Abstract
Ribonuclease P (RNase P) and RNase MRP are closely related ribonucleoprotein enzymes, which process RNA substrates including tRNA precursors for RNase P and 5.8 S rRNA precursors, as well as some mRNAs, for RNase MRP. The structures of RNase P and RNase MRP have not yet been solved, so it is unclear how the proteins contribute to the structure of the complexes and how substrate specificity is determined. Using electron microscopy and image processing we show that eukaryotic RNase P and RNase MRP have a modular architecture, where proteins stabilize the RNA fold and contribute to cavities, channels and chambers between the modules. Such features are located at strategic positions for substrate recognition by shape and coordination of the cleaved-off sequence. These are also the sites of greatest difference between RNase P and RNase MRP, highlighting the importance of the adaptation of this region to the different substrates.
INTRODUCTION
Ribonuclease P (RNase P) is an endonuclease that cleaves the 5′-leader sequence of pre-tRNAs [for review see (1)]. RNase P is conserved between all taxonomic kingdoms, demonstrating its early appearance in evolution. The complex consists of an RNA subunit [Rpr1 for Saccharomyces cerevisiae, (2)] that forms part of the catalytic core and protein components of variable size. Bacterial RNase P has a single protein component, whereas there are four to five protein subunits in archaea and at least nine in the eukaryotic system [Pop1, Pop3–Pop8, Rpp1, Rpr2 in S. cerevisiae for review see (3)]. In the eukaryotic complex, these proteins comprise more than half of the mass of the ribonucleoprotein (RNP) particle, which has an approximate total mass of 410 kDa.
In addition to RNase P, eukaryotes possess the mitochondrial RNA processing ribonuclease [RNase MRP, (4–5)]. RNase MRP has a related RNA core [Nme1 for S. cerevisiae, (6)] and shares eight protein subunits with RNase P [Pop1, Pop3–Pop8, Rpp1 in S. cerevisiae (7)]. Only two subunits are specific for RNase MRP, Rmp1 (8) (24 kDa) and Snm1 (9) (23 kDa), the latter of which is homologous to Rpr2 in RNase P. RNase MRP has a wider range of known substrates than does RNase P. As its name implies, it was first described as a mitochondrial complex that is involved in processing of an RNA primer required for initiation of DNA replication (4). Subsequent fluorescence studies revealed that RNase MRP, like RNase P, is mainly localized in the nucleolus (10–12) and is involved in processing of pre-mature 5.8 S rRNA (13–15). But it is also involved in regulation of cell cycle by cleavage of specific mRNAs (16). Similar to RNase P, RNase MRP is essential for the survival of the cell.
Many investigations have focused on elucidating the structure of the RNA core, which consists of a catalytic domain (C-domain) and a specificity domain (S-domain), which can bind pre-tRNA directly. Sequence analysis suggests that the catalytic RNA cores of RNase P and RNase MRP form similar secondary structures (17–20), and assemble into related 3D core structures (21–25). Structures have been determined for several protein subunits of RNase P; the archaeal homologs of Pop3, Pop4, Pop5, Rpp1 and Rpr2 (26–34), two eukaryotic proteins Pop6 and Pop7 (35), and the bacterial protein (36). This is complemented by a recent structure of the whole bacterial complex (24) with bound tRNA. However, the structures of the archaeal and eukaryotic particles, with more complex protein components, are still unknown. Thus, the exact role of the additional proteins in these complexes is still poorly understood, despite most of the individual protein structures being known.
Analysis of scans of gels of RNase P/MRP stained with SYPRO ruby provide additional information on the relative subunit composition suggesting that most protein subunits are present in multiple copies (8). These data are complemented by protein–protein interaction studies in different species (37–41), which among other interactions repeatedly report interactions between homologs of Pop4–Pop5, Pop4–Rpr2, Pop5–Rpp1 and Pop6–Pop7. Apart from the Pop4–Pop5 subcomplex, all of the predicted binary complexes have been crystallized either from archaea or from yeast (27,29,35).
Even with this wealth of information, there is still no consensus model for the architecture of eukaryotic RNase P and RNase MRP, due mainly to a lack of structural information on the holoenzymes. Therefore, we have used electron microscopy (EM) and single particle image processing to determine structures of both complexes at a resolution of ∼1.5–1.7 nm. These structures reveal the modular architecture of RNase P and MRP and show differences in the substrate-binding cavity.
MATERIALS AND METHODS
Yeast strains
Genomic integration of the HA tag (HIS3MX6-marker) was performed to create fusion proteins of Rpr2 (HA tag C-terminally) and Snm1 (HA tag N-terminally) as described (42) into the S. cerevisiae strain YSW1 (MATa POP4::TAPTAG::TRP1ks pep4:LEU2 nuc1::LEU2 sep1::URA3 trp-1his3–11,15 can-100 ura3–1 leu2–3,112) described previously (9). Snm1 was tagged N-terminally due to frequent C-terminal degradation of Snm1. Presence of the HA tag in the fusion proteins was confirmed by western blot. Genomic integrations of the green fluorescent protein (GFP) tag were performed on above described Rpr2-HA strain by tagging the C-terminus of Pop5, Pop6, Pop8 or Rpp1 with yeGFP tag (hphNT1 marker) and N-terminus of Rpr2-HA with promoter Gal1 : yeGFP tag (natNT2 marker) as described (43). Presence of the GFP tag in the fusion proteins was confirmed by western blot.
Affinity purification
Cells, grown in 12 l of YPD, were harvested at OD 3.0–3.5 and disrupted with glass beads in lysis buffer [50 mM Tris–HCl pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT, protease inhibitor FY (Serva), 0.0375% NP40] using a Pulverisette 5 (Fritsch, three cycles milling with power set to 350 for 4 min, 1 min break between cycles). TAP-tagged Pop4 protein was purified from cell lysate as described previously (44) until the step of the TEV eluate, which was then loaded on an HA affinity matrix (Roche) and incubated for 5 h at 4°C. The bound fraction was eluted via competition with the HA peptide (Roche) for 30 min at 37°C. The HA eluates were concentrated, analyzed using NuPAGE 4–12% gradient or 12% SDS–PAGE (Invitrogen) stained with colloidal Coomassie (Sigma) and verified by mass spectrometry. Furthermore, the separation of RNase P and RNase MRP was tested by quantitative RT–PCR.
For antibody labeling, the TEV eluate was split into two halves and one half was processed as described before. To the other half, anti-GFP monoclonal antibodies (Roche, 40 µg/ml final concentration) were added after 1 h incubation on the HA matrix and further incubated for 4 h.
Mass spectrometry
Mass spectrometry using tryptic digest from Coomassie-stained SDS–PAGE was performed as described previously (45). Proteins were identified using Mascot (Matrix Science) and the MSDB protein database.
Quantitative real-time reverse-transcription PCR
The HA eluates of Pop4–TAP Rpr2–HA and Pop4–TAP HA–Snm1 were treated with Proteinase K (10 µg/ml) to digest the proteins. The RNAs were then extracted by standard phenol/chloroform extraction (46). Contaminant DNA was digested by DNase I treatment.
Nme1 RNA (RNase MRP) and Rpr1 RNA (RNase P) were reverse-transcribed using Transcriptor First Strand cDNA Synthesis Kit (Roche) at 60°C to improve the efficiency of the reaction by unfolding the RNA. The primers used were 5′-TCCATTGGGTTACTCGATCC-3′ for Nme1 RNA and 5′-TGGAACAGCAGCAGTAATCG-3′ for Rpr1 RNA.
qPCR was performed using the SYBR®GREEN PCR Master Mix (Applied Biosystem) following the manufacturer's instructions. Reactions were performed in triplicate from three different purifications of each complex using an ABI 7500 according to these conditions: 50°C for 2 min and 95°C for 10 min followed by 40 amplification cycles (95°C for 15 s and 60°C for 1 min). A dissociation stage was performed to check the specificity of the reactions. Pairs of primers (5′-CGGTACTGGATTCCGTTTGT-3′ and 5′-AGCACGGGAAAGAGCAATC-3′ for Nme1 cDNA and 5′-CCGGGTTAATGTCGCTTTT-3′ and 5′-CGGTCGGTAAAGACTGGTTC-3′ for RPR1 cDNA) were designed by Primer3. The three pairs of primers were chosen with care taken that they did not anneal the regions of secondary structure of target molecules predicted by Mfold (47). Primer efficiencies were tested as described previously (48).
