodecapeptide in positions 60–71 of rpS26e. Moreover, the contacts of rpS18p with the mRNA in the 30S subunit have been observed only in a model initiation complex stabilized by Shine–Dalgarno interactions (40) that do not take place in eukaryotes.
Thus, our study provides new insights into molecular mechanisms of translation in eukaryotes. Motif 62-YxxPKxYxK-70 of rpS26e that is conserved in eukaryotes, is most probably involved in mRNA recruitment by eIF3 during initiation and in maintenance of the mRNA path from the area of codon–anticodon interactions to the exit site during translation. Interaction of this motif with the mRNA is probably mediated by K66 that is subjected to acetylation in the ribosome-free rpS26e, which in turn would protect this motif from interaction with cellular RNAs.
FUNDING
Russian Foundation for Basic Research (11-04-00597 to G.K.); the Program ‘Molecular and Cell Biology’ of the Presidium of the Russian Academy of Sciences (to G.K.). Funding for open access charge: the Program ‘Molecular and Cell Biology’ of the Presidium of the Russian Academy of Sciences (to G.K.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors gratefully thank Anne-Lise Haenni for critical reading of this article.
REFERENCES
- 1.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–292. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 2.Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 3.Steitz TA. A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol. 2008;9:242–253. doi: 10.1038/nrm2352. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 5.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 6.Klaholz BP, Pape T, Zavialov AV, Myasnikov AG, Orlova EV, Vestergaard B, Ehrenberg M, van Heel M. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature. 2003;421:90–94. doi: 10.1038/nature01225. [DOI] [PubMed] [Google Scholar]
- 7.Rawat U, Gao H, Zavialov A, Gursky R, Ehrenberg M, Frank J. Interactions of the release factor RF1 with the ribosome as revealed by cryo-EM. J. Mol. Biol. 2006;357:1144–1153. doi: 10.1016/j.jmb.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 8.Li W, Frank J. Transfer RNA in the hybrid P/E state: correlating molecular dynamics simulations with cryo-EM data. Proc. Natl Acad. Sci. USA. 2007;104:16540–16545. doi: 10.1073/pnas.0708094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandramouli P, Topf M, Menetret J-F, Eswar N, Cannone JJ, Gutell RR, Sali A, Akey CW. Structure of the mammalian 80S ribosome at 8.7 Å resolution. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor DJ, Devkota B, Huang AD, Topf M, Narayanan E, Sali A, Harvey SC, Frank J. Comprehensive molecular structure of the eukaryotic ribosome. Structure. 2009;17:1591–1604. doi: 10.1016/j.str.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armache J-P, Jarasch A, Anger AM, Villa E, Becker T, Bhushana S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. Localization of eukaryote-specific ribosomal proteins in a 5.5-Å cryo-EM map of the 80S eukaryotic ribosome. Proc. Natl Acad. Sci. USA. 2010;107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yusupova GZ, Yusupov MM, Cate JHD, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 13.Baram D, Yonath A. From peptide-bond formation to cotranslational folding: dynamic, regulatory and evolutionary aspects. FEBS Lett. 2005;579:948–954. doi: 10.1016/j.febslet.2004.11.063. [DOI] [PubMed] [Google Scholar]
- 14.Selmer M, Dunham CM, Murphy FM, IV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 15.Jenner L, Demeshkina N, Yusupova G, Yusupov M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat. Struct. Mol. Biol. 2010;17:1072–1079. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- 16.Demeshkina N, Repkova M, Ven'yaminova A, Graifer D, Karpova G. Nucleotides of 18S rRNA surrounding mRNA codons at the human ribosomal A, P and E sites, respectively: a cross-linking study with mRNA analogues carrying aryl azide group at either the uracil or the guanine residue. RNA. 2000;6:1727–1736. doi: 10.1017/s1355838200000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graifer D, Molotkov M, Styazhkina V, Demeshkina N, Bulygin K, Eremina A, Ivanov A, Laletina E, Ven'yaminova A, Karpova G. Variable and conserved elements of human ribosomes surrounding the mRNA at the decoding and upstream sites. Nucleic Acids Res. 2004;32:3282–3293. doi: 10.1093/nar/gkh657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molotkov M, Graifer D, Demeshkina N, Repkova M, Ven'yaminova A, Karpova G. Arrangement of mRNA 3′ of the A site codon on the human 80S ribosome. RNA Biol. 2005;2:63–69. doi: 10.4161/rna.2.2.1756. [DOI] [PubMed] [Google Scholar]
- 19.Stahl J, Kobetz ND. Affinity labelling of rat liver ribosomal protein S26 by heptauridylate containing a 5′-terminal alkylating group. Mol. Biol. Rep. 1984;9:219–222. doi: 10.1007/BF00775350. [DOI] [PubMed] [Google Scholar]
