Abstract
Binding of the catalytic divalent ion to the ternary DNA polymerase β/gapped DNA/dNTP complex is thought to represent the final step in the assembly of the catalytic complex and is consequently a critical determinant of replicative fidelity. We have analyzed the effects of Mg2+ and Zn2+ on the conformational activation process based on NMR measurements of [methyl-13C]methionine DNA polymerase β. Unexpectedly, both divalent metals were able to produce a template base-dependent conformational activation of the polymerase/1-nt gapped DNA complex in the absence of a complementary incoming nucleotide, albeit with different temperature thresholds. This conformational activation is abolished by substituting Glu295 with lysine, thereby interrupting key hydrogen bonds necessary to stabilize the closed conformation. These and other results indicate that metal-binding can promote: translocation of the primer terminus base pair into the active site; expulsion of an unpaired pyrimidine, but not purine, base from the template-binding pocket; and motions of polymerase subdomains that close the active site. We also have performed pyrophosphorolysis studies that are consistent with predictions based on these results. These findings provide new insight into the relationships between conformational activation, enzyme activity and polymerase fidelity.
INTRODUCTION
It is generally accepted that DNA polymerases utilize a ‘two-metal–ion’ mechanism for nucleotidyl transfer (1). For DNA synthesis, the catalytic ‘metal A’ is thought to lower the pKa of the 3′-OH of the growing primer terminus, while the nucleotide-binding ‘metal B’ coordinates the triphosphate moiety, facilitating binding of the incoming dNTP. The crystallographic structure of a pre-catalytic complex of DNA polymerase (pol) β with two Mg2+ ions provides compelling evidence for this mechanism (2,3). Both metals are coordinated by conserved aspartates (Asp190 and Asp192), and the catalytic metal is coordinated by an additional aspartate (Asp256). Since the ‘metal A’ binding site includes oxygen-ligands contributed by the incoming dNTP and primer terminus 3′-OH, the catalytic metal may be the last ligand to bind. The presence of the catalytic metal results in an altered sugar pucker of the primer terminus that has O3′ in a favorable position to attack the Pα of the incoming dNTP (2). The natural metal cofactor is Mg2+, but other transition metal ions, including Zn2+, have been shown to support catalysis by some polymerases (4), and studies involving other metal ions have proven useful to probe the polymerase catalytic mechanism (5–7).
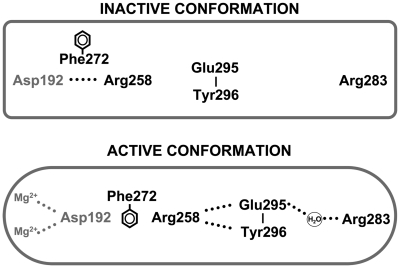
Although the role of the metals in catalysis has been described, their role in the conformational activation that couples correct base pairing with catalysis is not well understood. We are particularly interested in characterizing the activated state of pol β, since correct nucleotide insertion inherently dominates DNA polymerase fidelity (8). Crystallographic structures of binary DNA–polymerase complexes from several polymerase families indicate that a subdomain repositioning occurs upon binding a correct nucleotide (9). For right-handed DNA polymerases, the fingers subdomain closes to sandwich the nascent base pair between the protein sensor domain residues and the primer-terminal base pair. For pol β, the thumb subdomain plays an equivalent role and is referred to as the N-subdomain (10). Consequently, correct base pairing is signaled through an open to closed N-subdomain transition that results in a series of protein side-chain rearrangements that alter hydrogen-bonding interactions (Figure 1). This signaling cascade connects sensor residues of α-helix N (e.g. Arg283) to the catalytic site >10 Å away. These conformational changes facilitate formation of the metal–ion complex necessary to activate the primer 3′-OH and position the Pα of the incoming dNTP. The importance of this conformational activation is illustrated by the observation that mutagenesis of residues which participate in this signaling reduces nucleotide insertion (11,12).
Figure 1.
Altered side-chain interactions accompanying polymerase activation. In the open conformation, Arg283 does not interact with other key residues, but in the closed conformation, it interacts with the templating (coding) base, the upstream template nucleotide (not illustrated), and with Glu295 (indirectly). Thus, the position of the N-subdomain can be structurally transmitted to the active site through a series of interactions involving Arg283 (in α-helix N), and Asp192 which coordinates both active-site Mg2+ ions. This is also accompanied by altered interactions of Glu295/Tyr296 with Arg258 in the open (inactive) and closed (active) forms. Phe272 is postulated to transiently interfere with interactions between Asp192 and Arg258 permitting an interaction with Glu295/Tyr296. Residues of the N-subdomain are indicated in boldface text.
In a crystallographic study of pol β complexes in globally open conformations using various metals, Pelletier et al. (13) found that Mg2+ was unusual. All of the other metals evaluated led to an active conformation of the Asp192 side-chain without subdomain closing. Furthermore, transition metals that form tighter complexes with enzyme and substrate might induce activation without correct nascent base pairing. Thus, substitution of Mn2+ for Mg2+ has permitted crystallographic observation of pre-insertion structures of ternary complexes containing mutagenic intermediates (5,14). In this way, these ions could obviate the signal transduction pathway that connects the catalytic C-subdomain and the nascent base pair N-subdomain of pol β. The present studies were undertaken to characterize the role that the divalent metal ion plays in the conformational activation process.