The ratio of the RNA subunits of RNase P (Rpr1) and RNase MRP (Nme1) was calculated as follows:
[Rpr1]/[Nme1] = 2(Ct Rpr1 – Ct Nme1) = 2Δct
Cleavage assay
Six micrograms of premature tRNA were incubated with 3 µg affinity purified RNase P (approximate molar ratio 20:1) in buffer (50 mM Tris–HCl pH 8.0, 100 mM NH4Cl, 10 mM MgCl2 or 2 mM EDTA) for 30 min at 37°C (49). Reactions were stopped by adding 9 M urea, 1 mM EDTA, 1 mg/ml bromophenol blue in 1× TBE (89 mM Tris, 89 mM boric acid; 2 mM EDTA). The mixtures were denatured for 1 min at 95°C, and separated on a 12% polyacrylamide / 7 M urea gel. The gel was stained with 0.1% toluidine blue.
Pre-tRNA synthesis
Saccharomyces cerevisiae genomic DNA was used as the initial PCR template to amplify the tRNA Tyr gene (GenBank accession number J01380) with primers described in (50) and the tRNA Glu gene [tE(CUC)D GenBank accession number X06132.1] with primers ptRNA-Glu_F (5′-GTAATACGACTCACTATAGAGGTCAAACTATCACCAAGGTACA-3′) and ptRNA-Glu_R (5′-AAAAAGAAACTCCGAAGCGG-3′). The PCR products were transcribed using T7 Transcription kit (Fermentas, #K0412) to produce pre-tRNATyr and pre-tRNAGlu, respectively, following the manufacturer's instruction.
Electron microscopy
Grids were prepared by sandwich negative staining (51). For random conical tilt reconstruction, pairs of tilted (−55°) and untilted fields were recorded on a Philips CM200 FEG on a 2kx2k CCD (TVIPS GmbH) at a nominal magnification of 27 500 (5.2 Å/pixel) at room temperature. Micrographs were recorded under low dose conditions. Tracking of the area of interest was done manually.
For high-resolution image analysis, grids were prepared as outlined above and frozen in liquid nitrogen immediately after drying. Thereafter, grids were kept at liquid nitrogen temperature. Grids were transferred to the CM200 FEG with a Gatan 626 cryo-transfer holder. Micrographs were recorded on CCD at a nominal magnification of 66 000 (2.2 Å at specimen level) at 200 kV under low dose conditions. 2006 micrographs were collected for RNase MRP and 1861 micrographs for RNase P, respectively.
For antibody labeling, grids were prepared by sandwich negative staining (51), and microscopy and imaging were performed as described for random conical tilt reconstruction.
Image processing
Initial maps of RNase P and RNase MRP were calculated by random conical tilt reconstruction using Spider (52) for particle selection and windowing and Imagic 5 (53) for alignment, classification and reconstruction. The relative orientations of some of the random conical tilt maps were determined and used to calculate a new map from the tilted particles contributing to the successfully aligned maps. The maps were further refined by projection matching of the tilted and untilted images (Supplementary Figure S1 and Supplementary Methods).
These refined maps of RNase P and RNase MRP were used to generate starting references for iterative refinement by projection matching (Spider) of the larger datasets of negatively stained, frozen RNase P (42 009 particle images) and RNase MRP (59 400 particle images). For further processing, the effects of the contrast transfer function were reduced by appropriate phase reversal (Supplementary Methods). In addition, for each complex, the data was processed in 20 defocus groups; for each defocus group, a map was calculated by weighted back projection using Spider. The maps were added to give the final map (Supplementary Data).
Maps of RNase P and RNase MRP were similar but also had distinct differences. In order to test whether these differences were linked to variability in the image processing or to true differences between RNase P and RNase MRP, both maps were combined to a common starting reference for iterative refinement of both complexes. Images of RNase P and RNase MRP were processed separately. In the subsequent refinement, the current best map of each complex was used as a new reference. Refinement was continued until most orientations of particle images did not change and no further improvement in resolution was observed (see Supplementary Data for more details on iterative refinement). Refinement converged to two distinct maps, which showed similar key-differences as previously observed by starting with individual random conical tilt maps for RNase P and RNase MRP.
Furthermore, we assessed the heterogeneity of the particle populations for RNase P and RNase MRP separately following two approaches. (i) We used Xmipp Maximum-likelihood 3D-sorting (54) to sort particle images of either RNase P or RNase MRP into five classes. (ii) We used IMAGIC 4D, to generate 10 maps from particles randomly assigned to the 10 groups and then iteratively refined in projection matching and supervised classification. Both approaches showed small differences between the maps, but no indication for major intrinsic flexibility or heterogeneous composition of the particles in the population. This justified the refinement of particle images against one representative map in retrospect.
To check whether the orientations were assigned consistently, we sub-classified particle images, which were assigned with similar Euler angles using multivariate statistical analysis (see Supplementary Methods and Supplementary Figure S2). Visual inspection showed that the averages of the sub-classes had similar features as the reference projections (Figure 1D and Supplementary Figure S2), further highlighting that the majority of particle images was assigned with a correct overall orientation.
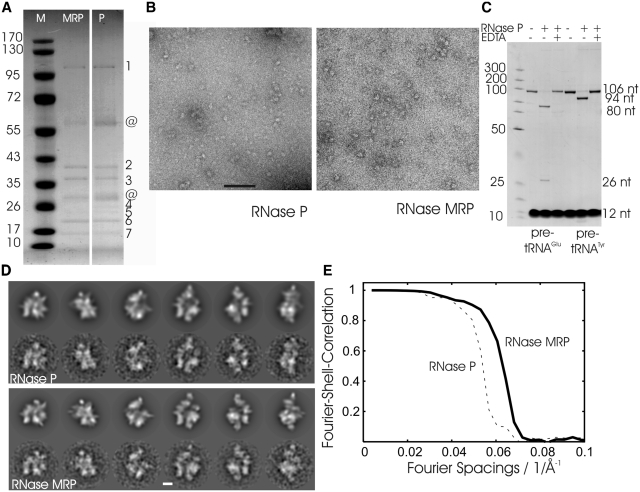
Figure 1.
Characterization of the RNase P/MRP preparation. (A) Coomassie-stained SDS–PAGE of RNase P and RNase MRP. Left: marker with given molecular masses (Page Ruler™ Prestained Protein Ladder; Fermentas) in kDa; centre: HA eluate of Pop4–TAP HA-Snm1 (RNase MRP); right: HA eluate of Pop4–TAP Rpr2–HA (RNase P). Bands identified by mass spectrometry are indicated by numbers: (1) Pop1; (2) Pop4; (3) Snm1, Rpp1 for RNase MRP and Rpp1 for RNase P; (4) Nothing for RNase MRP and Rpr2 for RNase P; (5) Pop3, Rmp1, Snm1 for RNase MRP and Pop3, Pop1 for RNase P; (6): Pop5, Pop6, Pop7; (7): Pop8; @ contaminations. (B) Micrographs of negatively stained RNase P (left panel) and RNase MRP (right panel) from representative preparations. Scale bar: 100 nm. (C) In vitro activity assay of RNase P using two different pre-tRNAs. Leader sequences (26 nt and 12 nt for pre-tRNAGlu and pre-tRNATyr, respectively) are cleaved off by RNase P to give rise to tRNAGlu (80 nt) and tRNATyr (94 nt) with mature 5′-ends. No cleavage of pre-tRNA molecules occurs in the absence of RNase P or in the presence of RNase P with EDTA added to the reaction showing the dependency of the catalysis on magnesium ions. The reaction mixtures were separated on 12% polyacrylamide / 7 M urea gels and stained with 0.1% toluidine blue. The strong bands at the bottom are from bromphenol blue in the sample buffer. The position of marker fragments (GeneRuler™, UltraLow Range DNA Ladder, Fermentas) in bp is shown on the left. (D) Selected projections of the 3D map of RNase P (upper panel, upper row) and RNase MRP (lower panel, upper row) and matching class-averages calculated by multivariate statistical analysis (lower rows). Scale bar: 5 nm. (E) Fourier-shell-correlation for maps of RNase P (dashed) and of RNase MRP (solid).
Resolution of the maps was estimated by Fourier shell correlation [FSC, (55)]. The FSC was 0.5 (56) at a spatial frequency of 1/1.7 nm−1 for RNase P and 1/1.5 nm−1 for RNase MRP [FSC = 0.14 (57) at 1/1.6 nm−1 for RNase P and 1/1.4 nm−1 for RNase MRP; Figure 1], respectively.
Image processing of antibody labeled particles
For image analysis in antibody labeling experiments, particle images were selected from micrographs using Boxer, which is part of EMAN (58). Subsequent alignment and classification were done with IMAGIC 5 (53) and were repeated until classes remained stable. To localize additional densities in the periphery of particles arising from bound antibodies, class averages were sub-classified using a mask focusing on the periphery of the particles.