- 20.Stahl J, Karpova GG. Investigation on the messenger RNA binding site of eukaryotic ribosomes by using reactive oligo(U) derivatives. Biomed. Biochim. Acta. 1985;44:1057–1064. [PubMed] [Google Scholar]
- 21.Malygin AA, Graifer DM, Bulygin KN, Zenkova MA, Yamkovoy VI, Stahl J, Karpova GG. Arrangement of mRNA at the decoding site of human ribosomes. 18S rRNA nucleotides and ribosomal proteins cross-linked to oligouridylate derivatives with alkylating groups at either the 3′ or the 5′-termini. Eur. J. Biochem. 1994;226:715–723. doi: 10.1111/j.1432-1033.1994.tb20100.x. [DOI] [PubMed] [Google Scholar]
- 22.Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CUT, Pestova TV. Specific functional interaction of nucleotides at key–3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20:624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CUT, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khairulina J, Graifer D, Bulygin K, Ven'yaminova A, Frolova L, Karpova G. Eukaryote-specific motif of ribosomal protein S15 neighbors A site codon during elongation and termination of translation. Biochimie. 2010;92:820–825. doi: 10.1016/j.biochi.2010.02.031. [DOI] [PubMed] [Google Scholar]
- 25.Bulygin KN, Khairulina YS, Kolosov PM, Ven'yaminova AG, Graifer DM, Vorobjev YN, Frolova LY, Kisselev LL, Karpova GG. Three distinct peptides from the N domain of translation termination factor eRF1 surround stop codon in the ribosome. RNA. 2010;16:1902–1914. doi: 10.1261/rna.2066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bulygin KN, Khairulina YS, Kolosov PM, Ven'yaminova AG, Graifer DM, Vorobjev YN, Frolova LY, Karpova GG. Adenine and guanine recognition of stop codon is mediated by different N domain conformations of translation termination factor eRF1. Nucleic Acids Res. 2011;39:7134–7146. doi: 10.1093/nar/gkr376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matasova NB, Myltseva SV, Zenkova MA, Graifer DM, Vladimirov SN, Karpova GG. Isolation of ribosomal subunits containing intact rRNA from human placenta. Estimation of functional activity of 80S ribosomes. Anal. Biochem. 1991;198:219–223. doi: 10.1016/0003-2697(91)90416-q. [DOI] [PubMed] [Google Scholar]
- 28.Repkova MN, Ivanova TM, Komarova NI, Meschaninova MI, Kuznetsova MA, Ven'yaminova AG. H-phosphonate synthesis of oligoribonucleotides containing modified bases. I. Photoactivatable derivatives of oligoribonucleotides with perfluoroarylazide groups in heterocyclic bases. Russ. J. Bioorg. Chem. 1999;25:612–622. [Google Scholar]
- 29.Smolenskaya IA, Bulygin KN, Graifer DM, Ivanov AV, Ven'yaminova AG, Repkova MN, Karpova GG. Localization of template in the decoding area by affinity modification of human ribosomes with photoactivated derivative of oligoribonucleotide pGUGUUU. Mol. Biol. (translated from Molekul. Biol.) 1998;32:233–241. [PubMed] [Google Scholar]
- 30.Hardy SJS, Kurland CG, Voynow P, Mora G. The ribosomal proteins of Escherichia coli. I. Purification of 30S ribosomal proteins. Biochemistry. 1969;8:2897–2905. doi: 10.1021/bi00835a031. [DOI] [PubMed] [Google Scholar]
- 31.Malygin A, Baranovskaya O, Ivanov A, Karpova G. Expression and purification of human ribosomal proteins S3, S5, S10, S19 and S26. Protein Expression Purif. 2003;28:57–62. doi: 10.1016/s1046-5928(02)00652-6. [DOI] [PubMed] [Google Scholar]
- 32.Demeshkina NA, Laletina ES, Meschaninova MI, Repkova MN, Ven'yaminova AG, Graifer DM, Karpova GG. mRNA codon environment at the P and E sites of human ribosomes deduced from photocrosslinking with pUUUGUU derivatives. Mol. Biol. (Moscow) (translated from Molekul. Biol.) 2003;37:132–139. [PubMed] [Google Scholar]
- 33.Molotkov MV, Graifer DM, Popugaeva EA, Bulygin KN, Meschaninova MI, Ven'yaminova AG, Karpova GG. mRNA 3’ of the A site bound codon is located close to protein S3 on the human 80S ribosome. RNA Biol. 2006;3:122–129. doi: 10.4161/rna.3.3.3584. [DOI] [PubMed] [Google Scholar]
- 34.Rodnina M, Wintermeyer W. Recent mechanistic insights into eukaryotic ribosomes. Curr. Opin. Cell Biol. 2009;21:435–443. doi: 10.1016/j.ceb.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 35.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 36.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 37.Louie DF, Resing KA, Lewis TS, Ahn NG. Mass spectrometric analysis of 40 S ribosomal proteins from Rat-1 fibroblasts. J. Biol. Chem. 1996;271:28189–28198. doi: 10.1074/jbc.271.45.28189. [DOI] [PubMed] [Google Scholar]
- 38.Malygin AA, Karpova GG. Site-specific cleavage of the 40S ribosomal subunit reveals eukaryote-specific ribosomal protein S28 in the subunit head. FEBS Lett. 2010;584:4396–4400. doi: 10.1016/j.febslet.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 39.Malygin AA, Karpova GG. Structural motifs of the bacterial ribosomal proteins S20, S18 and S16 that contact rRNA present in the eukaryotic ribosomal proteins S25, S26 and S27A, respectively. Nucleic Acids Res. 2010;38:2089–2098. doi: 10.1093/nar/gkp1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]