It is of fundamental importance to identify factors that affect metal binding and the structural, thermodynamic and kinetic consequences. However, the physical constraints imposed by crystallography limit the ability of the enzyme to respond to various stimuli and significantly populate alternative conformers. We have shown that DNA binding and catalytic activation in pol β can be conveniently monitored in solution with the NMR signals of 13C-labeled intrinsic methionine residues (15,16); the sensitivity of these resonances to conformational activation of the enzyme allows us to probe in solution the various factors that influence substrate binding and specificity (i.e. fidelity). Furthermore, conformational activation is also fundamental to the enzyme's reverse reaction, pyrophosphorolysis. This reaction plays an important role in removing blocked primer termini (e.g. 3′-azidothymidine) by HIV-1 reverse transcriptase (17) and pol γ (18), the mitochondrial replicative DNA polymerase. Mitochondrial toxicity represents a significant complication of long-term anti-HIV therapy with antiviral nucleosides (19).
In order to understand the effects of transition metal ions while avoiding the paramagnetic broadening caused by Mn2+, we performed NMR studies to probe the effect of divalent metals (Mg2+ or Zn2+), DNA sequence, and temperature on formation of the closed activated-state of a series of pol β/DNA complexes. This evaluation of the role of metal ions in conformational activation is central to an understanding of their effects on catalysis, fidelity and inhibitor sensitivity.
MATERIALS AND METHODS
Materials
The [methyl-13C]methionine-labeled pol β was prepared as described previously (15) by growth of the plasmid-containing Escherichia coli on a medium containing [methyl-13C]methionine (CIL, Cambridge, MA). The only modification of the protocol was the use of HiTrap Heparin HP (GE Healthcare) chromatography instead of ssDNA cellulose chromatography. The C267A, C239A and E295K pol β mutant enzymes were produced using the Quikchange kit (Stratagene). NMR samples contained 0.3–0.6 mM pol β in a D2O buffer consisting of 100 mM potassium phosphate (pH 6.7), 1 mM DTT, 0.1 mM AEBSF, 5 mM NaN3 and 50 µM DSS as an internal chemical shift standard. Oligonucleotides (Oligos Etc., Wilsonville, OR) were dissolved in D2O to make a stock solution of ∼10 mM. Non-hydrolyzable deoxynucleoside triphosphates were obtained from Jena Bioscience. The double-hairpin, 1-nt gap DNA substrates have the following sequences: 5′-PGGCGAAGCCTGGTGCGAAGCACC-3′ or 5′-PGGCGAAGCCGCGTGCGAAGCACG-3′ (underlined nucleotide is in the gap). The double-hairpin, 2-nt gap DNA substrate has the sequence: 5′-PGGCGAAGCCTGGTGCGAAGCAC-3′ (underlined nucleotides are in the gap).
The protein concentration was determined using an extinction coefficient of 20 088 M−1 cm−1 at 280 nm, and the DNA concentrations were determined using their 260 nm extinction coefficients.
NMR spectroscopy
NMR experiments were performed on a Varian UNITY INOVA 600 MHz NMR spectrometer, using a 5 mm Varian (600 MHz) 1H{13C,15N} triple-resonance probe, equipped with actively shielded Z-gradients. The 1H-13C HSQC spectra were acquired using Varian's gChsqc sequence (20). The data were acquired as a 230 (t1) × 512 (t2) complex matrix, with acquisition times of 63.9 (t1) and 64.0 ms (t2), 128 scans per increment, and a 1.0 s delay between scans. The spectra were processed using NMRPipe version 2.1 (21) and analyzed using NMRView version 5.0.4 (22). All spectra were processed using squared cosine bell apodization functions in all dimensions and forward-backward linear prediction in the indirect 13C-dimension (23).
Kinetic assays
A 34-mer oligonucleotide DNA substrate containing a 1-nt gap was prepared by annealing three gel-purified oligonucleotides (IDT, Coralville, IA) to create a 1-nt gap at position 16. Each oligonucleotide was resuspended in 10 mM Tris–HCl, pH 7.4 and 1 mM EDTA, and the concentration was determined from their UV absorbance at 260 nm. The annealing reactions were carried out in a PCR thermocycler by incubating a solution of 10 µM primer with 12 µM each of downstream and template oligonucleotides at 95°C for 5 min followed by 30 min at 65°C, and then slow cooling (1°C/min) to 10°C. The sequence of the gapped DNA substrate was: primer, 5′-CTGCAGCTGATGCGC-3′; downstream oligonucleotide, 5′-GTACGGATCCCCGGGTAC-3′; and template, 3′-GACGTCGACTACGCGXCATGCCTAGGGGCCCATG-5′ (X is the template residue in the gap). The primer was 5′-labeled with [γ-32P]ATP using Optikinase (USB Corp.) and the free radioactive ATP was removed with either MicroSpin G-25 or Biospin 6 columns. The downstream oligonucleotide was synthesized with a 5′-phosphate.