Fitting of atomic models
Chimera (59) was used to represent results graphically and to fit atomic models. Some components [Pop6–Pop7–P3 subcomplex (35) and the archaeal homolog of the 2:2 Pop5–Rpp1 subcomplex (29)] were fitted automatically to the segmented map using the Segger option of Chimera (fit to groups of regions). Other components [model of the Schizosaccharomyces pombe RNA (60), the archaeal homolog of the Rpr2–Pop4 subcomplex (27) and the L7Ae protein, which is homologous to Pop3 (34)] were placed manually, taking constrains from published interaction studies and our antibody labeling into account. The manual fits were further optimized with the fit to map option of Chimera, which performs a local optimization. The fit of these components was further validated by calculating local cross-correlations between the map and the fitted components (Supplementary Figure S3).
RESULTS
RNase P and RNase MRP share a highly similar protein subunit composition. Therefore, a two-step affinity purification strategy was employed to separate both complexes. A strain with genomically integrated TAP tagged Pop4 subunit was used for tagging either Rpr2 (unique to RNase P) or Snm1 (unique to RNase MRP) with an HA tag. Affinity tags on different subunits led to efficient separation of RNase P and RNase MRP as determined by SDS–PAGE followed by mass spectrometry (Figure 1A) and further confirmed by quantitative RT–PCR (see also Supplementary Figure S4). Preparations of negatively stained RNase P and RNase MRP show a homogeneous spread of elongated particles of similar size (Figure 1B). The purified RNase P was active in cleaving pre-tRNA in the presence of magnesium ions indicating the recovery of a functional enzyme (Figure 1C).
For structure determination by EM, RNase P and RNase MRP were prepared by cryo-negative staining, which keeps residual structural water and reduces flattening compared to conventional negative staining. A total of 42 104 individual particle images for RNase P and of 59 104 for RNase MRP were selected for further processing. The final reconstructions were calculated from 24 373 (RNase P) and 32 232 (RNase MRP) particle images, respectively. The longest axes of the complexes were ∼20 nm. Visual inspection showed that projections of the reconstructions agreed well with class averages calculated by multivariate statistical analysis (Figure 1D) and showed no major heterogeneity in sub-classes (Supplementary Figure S2) highlighting the validity of the 3D maps. The maps had a resolution of 1.7 nm for RNase P and 1.5 nm for RNase MRP (Figure 1E). However, considering that the particles were stained, these maps only reproduce the stain-accessible surfaces of the particles but not the internal density distribution of protein and RNA.
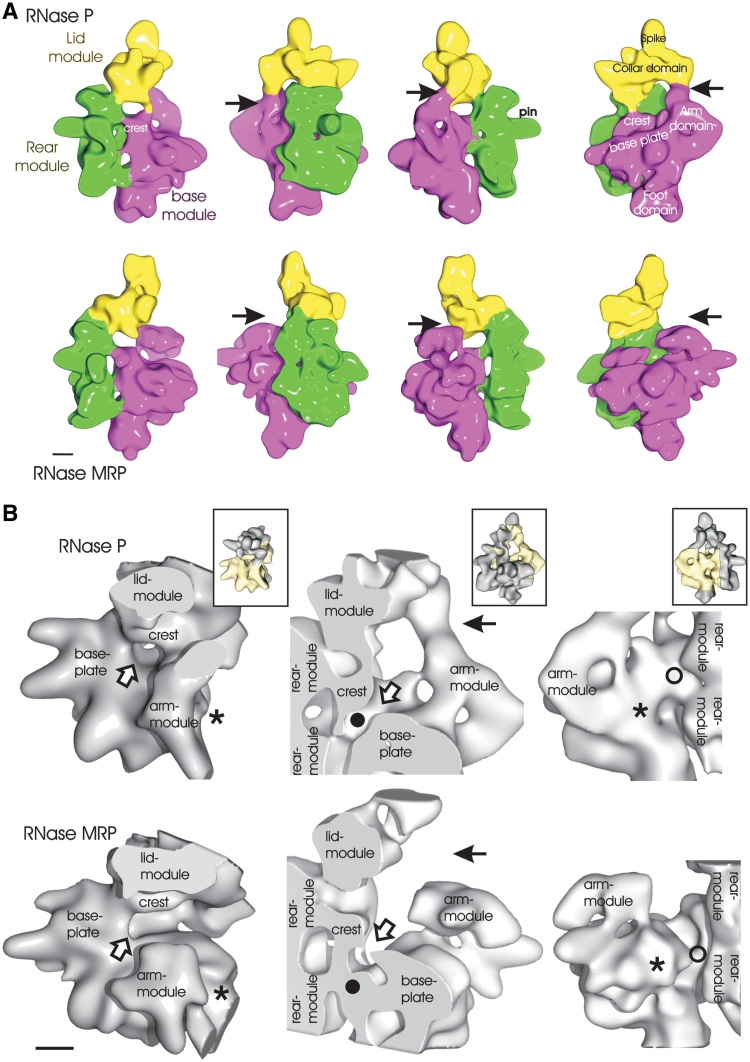
RNase P and RNase MRP consist of three modules
Image reconstruction of RNase P and RNase MRP revealed that both enzymes share a similar architecture (Figure 2). RNase P/MRP consists of three modules, which form a cavity in their centre. The boundaries of the modules were defined by their limited contacts with neighboring modules. We refer to these modules as the base, rear and lid modules (see Figure 2A for labeling).
Figure 2.
Surface representation of RNase P and RNase MRP. (A) RNase P is shown in the top panel and RNase MRP in the bottom panel. The maps are rotated anti-clockwise in 90° steps around the long molecular axis. The different modules are colored, and the names of the modules are shown in the same color: base module (magenta); rear module (green); lid module (yellow). Different landmarks in the map are indicated. The arrow points at the interface between arm domain and lid module, which are in contact in RNase P and are separated in RNase MRP. (B) Close-up views of selected regions of RNase P (top) and RNase MRP (bottom), which differ most in the two complexes. In the inserts the whole complex is shown in the same orientation as the respective close-up and the part of the close-up is highlighted in yellow. Left panel: the entry to the channel (open arrow) is enclosed in RNase P and open to one side in RNase MRP (open arrow). RNase MRP has additional density close to the arm module (asterisk), which is missing in RNase P (asterisk). Central panel: RNase P and RNase MRP are cut at the same position through the centre of the channel. The channel is open in RNase P (filled circle) and blocked in RNase MRP (filled circle). The arm module is connected to the lid module in RNase P and separated in RNase MRP (arrow). Right panel: the channel is protected to the outside in RNase P (unfilled circle) and accessible in RNase MRP (unfilled circle). The additional density of RNase MRP (asterisk) is close to the arm module and gives rise to a protrusion that is missing in RNase P (asterisk). Scale bar: 2 nm.
Base module
The base module consists of the base plate, a flat trapezoidal entity with alternating ridges and grooves, a globular foot domain underneath it and an arm domain that contacts the lid module in RNase P but not in RNase MRP (Figure 2A). The base plate forms the largest surface of the central cavity and rises to a crest close to the rear module. In RNase P, the crest occupies both sides of the entry to a channel that is connected to chambers between the base module and the rear module (Figure 2B, channel entry: open arrows). In contrast, in RNase MRP the crest is located to one side of the channel entrance, resulting in more elongated access to the chambers between the base and rear module (Figure 2B, open arrow). In RNase P, the channel traverses the complex in a straight line, whereas it is blocked in RNase MRP (Figure 2B, block: filled circle). This coincides with the channel being accessible from the outside (Figure 2B, unfilled circle) in RNase MRP and being enclosed in RNase P.
The arm domain emerges at the other side of the base plate. In RNase P the arm domain is thinner and more elongated and contacts the lid module (Figure 2A and B, solid arrow), whereas in RNase MRP the arm is more globular and is not connected to the lid module. RNase MRP has additional density close to where the arm module emerges from the base plate, which is absent from RNase P (Figure 2B, asterisk).
Lid module
The lid module is most variable between different reconstructions, which might be caused by flexibility or high susceptibility to degradation. The module consists of two domains, which we refer to as the collar and the spike domains (Figure 2A for labeling). In RNase P, the collar domain connects the lid module to the arm domain of the base module and to the rear module. In contrast, the lid module in RNase MRP is only connected to the rear module.
Rear module
The rear module consists of two similar domains, which are related by ∼2-fold symmetry, suggesting a dimeric building block. The lower half of the dimeric core contributes to the channel, whereas the upper half anchors the lid module. The rear module is connected to the base module at several defined contact points. The upper half contacts the crest whereas the lower half contacts mainly the lower part of the base plate.
Localization of the subunits in RNase P/MRP
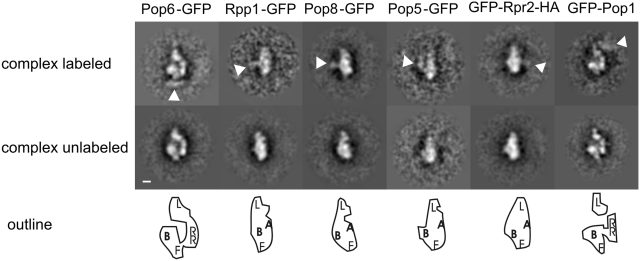
To build a pseudo-atomic model of RNase P/MRP, we wanted to fit the various high-resolution structures of homologous subunits and subcomplexes from different species as well as a structural model for the RNA subunit of S. pombe (60) into our EM maps. However, at the current resolution there were ambiguities where to place some of the high-resolution structures. We therefore localized several subunits of RNase P/MRP by EM and image analysis of complexes with antibody labeled subunits. For labeling we either fused GFP N-terminally (Pop1, Rpr2–HA) or C-terminally (Pop6, Pop8, Pop5, Rpp1) to the subunit of interest. In our hands the GFP tag was too small to be directly detectable in difference imaging. To increase the size we bound a commercially available monoclonal antibody to GFP. Labeling rates for the antibodies were typically <10%, which is in the range of labeling efficiency that we obtain for other complexes (61–63). Labeled complexes were observed in preferred orientations, which is possibly due to the additional GFP moiety on one of the protein subunits of the complex. The preferred orientation makes mapping of the epitopes in 3D impossible. However, labeled complexes could be identified by classification techniques and the approximate position of the GFP–antibody complex in the particles could be determined (Figure 3). The 2D distribution of labels localized Rpp1, Pop5, Pop8 and Rpr2 to the centre of the particle. The positions of Pop8 and Rpp1 were virtually superimposed. However, due to the lack of 3D information it was impossible to distinguish whether these subunits were based in the rear-module or close to the base plate. Pop6 was located in the foot domain and Pop1 mapped to the lid module.
Figure 3.
Localization of RNase P subunits by antibody labeling. Sub-class averages with an extra density are shown in the upper row, in contrast to sub-class averages with no extra densities shown in the middle row. White arrowheads point to the position of the extra density due to the bound antibody. Scale bar: 2 nm. The bottom row shows the outline of the class averages of the unlabeled complex above. For clarity, the outline is enlarged by a factor of 2. The approximate positions of the domains are indicated by letters: A: arm domain, L: lid module; F: foot domain; B: base plate; RR: rear module [only shown in side views (panel 1 and 6) where it does not overlap with the arm module and base plate].
Pop1 has a molecular mass of 101 kDa and large enough to account for the complete lid module. Indeed, the map of the lid module was 100 000 Å3 in RNase P, which is somewhat smaller than expected for a 100-kDa protein. Mass spectrometry on typical purifications showed that Pop1 was partially degraded. This was also frequently observed in western blots of C-terminally labeled Pop1–GFP using anti-GFP (data not shown). This instability of Pop1 is consistent with the structural variability that we observe for the lid module and the underestimation of the volume.
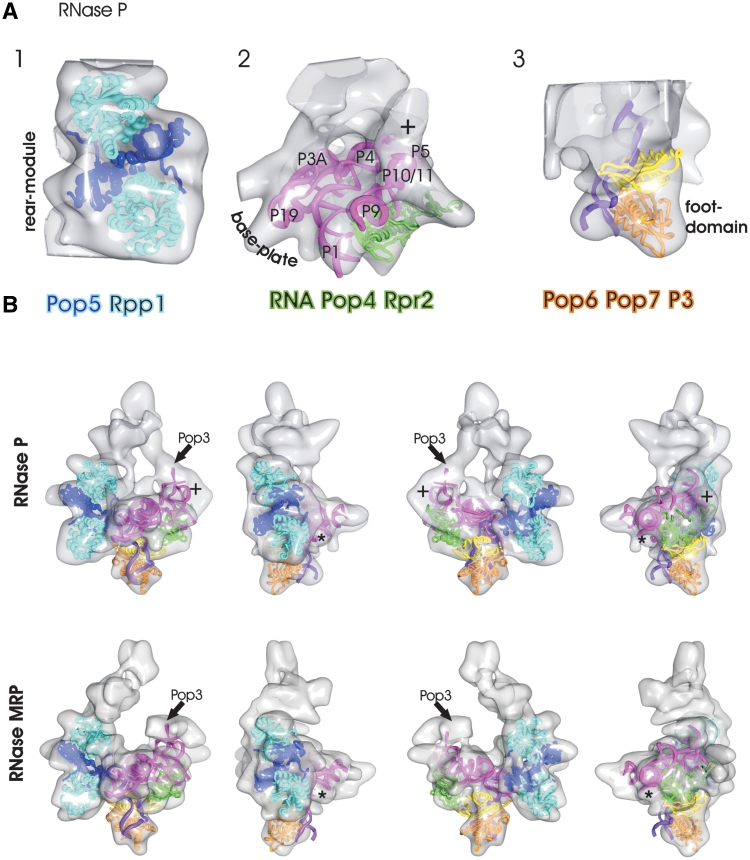
Pop5 and Rpp1 form the rear module
The archaeal homologs of Rpp1 and Pop5 form a 2:2 complex in crystals (29). At low resolution, the structure of this subcomplex closely resembles the dimeric core of the rear module (Figure 4A, panel 1). The rear module also gives the best fit in automatic fitting of the crystal structure of the Rpp1–Pop5 complex to segments of the segmented map (cross-correlation = 0.89 to the respective segment). This position is also consistent with our localization experiments that indicated that Rpp1 and Pop5 were located in the centre of the particle. Therefore, the Rpp1–Pop5 complex was fitted into the rear module. In this fit, the positively charged side of Pop5 faces the chamber between the base module and the rear module. The Rpp1–Pop5 complex accounts for most of the density in the rear module apart from the pin (see Figure 2A for definition). This feature, close to the local symmetry axis, suggests some deviation from the 2:2 organization. Possibly Pop8, which is mapped with antibody to the same position as Rpp1 (Figure 3) has replaced one of the related Pop5 proteins.
Figure 4.
Surface representations of the RNase P with fitted subunits: (A) Panel 1: heterotetramer [2CZV (29)] of the archaeal homologs of Pop5 (dark blue) and Rpp1 (light blue) are fitted to the rear module. Panel 2: model of S. pombe RNA (60) (magenta) and archaeal homologs of the Pop4–Rpr2 dimer [2ZAE, green (27)] fitted to the base plate. Panel 3: Pop6 (orange), Pop7 (yellow) and P3 loop (purple) [3IAB, (35)] fitted to the foot domain. (B) Fitted subunits (see A for color coding) superimposed to RNase P (top panel) and to RNase MRP (bottom panel). The views are rotated in 90°-steps around the y-axes. The ‘plus’ highlights unaccounted density at the end of the P10/P11 stem where 50 bases of the P12 stem are missing in the RNA model. The ‘asterisk’ highlights an unaccounted rod-shaped density at the start of the P3 bulge loop where 30 bases are missing in the RNA model. The arrow highlights the approximate position of Pop3. The local cross correlation between the maps and the fitted models are shown in Supplementary Figure S3.
The RNA subunit forms the base plate
Binding of Pop5 and Rpp1 to the RNA subunit leads to a protection of the RNA against nucleases around the conserved catalytic region formed by stems P3, P4 and P5 (64). This puts the RNA subunit with its catalytic centre into close proximity to the rear module (see also Supplementary Figure S5 for overview), which is consistent with placing it into the base plate. The modeled RNA subunit of RNase P from S. pombe (60), which is considerably smaller than the RNA subunit of S. cerevisiae, has a similar trapezoidal shape as the base plate. The modeled RNA subunit shows a pattern of alternating ridges and grooves that closely resembled the features of the base plate. Placing the RNA model into the density of the base plate with stems P3A, P4 and P5 facing the rear module brings the catalytic core of the RNA subunit into close proximity to the channel entry and the crest (Figure 4A, panel 2). At the opposite side of the base plate, the P10/11, P9, P1 and P19 stems in the RNA model (60) approximately match the ridges.