Steady-state kinetic parameters for dCTP insertion opposite guanine in a 1-nt DNA gap were determined by initial velocity measurements as described previously (10). Enzyme activities were determined using a standard reaction mixture containing 50 mM Tris–HCl, pH 7.4 (37°C), 100 mM KCl, 5 mM MgCl2 and 200 nM 1-nt gapped DNA. Enzyme concentrations and reaction time intervals were chosen so that substrate depletion or product inhibition did not influence initial velocity measurements. Reactions were stopped with EDTA and mixed with an equal volume of formamide dye, and the products separated on 16% denaturing polyacrylamide gels. The dried gels were analyzed by phosphorimagery to quantify product formation. Steady-state kinetic parameters for Zn2+-dependent DNA synthesis were determined at room temperature as described above, except that ZnCl2 replaced MgCl2. Additionally, the divalent metal concentrations for the activities tabulated in Supplementary Table S1 were 2.5 mM.
Pyrophosphorolysis was quantified by following the removal of the primer terminus upon addition of 150 µM PPi under steady-state reaction conditions. Unless noted otherwise, enzyme activities were determined using a standard reaction mixture (30 µl) containing 50 mM Tris–HCl, pH 7.4 (room temperature or 37°C), 100 mM KCl, 5 mM MgCl2 and 200 nM 1-nt gapped DNA. Assays were initiated by mixing enzyme with DNA/PPi/metal. Enzyme concentrations (2–5 nM) and reaction time intervals were chosen so that substrate depletion or product inhibition did not influence product formation. Reactions were stopped with 15 µl of 0.5 M EDTA and mixed with an equal volume of formamide dye. The substrates and products were separated on 16–18% denaturing (8 M urea) polyacrylamide gels and quantified in the dried gels by phosphorimagery.
RESULTS
NMR observation of conformational activation
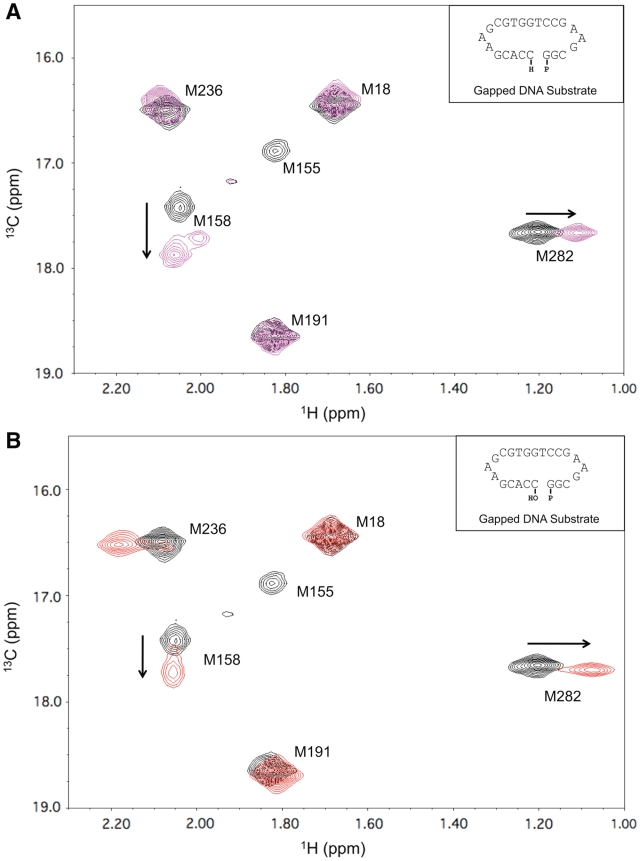
As shown previously (15,16), the methyl resonances of [methyl-13C]methionine-labeled pol β provide useful probes of DNA binding (Met18, Met236) and conformational activation (Met155, Met158, Met236, Met282). Met236 is the only methionine residue that makes direct contact with the substrate, and its resonance’s shift is highly variable. In contrast, the shift and linewidth characteristics of the other methionine methyl resonances are dependent on indirect, conformationally-mediated processes and exhibit characteristic perturbations upon binding gapped DNA and upon formation of an abortive ternary complex (15,16). Figure 2A shows the changes observed in the HSQC spectrum of labeled pol β upon formation of a ternary pol β/gapped DNA/Mg2+/dNTP complex. Addition of Mg-dATP to a dideoxy-terminated 1-nt gapped DNA substrate with a templating thymine perturbs the resonances of Met155 and Met158, located in the catalytic C-subdomain near the active-site metals, and Met282 of the N-subdomain. The resonance shift of Met236, positioned near the primer terminus, is altered due to changes in the position of the DNA and enzyme conformation that result from dNTP binding. The Met282 residue, positioned at the center of α-helix N, is particularly valuable as an indicator of the helix repositioning that occurs upon formation of a complex with a complementary dNTP (16). Addition of Mg2+ to pol β or to pol β•DNA at 25°C produces negligible shift perturbations (data not shown), and crystallographic data indicate that catalytic metal binding is not necessary for dNTP-induced conformational activation (2). In order to obtain further insight into the role of the metal ion in producing an active conformation, we added ZnCl2 to the binary DNA complex. Unexpectedly, this resulted in spectral perturbations that are very similar to those observed for the ternary abortive complex (Figure 2A and B), suggesting that comparable conformational changes have occurred in response to the addition of Zn2+ alone. Thus, it is possible to induce a nucleotide-free activation of pol β that appears to resemble that induced by the presence of a complementary incoming dNTP (15). Kinetic evaluation of pol β gap-filling DNA synthesis indicates that Zn2+ is a good substitute for Mg2+ (Supplementary Table S1).