The RNA model lacks a further 44 bases including the P3 bulge loop (see also Supplementary Figure S5). According to the fitting of the RNA model into the density map, the stem (P3A) leading towards the missing P3 bulge loop points towards the rear module of RNase P and RNase MRP, where an unaccounted density loops back towards the foot domain (Figure 4B, denoted by asterisk). It is likely that at least part of this density is accounted for by the stem leading to the P3 bulge loop.
The RNA model also lacks 50 bases of the P12 stem at the end of the P10/P11 stem (see also Supplementary Figure S5). According to the fit, the P12 stem points directly towards the arm domain (Figure 4A, denoted by plus), suggesting that RNA forms at least part of the arm domain. In agreement with this assumption, fitting the specificity domain of the RNA subunit of the bacterial complex (24) into our EM-map showed the close resemblance of the shapes (Supplementary Figure S6) despite the differences in secondary structure.
Pop4 and Rpr2/Snm1 stabilizes the S-domain of the RNA subunit
Pop4 tightly interacts with Rpr2 (38,40), and a solution structure of the archaeal homolog of this complex has been determined (64). Binding of this complex to the RNA subunit mainly protects regions in the stems of the specificity domain (S-domain, P9, P10, P11 and P12) (64). Our fitting of the RNA model of S. pombe places this close to the arm module (Supplementary Figure S7 position 4), where there is unaccounted density underneath the base plate (Figure 4A, panel 2). This is consistent with our localization experiments of Rpr2 but incompatible with studies that report an interaction of Pop5 with Pop4 (37–38,41). A Pop5–Pop4 interaction would place the Pop4–Rpr2 subcomplex close to the rear module, but we did not observe sufficient density in this region to accommodate the structure of the Pop4–Rpr2 homologs. The Pop4–Rpr2 complex is therefore more likely to be positioned close to the arm domain. Our labeling experiments do not allow us to distinguish unequivocally between positions close to the rear module or adjacent to the arm domain.
Pop6 and Pop7 form the foot domain
The P3 stem of the RNA subunit binds the Pop6–Pop7 heterodimer (65). Our labeling studies localized Pop6 of this subcomplex in the foot domain, which would be consistent with the emerging P3 stem. Recently, the structure of the whole subcomplex consisting of Pop6, Pop7 and the P3 stem has been solved (66). Automatic fitting of the Pop6–Pop7–P3 complex to the segmented map (Chimera segger option) also gave the best match for the foot domain (correlation 0.84 to the respective segment) further highlighting the validity of the assignment. Fitting this subcomplex into the foot domain showed that the Pop6–Pop7–P3 subcomplex accounts for it completely (Figure 4A, panel 3). We obtained the best fit with the P3 stem pointing towards the rear module (Supplementary Figure S7, position 1) close to the missing 44 nt in the RNA model, supporting our fitting of the modeled RNA subunit of S. pombe to the base plate.
DISCUSSION
We have determined the structures of eukaryotic RNase P and RNase MRP at intermediate resolution and mapped a number of subunits in these complexes. Superimposition of known protein structures and a computational model of the eukaryotic RNA subunit onto the EM structure allowed us to build pseudo-atomic models of the RNase P and MRP complexes. The model is consistent with foot-printing data between subunits and the RNA subunit (64,67), our labeling experiments and the shape of the high-resolution structures. The supporting evidence is summarized in Supplementary Table S1 and reported interactions between protein and the RNA-subunit are highlighted in the pseudo-atomic model in Supplementary Figure S7. For RNase P this model accounts for ca. 75% of the observed volume (Figure 4B), lacking only Pop1 in the lid module and individual domains of the RNA and some proteins. The model, together with our maps of RNase P and RNase MRP, offers important insights into the role of the protein components in the particles. Furthermore, the structural differences between RNase P and RNase MRP suggest a mechanism for substrate recognition by RNase P and its differences from RNase MRP.
Proteins in RNase P/MRP support the structure of the RNA
During evolution from bacteria to archaea and eukaryotes, the RNA subunit of RNase P/MRP became smaller, while additional protein components were gained. This led to the assumption that a major function of the protein subunits is stabilizing the folding of the RNA. Our model suggests that this is true for the Pop4 and Rpr2 subunits. These were acquired in archaea and retained in eukaryotes and bind underneath the arm domain, which is formed by the S-domain of the RNA subunit. In RNase MRP, Rpr2 is replaced by the larger Snm1 subunit. The S-domain also differs between RNase P and RNase MRP, much more than the rest of the RNA subunit, suggesting that binding to this area involves a specifically adapted subunit. It has been reported that Snm1 interacts with Rmp1 (68), another protein that is unique to RNase MRP and has no counterpart in RNase P. Consistent with Snm1 being larger than its homolog Rpr2 and the additional presence of Rmp1, we observe extra density close to the arm region in RNase MRP (Figure 2B, asterisk) next to the predicted location of Rpr2.
Pop3 is another protein that potentially binds to the arm domain. It is homologous to the ribosomal L7Ae/L30e protein (69), which is incorporated into the archaeal enzyme (70) and elevates the optimal temperature for catalytic activity (71). Pop3 binds to kink-turn motifs, which are predicted for the P12 stem of the RNA subunits of RNase P and MRP from various species (72). Binding of Pop3 to the P12 stem is supported by cross-link data (73) and GST-pull downs (41). An interaction with P12 would place Pop3 in the arm regions of RNase P and MRP, which are large enough to accommodate Pop3 at similar positions with respect to the base plate (see Figure 4B, arrow). However, the resolution of the maps is insufficient to unambiguously determine the exact orientation of Pop3. At the assigned position, Pop3 would be strategically placed for mediating substrate recognition and substrate binding at a site distant from the catalytic core. Such a role in enhancing and stabilizing substrate binding would be compatible with the requirement for Pop3 binding in catalysis at elevated temperatures (71).
The protein subunits Pop6 and Pop7 form a heterodimer and are specific to eukaryotes. Pop6–Pop7 binds to the P3 stem of the RNA subunit in both RNase P and RNase MRP (65) and account for the foot domain. The Pop6–Pop7 heterodimer holds the P3 stem in an orientation where it kinks back and is positioned directly underneath the base plate. This location is similar to the P15.2 stem of the bacterial B-type RNA (23) and the P18 stem of the bacterial A-type RNA (25), which also loop underneath the core of the RNA subunit. This suggests that the P15.2/P18 stem of bacteria might have been replaced by the P3 stem, together with the associated Pop6–Pop7 heterodimer, in eukaryotes. A channel is formed between the foot domain and the rear module, providing a protective environment that restricts the conformational space for the RNA substrate that is cleaved off.
Proteins contribute to substrate binding
Other subunits of RNase P/MRP are less tightly intermingled with the RNA subunit and are, therefore, likely to contribute to functions other than direct stabilization of RNA folding. Among these subunits are Pop5 and Rpp1, which form a heterotetramer (29) that comprises the rear module. Pop8 belongs to the same family as Pop5 (72) suggesting that in yeast one of the two Pop5 proteins in the heterotetramer is replaced by the smaller Pop8. This would be consistent with the slight asymmetry observed in the rear module and with our localization of Pop5 and Pop8 at similar positions adjacent to Rpp1.
The rear module is comprised of Pop5 and Rpp1, which were acquired in archaea. The rear module binds to the RNA subunit, forming extensive cavities and channels between the rear and base modules (base plate and foot-domain). As for the foot domain, the cavities are strategically positioned to contribute to coordinating the cleaved RNA sequence. Pop5–Rpp1 in eukaryotes and the protein in bacteria fulfill a similar function in enhancing catalysis (67). This suggests that the rear module is a replacement for the bacterial protein, but provides more sophisticated coordination of the cleaved-off leader sequence.
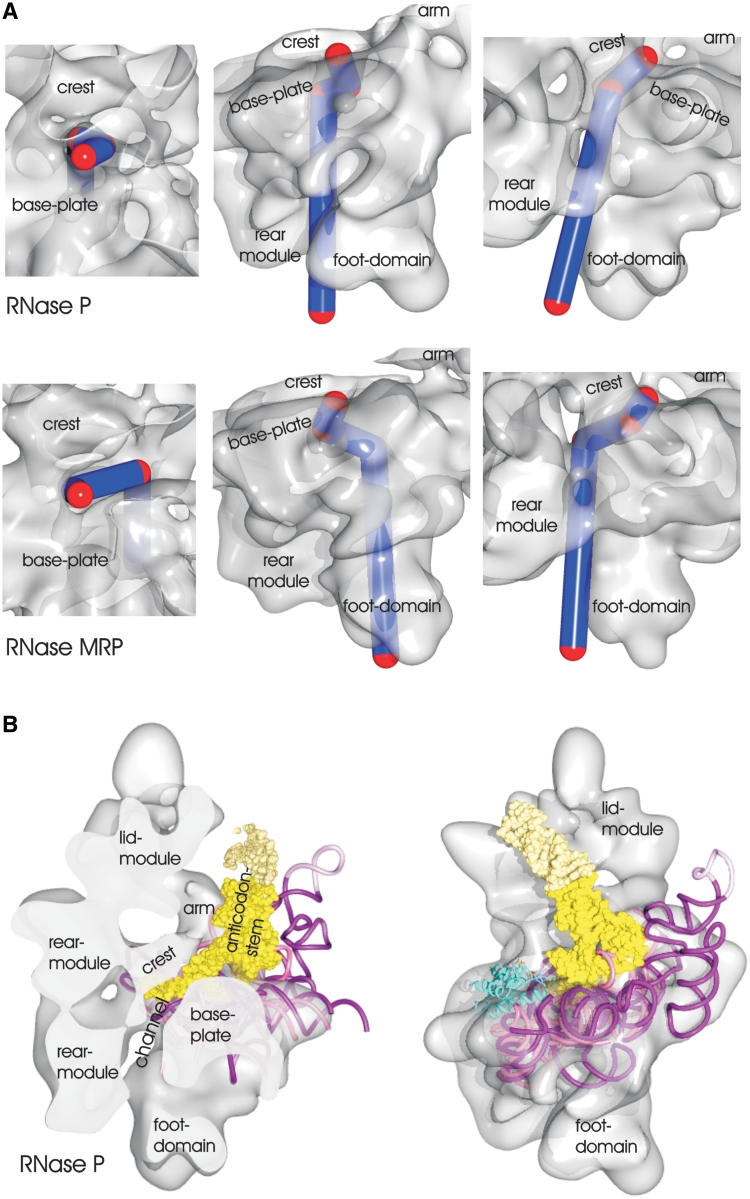
Interestingly, the system of channels and cavities is different in RNase P and RNase MRP. Whereas RNase P has an enclosed channel, RNase MRP provides a slit that is open towards the side of the S-domain. Accordingly, the possible exit path for the cleaved-off sequence varies between RNase P and RNase MRP (Figure 5A).
Figure 5.
(A) Substrate channel in RNase P (upper row) and RNase MRP (lower row). Turning points in the channels are marked by red spheres (diameter 10 Å) and linked by blue lines. Different views highlight the different routes of the channels close to the catalytic domain. (B) Surface representation of the map of RNase P with the structure of the bacterial RNase P with bound tRNA (24) superimposed. The bacterial RNA subunit is shown in purple with the bound tRNA as yellow surface and the bacterial protein in cyan. The core-domain of the bacterial RNA subunit was used for fitting and is superimposed to the fitted model of the S. pombe RNA subunit (magenta). The fit also shows the likely position of the bound tRNA, which is part of the crystal structure but not included in the EM-map. The crystal structure of the bacterial complex contains engineered crystal contacts, which are shown in light yellow for the tRNA and in light-magenta for the RNA subunit. Left panel: the map and the fitted structure are clipped at the central channel of RNase P. According to the fit of the bacterial RNA subunit to the map, the 3′-end of the pre-tRNA enters the channel. Right panel: the view is rotated by 90° around the long axis in respect to the view on the right.
The Pop1 subunit does not directly contact the RNA but forms the lid module and delineates the substrate-binding site. In RNase P, the lid module contacts the arm, forming an enclosed substrate-binding site with a specific shape that resembles a tRNA. In RNase MRP, the lid module does not contact the arm module. This could give rise to a greater flexibility, allowing the substrate-binding cavity to accept a wider range of substrates.
The tRNA is coordinated by the arm and lid modules
Superimposing the structure of a bacterial RNAse P complex with bound tRNA (24) onto our model of RNase P (Figure 5B) shows the likely contributions of the arm domain and the lid module to substrate binding for tRNA. The tRNA in the fitted ternary complex is oriented such that the 3′-end of the tRNA is pointing towards the rear module and enters the channel between the rear module and the base module (Figure 5B). The fit suggests that the channel entrance in the eukaryotic RNase P is some 2 nm away from the residues identified as being part of the active site in the bacterial enzyme. In the bacterial enzyme the 5′-leader sequence is coordinated by the bacterial protein, which has no counterpart in the EM-density map of the eukaryotic RNase P. This suggests that the 5′-leader sequence is coordinated in a different way. Possibly, it also enters the channel and interacts with the extensive system of channels and cavities between base module and rear module.
Further from the catalytic site the structural match is less good, but it is conceivable that the arm domain, where we placed Pop3, coordinates the anticodon stem of the pre-tRNA opposite to the catalytic site. In addition, the lid module restricts the maximal possible length of the anticodon stem of the tRNA (Figure 5B). This is consistent with a substrate binding pocket that not only recognizes specific sequences but also identifies the characteristic L-shape of the tRNA.
In the bacterial complex, the S-domain, which we think forms part of the arm domain in the eukaryotic complex, points away from the anticodon stem of the tRNA (24) and is not accommodated by the EM-density (Figure 5B). This suggests that the S-domain in the bacterial complex has a more open conformation than in the eukaryotic complex. However, it is difficult to understand, why such an extensive structure is acquired in the S-domain, if it does not directly contribute to substrate binding or catalysis. We think that in the bacterial complex the S-domain was possibly stabilized in the open conformation by the crystal contacts, which were engineered between the tip of the anticodon arm of the tRNA and the end of the P12-stem of the RNA-subunit. Thus the bacterial RNA-subunit could have a greater flexibility adopting an open conformation to facilitate the initial, loose substrate binding and a closed conformation, similar to what we observe in the eukaryotic complex, for further strengthening the substrate binding (induced fit).
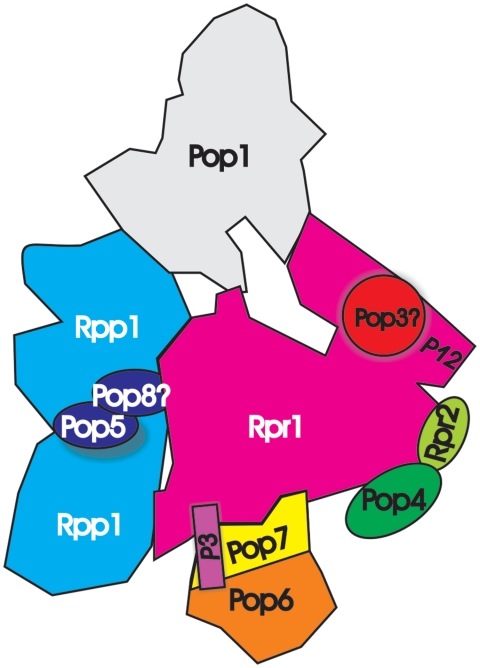
According to our model of RNase P (Figure 6), the components that were specifically gained in eukaryotes (Pop1, Pop6 and Pop7) are located at the ends of the complex, where they contribute to substrate recognition and binding. This evolution might have been driven by the need to cope with the increased complexity of RNA species in the cell. We speculate that the gains provide a selection for specific shapes, which sample a larger proportion of the surface of potential substrates than in the bacterial complex.
Figure 6.
Cartoon of the subunit organization of RNase P. Subunits where structures could be fitted are shown in the same color as in Figure 4. Regions, which are not accounted by fitted subunits, are shown in grey.
Architecture of the complex and binary interactions
We present a map of the architecture of the complex (Figure 6), which accounts for most of the observed density. However, our model agrees only partly with the numerous interaction studies (37–38,41,68,74), since many predicted binary protein–protein interactions cannot be confirmed. However, the published data indicate that these small (20–40 kDa) proteins make five or more protein–protein contacts, and it is unclear if this is really the case. According to our model, no protein subunit in the complex is in direct contact with more than three other proteins. This highlights the difficulties in deducing the architecture of complexes from interaction studies alone. Furthermore, apart from Rpp1 and possibly Pop5 we have no evidence that any of the subunits of RNase P/MRP occurs in multiple copies, in contrast to estimates of the stoichiometry using SYPRO ruby (8). A potential explanation for these discrepancies might be the formation of dimers that occur in the bacterial RNase P under certain conditions (75–76) or high-order structures in the cell.