Figure 2.
1H-13C HSQC spectra of [methyl-13C]methionine pol β. (A) 1H-13C HSQC spectra of binary (black) and ternary (magenta) substrate complexes of 150 µM labeled pol β. The binary complex was formed by adding 10% excess of dideoxy-terminated gapped DNA substrate, and the ternary complex formed with the addition of 5 mM MgCl2 and 200 µM dATP. (B) Comparison of the spectra of binary pol β•gapped DNA complexes in the absence (black) and presence (red) of 2.5 mM ZnCl2. Data were collected at 25°C in 100 mM potassium phosphate buffer, pH 6.7, containing 1 mM DTT and 5 mM sodium azide. The DNA sequences of the double hairpin substrates are indicated in the insets.
We note here that, based on NOE and nudge mutation data summarized in the Supplementary Data, assignments of Met158 and Met191 resonances are now reversed relative to our previous studies (Supplementary Figure S1). As stated in our previous paper (15), ‘in the absence of specific assignments for the other proton resonances of the protein, definitive assignment information cannot be obtained’. The methyl groups of methionine residues 155, 158 and 191 are all located within several angstroms of each other and, since we have not as yet developed a specific model for relating changes in these resonances to the conformational activation process, these have been interpreted phenomenologically as a set of conformational activation-dependent resonance perturbations. This is not the case for Met18, Met236 and Met282, for which the resonance perturbations have been interpreted on the basis of specific conformational and ligand-binding effects (15,16). Hence, the interchange of the Met158 and Met191 assignments has no effect on any of our earlier conclusions.
To further understand the basis for the Zn2+-induced activation, we investigated the dependence of the activation on the length and chemistry of the gap. A comparison of the spectra obtained with pol β complexes formed with 2′-deoxy- or 2′,3′-dideoxy-terminated primers showed that coordination of a metal ion with the 3′-OH of the primer terminus, observed in the crystallographic ternary complex (2), was not required for Zn2+-dependent activation (Supplementary Figure S2). It also was found that a 2-nt gap substrate did not support Zn2+-induced activation (Supplementary Figure S3).
Zn2+-induced DNA translocation
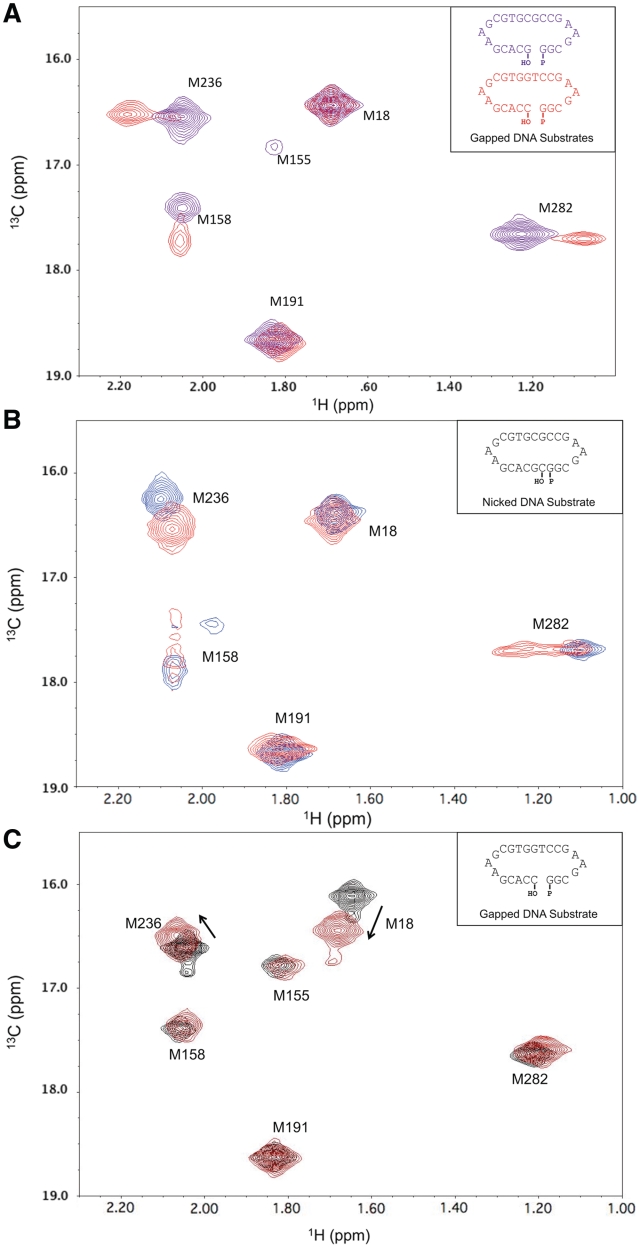
The dependence of the Zn2+-induced conformational activation on DNA gap size suggested an explanation for the effect. We hypothesized that the activation might involve substrate translocation such that the base pair involving the primer terminus would occupy the active site and the unpaired templating base would become extra-helical, thereby permitting α-helix N to interact with the primer terminal base pair in the active site. Such an effect could be promoted by Zn2+, which would interact with the bridging, 5′-phosphate group of the primer terminus. This interpretation is also consistent with the Zn2+-induced Met236 resonance shift (Figure 2B, red spectrum), since DNA translocation would be expected to significantly perturb the environment of this residue which is located near the primer