CONCLUSION
In conclusion, we have found that RNase P and RNase MRP show a modular architecture. Modules acquired in archaea and eukaryotes can be understood as evolutionary gains that contribute to substrate recognition and coordination. In eukaryotes, subtle changes in the interplay between the modules in RNase P and RNase MRP alter the shape of the substrate-binding cavities and the channel. This is in agreement with their different substrate specificities and highlights the importance of coordination of the cleaved leader sequence of the substrate.
ACCESSION NUMBERS
emd-1929, emd-1928.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online: Supplementary Table 1, Supplementary Figures 1–7, Supplementary methods and supplementary references [77–80].
FUNDING
The EU-grant ‘3D repertoire,’ contract number LSHG-CT-2005-512028; the Deutsche Forschungsgemeinschaft (DFG, grant number BO1150); the Scottish Universities Life Sciences Alliance (SULSA) and a Wellcome Trust equipment grant (grant number WT087658MA) for the work carried out at the Electron Cryo Microscopy facility at the University of Edinburgh; the Darwin Trust of Edinburgh to B.B; Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081). Funding for open access charge: DFG.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Stephanie Kronenberg for her contribution in the early steps of the purification, Stephan Kästner and Margot Scheffer for their contribution in data collection of random conical tilt data and in the initial steps of the image processing, and Tony Crowther and David Tollervey for comments on the manuscript. The authors thank the Proteomic Core Facility at EMBL-Heidelberg and the Lechner-group at University Heidelberg (BZH) for mass spectrometry and the Genomic Core Facility at EMBL-Heidelberg for support of the RT–qPCR.
REFERENCES
- 1.Nakanishi K, Nureki O. Recent progress of structural biology of tRNA processing and modification. Mol. Cell. 2005;19:157–166. [PubMed] [Google Scholar]
- 2.Lee JY, Rohlman CE, Molony LA, Engelke DR. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol. Cell. Biol. 1991;11:721–730. doi: 10.1128/mcb.11.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker SC, Engelke DR. Ribonuclease P: the evolution of an ancient RNA enzyme. Crit. Rev. Biochem. Mol. Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang DD, Clayton DA. A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. EMBO J. 1987;6:409–417. doi: 10.1002/j.1460-2075.1987.tb04770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karwan R, Bennett JL, Clayton DA. Nuclear RNase MRP processes RNA at multiple discrete sites: interaction with an upstream G box is required for subsequent downstream cleavages. Genes Dev. 1991;5:1264–1276. doi: 10.1101/gad.5.7.1264. [DOI] [PubMed] [Google Scholar]
- 6.Schmitt ME, Clayton DA. Yeast site-specific ribonucleoprotein endoribonuclease MRP contains an RNA component homologous to mammalian RNase MRP RNA and essential for cell viability. Genes Dev. 1992;6:1975–1985. doi: 10.1101/gad.6.10.1975. [DOI] [PubMed] [Google Scholar]
- 7.Chamberlain JR, Lee Y, Lane WS, Engelke DR. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salinas K, Wierzbicki S, Zhou L, Schmitt ME. Characterization and purification of Saccharomyces cerevisiae RNase MRP reveals a new unique protein component. J. Biol. Chem. 2005;280:11352–11360. doi: 10.1074/jbc.M409568200. [DOI] [PubMed] [Google Scholar]
- 9.Schmitt ME, Clayton DA. Characterization of a unique protein component of yeast RNase MRP: an RNA-binding protein with a zinc-cluster domain. Genes Dev. 1994;8:2617–2628. doi: 10.1101/gad.8.21.2617. [DOI] [PubMed] [Google Scholar]
- 10.Bertrand E, Houser-Scott F, Kendall A, Singer RH, Engelke DR. Nucleolar localization of early tRNA processing. Genes Dev. 1998;12:2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobson MR, Cao LG, Wang YL, Pederson T. Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J. Cell. Biol. 1995;131:1649–1658. doi: 10.1083/jcb.131.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiss T, Filipowicz W. Evidence against a mitochondrial location of the 7-2/MRP RNA in mammalian cells. Cell. 1992;70:11–16. doi: 10.1016/0092-8674(92)90528-k. [DOI] [PubMed] [Google Scholar]
- 13.Lygerou Z, Allmang C, Tollervey D, Seraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt ME, Clayton DA. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindahl L, Bommankanti A, Li X, Hayden L, Jones A, Khan M, Oni T, Zengel JM. RNase MRP is required for entry of 35S precursor rRNA into the canonical processing pathway. RNA. 2009;15:1407–1416. doi: 10.1261/rna.1302909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill T, Cai T, Aulds J, Wierzbicki S, Schmitt ME. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: novel method of mRNA degradation. Mol. Cell. Biol. 2004;24:945–953. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piccinelli P, Rosenblad MA, Samuelsson T. Identification and analysis of ribonuclease P and MRP RNA in a broad range of eukaryotes. Nucleic Acids Res. 2005;33:4485–4495. doi: 10.1093/nar/gki756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen JL, Pace NR. Identification of the universally conserved core of ribonuclease P RNA. RNA. 1997;3:557–560. [PMC free article] [PubMed] [Google Scholar]
- 19.Lindahl L, Fretz S, Epps N, Zengel JM. Functional equivalence of hairpins in the RNA subunits of RNase MRP and RNase P in Saccharomyces cerevisiae. RNA. 2000;6:653–658. doi: 10.1017/s1355838200992574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Frank DN, Pace N, Zengel JM, Lindahl L. Phylogenetic analysis of the structure of RNase MRP RNA in yeasts. RNA. 2002;8:740–751. doi: 10.1017/s1355838202022082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krasilnikov AS, Xiao Y, Pan T, Mondragon A. Basis for structural diversity in homologous RNAs. Science. 2004;306:104–107. doi: 10.1126/science.1101489. [DOI] [PubMed] [Google Scholar]
- 22.Krasilnikov AS, Yang X, Pan T, Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 23.Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. Crystal structure of a bacterial ribonuclease P RNA. Proc. Natl Acad. Sci. USA. 2005;102:13392–13397. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragon A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468:784–789. doi: 10.1038/nature09516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584–587. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- 26.Amero CD, Boomershine WP, Xu Y, Foster M. Solution structure of Pyrococcus furiosus RPP21, a component of the archaeal RNase P holoenzyme, and interactions with its RPP29 protein partner. Biochemistry. 2008;47:11704–11710. doi: 10.1021/bi8015982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honda T, Kakuta Y, Kimura K, Saho J, Kimura M. Structure of an archaeal homolog of the human protein complex Rpp21-Rpp29 that is a key core component for the assembly of active ribonuclease P. J. Mol. Biol. 2008;384:652–662. doi: 10.1016/j.jmb.2008.09.056. [DOI] [PubMed] [Google Scholar]
- 28.Kakuta Y, Ishimatsu I, Numata T, Kimura K, Yao M, Tanaka I, Kimura M. Crystal structure of a ribonuclease P protein Ph1601p from Pyrococcus horikoshii OT3: an archaeal homologue of human nuclear ribonuclease P protein Rpp21. Biochemistry. 2005;44:12086–12093. doi: 10.1021/bi050738z. [DOI] [PubMed] [Google Scholar]
- 29.Kawano S, Nakashima T, Kakuta Y, Tanaka I, Kimura M. Crystal structure of protein Ph1481p in complex with protein Ph1877p of archaeal RNase P from Pyrococcus horikoshii OT3: implication of dimer formation of the holoenzyme. J. Mol. Biol. 2006;357:583–591. doi: 10.1016/j.jmb.2005.12.086. [DOI] [PubMed] [Google Scholar]
- 30.Numata T, Ishimatsu I, Kakuta Y, Tanaka I, Kimura M. Crystal structure of archaeal ribonuclease P protein Ph1771p from Pyrococcus horikoshii OT3: an archaeal homolog of eukaryotic ribonuclease P protein Rpp29. RNA. 2004;10:1423–1432. doi: 10.1261/rna.7560904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sidote DJ, Hoffman DW. NMR structure of an archaeal homologue of ribonuclease P protein Rpp29. Biochemistry. 2003;42:13541–13550. doi: 10.1021/bi030170z. [DOI] [PubMed] [Google Scholar]
- 32.Takagi H, Watanabe M, Kakuta Y, Kamachi R, Numata T, Tanaka I, Kimura M. Crystal structure of the ribonuclease P protein Ph1877p from hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biochem. Biophys. Res. Commun. 2004;319:787–794. doi: 10.1016/j.bbrc.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 33.Wilson RC, Bohlen CJ, Foster MP, Bell CE. Structure of Pfu Pop5, an archaeal RNase P protein. Proc. Natl Acad. Sci. USA. 2006;103:873–878. doi: 10.1073/pnas.0508004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamma T, Ferre-D'Amare AR. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 A resolution. Structure. 2004;12:893–903. doi: 10.1016/j.str.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 35.Perederina A, Esakova O, Quan C, Khanova E, Krasilnikov AS. Eukaryotic ribonucleases P/MRP: the crystal structure of the P3 domain. EMBO J. 2010;29:761–769. doi: 10.1038/emboj.2009.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stams T, Niranjanakumari S, Fierke CA, Christianson DW. Ribonuclease P protein structure: evolutionary origins in the translational apparatus. Science. 1998;280:752–755. doi: 10.1126/science.280.5364.752. [DOI] [PubMed] [Google Scholar]