terminus. It was further reasoned that the tendency of the templating base to adopt an extra-helical orientation could be dependent on its ability to stack within the DNA helix. Accordingly, pyrimidines should exhibit a greater propensity for adopting an extra-helical conformation than purines. Consistent with this hypothesis, use of a substrate with a templating guanine rather than thymine was found to block Zn2+-induced conformational activation almost completely (Figure 3A, purple spectrum). This finding supports the model in which the conformational activation produced by Zn2+ in the presence of a 1-nt gapped DNA results from a translocation of the primer terminus into the nascent base-pair-binding pocket, displacement of the templating base into an extra-helical position, and repositioning (i.e. closing) of the N-subdomain to interact with the primer terminus and its templating base.
Figure 3.
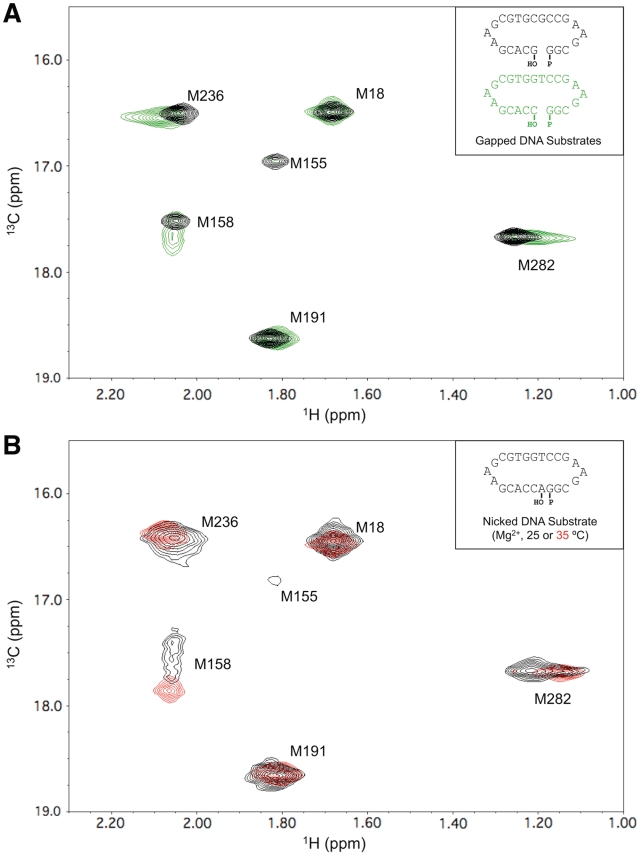
Role of altered DNA sequence and protein on conformational activation. (A) 1H-13C HSQC spectra of labeled pol β/1-nt gapped DNA binary complexes with a templating thymine (red) or guanine (purple) in the presence of 2.5 mM ZnCl2. (B) Spectra of the complex of labeled pol β with 10% excess of nicked DNA substrate in the presence of 5 mM MgCl2 (red) or 2.5 mM ZnCl2 (blue). The nicked complexes were formed by allowing pol β to catalyze the insertion of the correct nucleoside triphosphate into a 1-nt gapped substrate. (C) Spectra of labeled E295K pol β (black) and the binary DNA complex in the presence of 2.5 mM ZnCl2 (red). Spectra were collected at 25°C in 100 mM potassium phosphate buffer, pH 6.7, containing 1 mM DTT and 5 mM sodium azide. The DNA sequences of the double hairpin substrates are indicated in the insets.
Interaction of metal ions with nicked DNA
A second implication of the above hypothesis is that a pol β/nicked DNA complex should also be susceptible to Zn2+-induced activation, since the active site is again occupied by a base pair. A crystallographic structure of the binary pol β/nicked DNA complex (PDB ID 1BPZ) (24), shows the enzyme in an inactive open conformation without metals. Consistent with this expectation, the 1H-13C HSQC spectrum of a binary methionine-labeled pol β/nicked DNA complex, with a chelator present to sequester the divalent metals, showed none of the perturbations associated with conformational activation. However, when Zn2+ was not chelated, the spectral perturbations were similar to those observed for the complex containing a 1-nt gapped DNA with templating thymine (Figure 3B, blue spectrum). This result indicates that Zn2+ is able to interact with the active-site aspartyl residues and, in combination with the presence of a base pair in the active site, induce a closed, conformationally activated structure without the need for DNA translocation. Addition of MgCl2 to the nicked-substrate complex produced a different spectral response, suggesting a mixture of slowly inter-converting activated and non-activated structures (Figure 3B, red spectrum), indicating that the differences observed between the responses to Zn2+ and Mg2+ may be more quantitative than qualitative.
NMR studies of E295K pol β
A mutation in a signal transduction cascade residue (Glu295) of pol β has been identified in some human gastric carcinomas and was found to disrupt enzyme activity and base excision repair (12,25). The HSQC spectra of the methionine-labeled E295K mutant apoenzyme and its binary complex with gapped DNA indicate that the mutant enzyme is properly folded and demonstrates the expected shifts in Met18 and Met236 resonances upon binding DNA (Supplementary Figure S4). In contrast, the typical spectral changes observed for the resonances of Met155, Met158, Met282 and, to a more variable extent, Met236 were not observed upon formation of either a ternary E295K pol β/1-nt gapped DNA/Mg-dNTP complex or a binary E295K pol β/1-nt gapped DNA/Zn2+ complex (Figure 3c). This demonstrates that Glu295 plays a critical role in the signal transduction pathway that couples base-pair sensing to catalytic activation, and shows that the mechanism of Zn2+ activation also utilizes this pathway.
Pyrophosphorolysis
The NMR evidence for the interaction of Zn2+ and, in some instances, Mg2+ with the 5′-phosphate of the primer terminus in both nicked and 1-nt gapped DNA complexes has direct implications for the reverse ‘depolymerization’ reaction, pyrophosphorolysis. In this reaction, the catalytic metal ion must interact with the primer terminus 5′-phosphate, which ultimately becomes the α-phosphate of the dNTP product (3). Pyrophosphorolysis would be expected to require conformational changes that occur prior to and following the chemical step (26).
We have previously documented a pyrophosphorolysis reaction for pol β with nicked DNA in kinetic studies performed at 37°C (27), however, attempts to catalyze the reverse reaction in the presence of Zn2+/PPi were unsuccessful, perhaps as a result of the lower ability of Zn2+ (compared with Mg2+) to support the chemistry of the transition state, and/or as a result of the low solubility of Zn2+/PPi. Additionally, pyrophosphorolysis is not readily observed at room temperature with physiological concentrations of Mg2+/PPi (28); this is consistent with the NMR studies indicating that Mg2+ by itself cannot induce the activated complex. Additionally, the lack of pyrophosphorolysis presumably indicates that even in the presence of PPi, Mg2+ cannot induce an active conformation with a 1-nt gapped DNA substrate.
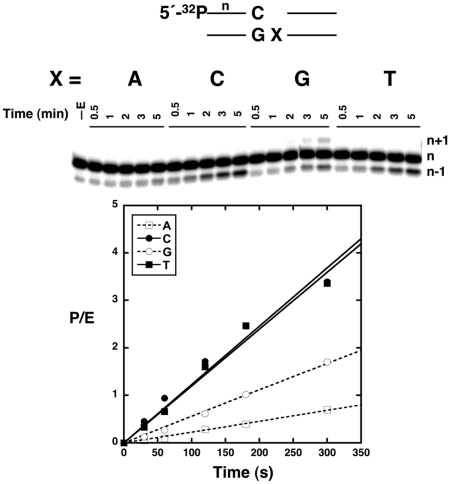
Since the reactions described above were performed at 25°C, we determined whether increasing the reaction temperature to 37°C might promote conformational activation and the ability of the templating base to assume an extra-helical position, presumably required to provide a substrate for pyrophosphorolysis. In contrast with the results at 25°C, the reverse reaction was readily observed with 1-nt gapped DNA as indicated by the shortened primer (n–1 product) (Figure 4). As expected if helical stacking influences conformational activation, the reverse reaction was much weaker with templating purines than with pyrimidines. Also note that a weak n + 1 product was produced when guanine was the templating base; this confirms that dCTP was a product of the observed reaction since the primer terminal nucleotide is cytidine. Even though very low concentrations of dCTP are produced over the short time span of the experiment, dCTP can fill the gap on a 1-nt gapped substrate with G as the templating base but not with other templating bases, since these would require an incorrect insertion.
Figure 4.
Influence of the identity of the templating base on pol β-dependent pyrophosphorolysis. Assays were initiated by mixing pol β with DNA/PPi/Mg2+ at 37°C. The templating nucleotide (X) in the 1-nt gapped DNA was varied. The first lane (–E) shows 1-nt gapped DNA (with templating A) in the absence of enzyme. The 5′-[32P]-labeled primer (n) is indicated and a diagram of the gapped DNA is shown. Pyrophosphorolysis results in removal of the primer terminus resulting in a shorter (n–1) product band. A small amount of n+1 product is observed when G is the templating nucleotide due to insertion of the resulting dCTP (the primer terminus is cytidine). The time dependence of product formation (P/E, enzyme turnovers) is also plotted. From a least-squares linear regression analysis, the observed activities (slopes) are 0.0023/s (A, R2 = 0.99), 0.0123/s (C, R2 = 0.97), 0.0056/s (G, R2 = 0.99) and 0.0120/s (T, R2 = 0.98).