- 37.Hall TA, Brown JW. Interactions between RNase P protein subunits in archaea. Archaea. 2004;1:247–254. doi: 10.1155/2004/743956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houser-Scott F, Xiao S, Millikin CE, Zengel JM, Lindahl L, Engelke DR. Interactions among the protein and RNA subunits of Saccharomyces cerevisiae nuclear RNase P. Proc. Natl Acad. Sci. USA. 2002;99:2684–2689. doi: 10.1073/pnas.052586299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang T, Altman S. Protein-protein interactions with subunits of human nuclear RNase P. Proc. Natl Acad. Sci. USA. 2001;98:920–925. doi: 10.1073/pnas.021561498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kifusa M, Fukuhara H, Hayashi T, Kimura M. Protein-protein interactions in the subunits of ribonuclease P in the hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biosci. Biotechnol. Biochem. 2005;69:1209–1212. doi: 10.1271/bbb.69.1209. [DOI] [PubMed] [Google Scholar]
- 41.Welting TJ, van Venrooij WJ, Pruijn GJ. Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res. 2004;32:2138–2146. doi: 10.1093/nar/gkh539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 43.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 44.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 45.Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd edn. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 47.Zuker M, Mathews DH, Turner DH. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: Barciszewski J, Clark BFC, editors. RNA biochemistry and biotechnology. Dordrecht: Kluwer Academic Publishers; 1999. pp. 11–43. [Google Scholar]
- 48.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 49.Yang L, Altman S. A noncoding RNA in Saccharomyces cerevisiae is an RNase P substrate. RNA. 2007;13:682–690. doi: 10.1261/rna.460607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ziehler WA, Day JJ, Fierke CA, Engelke DR. Effects of 5′ leader and 3′ trailer structures on pre-tRNA processing by nuclear RNase P. Biochemistry. 2000;39:9909–9916. doi: 10.1021/bi000603n. [DOI] [PubMed] [Google Scholar]
- 51.Golas MM, Sander B, Will CL, Luhrmann R, Stark H. Molecular architecture of the multiprotein splicing factor SF3b. Science. 2003;300:980–984. doi: 10.1126/science.1084155. [DOI] [PubMed] [Google Scholar]
- 52.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 53.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 54.Scheres SH, Gao H, Valle M, Herman GT, Eggermont PP, Frank J, Carazo JM. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat. Methods. 2007;4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 55.Harauz G, Van Heel M. Exact filters for general geometry 3-dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- 56.Bottcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 57.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 58.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 59.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 60.Marquez SM, Chen JL, Evans D, Pace NR. Structure and function of eukaryotic Ribonuclease P RNA. Mol. Cell. 2006;24:445–456. doi: 10.1016/j.molcel.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernandez-Tornero C, Bottcher B, Riva M, Carles C, Steuerwald U, Ruigrok RW, Sentenac A, Muller CW, Schoehn G. Insights into transcription initiation and termination from the electron microscopy structure of yeast RNA polymerase III. Mol. Cell. 2007;25:813–823. doi: 10.1016/j.molcel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 62.Diepholz M, Venzke D, Prinz S, Batisse C, Florchinger B, Rossle M, Svergun DI, Bottcher B, Fethiere J. A different conformation for EGC stator subcomplex in solution and in the assembled yeast V-ATPase: possible implications for regulatory disassembly. Structure. 2008;16:1789–1798. doi: 10.1016/j.str.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 63.Ulbrich C, Diepholz M, Bassler J, Kressler D, Pertschy B, Galani K, Bottcher B, Hurt E. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell. 2009;138:911–922. doi: 10.1016/j.cell.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 64.Xu Y, Amero CD, Pulukkunat DK, Gopalan V, Foster MP. Solution structure of an archaeal RNase P binary protein complex: formation of the 30-kDa complex between Pyrococcus furiosus RPP21 and RPP29 is accompanied by coupled protein folding and highlights critical features for protein-protein and protein-RNA interactions. J. Mol. Biol. 2009;393:1043–1055. doi: 10.1016/j.jmb.2009.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perederina A, Esakova O, Koc H, Schmitt ME, Krasilnikov AS. Specific binding of a Pop6/Pop7 heterodimer to the P3 stem of the yeast RNase MRP and RNase P RNAs. RNA. 2007;13:1648–1655. doi: 10.1261/rna.654407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perederina A, Krasilnikov AS. The P3 domain of eukaryotic RNases P/MRP: making a protein-rich RNA-based enzyme. RNA Biol. 2010;7:534–539. doi: 10.4161/rna.7.5.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Honda T, Hara T, Nan J, Zhang X, Kimura M. Archaeal homologs of human RNase P protein pairs Pop5 with Rpp30 and Rpp21 with Rpp29 work on distinct functional domains of the RNA subunit. Biosci. Biotechnol. Biochem. 2010;74:266–273. doi: 10.1271/bbb.90550. [DOI] [PubMed] [Google Scholar]
- 68.Aspinall TV, Gordon JM, Bennett HJ, Karahalios P, Bukowski JP, Walker SC, Engelke DR, Avis JM. Interactions between subunits of Saccharomyces cerevisiae RNase MRP support a conserved eukaryotic RNase P/MRP architecture. Nucleic Acids Res. 2007;35:6439–6450. doi: 10.1093/nar/gkm553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dlakic M. 3D models of yeast RNase P/MRP proteins Rpp1p and Pop3p. RNA. 2005;11:123–127. doi: 10.1261/rna.7128905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cho IM, Lai LB, Susanti D, Mukhopadhyay B, Gopalan V. Ribosomal protein L7Ae is a subunit of archaeal RNase P. Proc. Natl Acad. Sci. USA. 2010;107:14573–14578. doi: 10.1073/pnas.1005556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fukuhara H, Kifusa M, Watanabe M, Terada A, Honda T, Numata T, Kakuta Y, Kimura M. A fifth protein subunit Ph1496p elevates the optimum temperature for the ribonuclease P activity from Pyrococcus horikoshii OT3. Biochem. Biophys. Res. Commun. 2006;343:956–964. doi: 10.1016/j.bbrc.2006.02.192. [DOI] [PubMed] [Google Scholar]
- 72.Rosenblad MA, Lopez MD, Piccinelli P, Samuelsson T. Inventory and analysis of the protein subunits of the ribonucleases P and MRP provides further evidence of homology between the yeast and human enzymes. Nucleic Acids Res. 2006;34:5145–5156. doi: 10.1093/nar/gkl626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pluk H, van Eenennaam H, Rutjes SA, Pruijn GJ, van Venrooij WJ. RNA-protein interactions in the human RNase MRP ribonucleoprotein complex. RNA. 1999;5:512–524. doi: 10.1017/s1355838299982079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Welting TJ, Kikkert BJ, van Venrooij WJ, Pruijn GJ. Differential association of protein subunits with the human RNase MRP and RNase P complexes. RNA. 2006;12:1373–1382. doi: 10.1261/rna.2293906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barrera A, Fang X, Jacob J, Casey E, Thiyagarajan P, Pan T. Dimeric and monomeric Bacillus subtilis RNase P holoenzyme in the absence and presence of pre-tRNA substrates. Biochemistry. 2002;41:12986–12994. doi: 10.1021/bi020416k. [DOI] [PubMed] [Google Scholar]
- 76.Buck AH, Dalby AB, Poole AW, Kazantsev AV, Pace NR. Protein activation of a ribozyme: the role of bacterial RNase P protein. EMBO J. 2005;24:3360–3368. doi: 10.1038/sj.emboj.7600805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Radermacher M, Wagenknecht T, Verschoor A, Frank J. 3-dimensional reconstruction from a single exposure, random conical tilt series applied to the 50S-risomal subunit of escherichia-coli. J. Microscopy. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 78.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 79.van Heel M, Harauz G, Orlova EV. A new generation of the IMAGIC image processing system. J. Struct. Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 80.van Heel M, Frank J. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy. 1981;6:187–194. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.