High temperature NMR studies of C267A pol β
Although the NMR studies were qualitatively consistent with the pyrophosphorolysis results, it was unclear from the NMR studies why this reaction could not be observed at 25°C; therefore, we sought to obtain NMR data at 35°C. However, enzyme stability can be problematic when obtaining NMR spectra over extended time periods at elevated temperatures, and we encountered a significant degree of precipitation and/or the presence of soluble, partially unfolded pol β, depending on the conditions of the study. More specifically, a variable resonance previously suggested as arising from incompletely proteolyzed Met1 (15) is now attributed to partially unfolded but soluble pol β. After a series of studies, we found that replacing Cys267 with alanine permitted longer collection times at elevated temperature (35°C) without significant degradation of the protein, as evidenced by the stability of the NMR spectra and the absence of observable precipitate. Alanine substitution at Cys267 does not dramatically alter the structure or behavior of the enzyme as demonstrated by the similar steady-state kinetic behavior for 1-nt gap filling with the mutant and wild-type enzymes (Supplementary Table S2), as well as the minimal perturbations observed for the methionine 1H-13C HSQC spectrum of labeled pol β.
In contrast with the results obtained at 25°C, HSQC spectra of [methyl-13C]methionine C267A pol β obtained at 35°C in the presence of MgCl2 indicated a significantly greater degree of conformational activation with a templating thymidine as judged by the broadening of Met155 and the shift in Met158 resonances (Figure 5A). These spectral changes are similar to those observed at 25°C with nicked DNA in the presence of Mg2+ (Figure 3B). The elongated shape of the Met282 resonance observed at 35°C for C267A pol β complexed with gapped DNA containing a templating thymine base presumably arises due to intermediate exchange between the open and closed conformational states of the enzyme. Simulations using a range of estimated parameters yields a lifetime for the closed (activated) conformation of ∼2–5 ms (Supplementary Figure S5). These values are near those expected from observed fluorescence transients that monitor subdomain motions (29).
Figure 5.
Effect of temperature on conformational activation. (A) 1H-13C HSQC spectra of the binary complex of labeled C267A pol β and 1-nt gapped DNA with a templating thymine (green) or templating guanine (black). Spectra were obtained at 35°C in 100 mM potassium phosphate buffer, pH 6.7, containing 5 mM MgCl2, 1 mM DTT and 5 mM sodium azide. (B) spectra of binary DNA complexes of C267A pol β with a nicked DNA substrate in the presence of 5 mM MgCl2 at 25°C (black) or 35°C (red). The DNA sequences of the double hairpin substrates are indicated in the insets.
We further evaluated the effect of temperature by comparing results at 25°C and 35°C for pol β complexes with nicked DNA in the presence of Mg2+ (Figure 5B). Consistent with the above results, the spectra obtained at the higher temperature show a more complete conformational activation of the enzyme. Thus, the intermediate degree of conformational activation observed at 25°C becomes more complete at 35°C; this is consistent with the earlier demonstration of pyrophosphorolysis of nicked DNA at 37°C (27).
DISCUSSION
Conformational activation
High fidelity DNA synthesis requires efficient nucleotide insertion when proper base pairing occurs and the base pairing events are physically separated from the site of chemistry by ∼10 Å. For pol β, bridging this distance is accomplished through a series of altered side-chain conformations associated with repositioning of the N-subdomain (i.e. fingers) relative to the C-subdomain (palm) upon binding a nucleoside triphosphate. Enzyme conformational activation is globally characterized by an open to closed structural transition inducing many subtle conformational adjustments. A defining step of this signaling cascade is the release of Asp192 from its salt bridge interaction with Arg258, allowing Asp192 to coordinate both the catalytic metal and nucleotide-bindng metal (Figure 1). Metal B binds in complex with dNTP (forward reaction) or PPi (reverse reaction). It has been suggested that, following nucleotide insertion, the ternary product complex undergoes a conformational change that facilitates metal/PPi release (30,31). The NMR studies presented here demonstrate that metal ions can play an active role in the induction of the conformationally-activated state in the absence of nucleotide ligand.
Metal interactions with the primer terminus
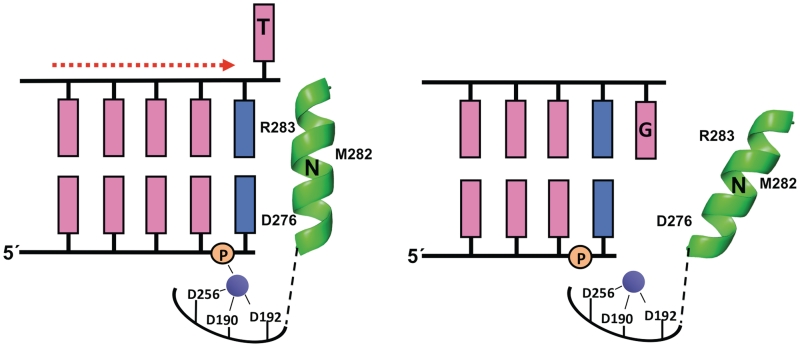
The NMR solution studies described here demonstrate that in the presence of 1-nt gapped DNA, Zn2+ can induce an active closed conformation that requires positioning of the primer terminus into the nascent base-pair-binding pocket (Figure 2B). Furthermore, they suggest that this results not only from the ability of the Zn2+ ion to interact with Asp192, but also from the ability of Zn2+ to interact with the 5′-phosphate group of the primer terminal nucleotide. Crystallographic structures of product complexes (post-insertion complex before translocation) of pol β (32) and pol λ (33)—also an X-family DNA polymerase—indicate that the lone active-site metal (B) coordinates PPi, two active-site aspartates (190 and 192), and the 5′-phosphate of the primer terminus. We suggest that neither the crystallographic structure of the metal-free nicked DNA complex nor the product complexes noted above correspond to the complexes observed in the solution NMR studies presented here, since they lack the catalytic metal A required for chemistry (DNA synthesis or pyrophosphorolysis). Because the metal B site is partially formed by non-bridging oxygens on the incoming dNTP (DNA synthesis) or PPi (pyrophosphorolysis), the metal activation observed here probably reflects binding to the catalytic metal A-site. In this context, primer relocation into the dNTP binding site completes the coordination sphere for the catalytic metal site and suggests a low energy barrier for translocation of the primer terminus when the templating base in the DNA gap is a pyrimidine (Figure 6).
Figure 6.
Zinc-induced DNA translocation. Schematic representation of pol β complexed with a 1-nt gapped DNA containing either a templating thymine (left) or a templating guanine (right). The vertical orientation of α-helix N (shown in green) where it (e.g. residues Asp276; and Arg283) interacts with the base pair in the nascent base-pair-binding pocket represents the activated state, while the tilted orientation of α-helix N that does not contact the empty nascent base-pair-binding pocket corresponds to the relaxed (‘open’) state. The primer terminus and its complementary template nucleotide base are shown in blue with the remaining bases in pink. The 5′-phosphate of the primer nucleotide is also indicated. The templating thymine is proposed to adopt an extrahelical orientation in the presence of Zn2+ (purple), while the templating guanine exhibits a preference for the base-stacked orientation. The catalytic metal site is indicated with coordinating ligands (Asp190, Asp192, Asp256).
Metal induced activation
The above observations raise more general questions about the ability of Zn2+ and other ions to promote translocation or stabilization of the DNA in the active site by interacting with the 5′-phosphate group of the primer terminus. We therefore investigated the effect of both Zn2+ and Mg2+ (Figures 3B and 5B) on a nicked DNA substrate. The NMR spectrum of the methionine-labeled pol β in the presence of Zn2+ and nicked DNA showed a pattern characteristic for conformational activation: broadening of the Met155 resonance, and broadening and characteristic shifts of the Met158 and Met282 resonances (Figures 3B and 5B). Thus, the Zn2+ ion stabilizes the active-site structure, resulting in a reversal of the signal transduction cascade so that the N-subdomain favors a closed conformation. The substitution of Mg2+ for Zn2+ produces a similar effect at 25°C, but with multiple resonances distributed between the non-activated and activated shift values (Figure 3B). The Met282 resonance observed in the pol β/nicked DNA/Mg2+ complex was broad, consistent with exchange between open and closed structures. The broadening might result from a conformational mixture characterizing the pol β/DNA/Mg2+ complex, or a mixture of pol β/DNA/Mg2+ and pol β/DNA complexes. Thus, the dynamic process being monitored could arise from slow conformational exchange and/or a slow Mg2+ association/dissociation process. Furthermore, studies utilizing nicked DNA at a higher temperature (35°C, Figure 5B) indicated that, from a conformational standpoint, the differences between Mg2+ and Zn2+ tended to be more quantitative than qualitative. Magnesium ions become more effective at inducing conformational activation at higher temperatures when the nascent base-pair-binding pocket is occupied (i.e. nicked DNA). For both metals, these effects involve reversal of information flow through the signal transduction pathway, which is most clearly supported by the studies with E295K pol β that blocks signal transmission (Figures 1 and 3C). Additionally, interaction with the 5′-phosphate group of the primer terminus appears to be an important component leading to translocation of 1-nt gapped DNA so that the primer terminus occupies the nascent base-pair-binding pocket (N-site, see below).
Primer terminus translocation
During catalytic cycling, the primer terminus must translocate from the site of nucleotide incorporation to the primer site, opening the dNTP-binding pocket for insertion of the next nucleotide. These have been referred to as the N- and P-sites, respectively (34). The factors that influence the translocation of the primer-terminus between the P- and N-sites are poorly understood.
In the absence of an incoming nucleotide, the position of the primer terminus of 1-nt gapped DNA is dependent on the presence of a divalent metal and the identity of the template base in the gap. For the primer terminus to occupy the N-site, the DNA gap must either move downstream or collapse (i.e. the nucleotide in the gap is extruded to an extra-helical position). The studies reported here demonstrate that the latter scenario is the most likely explanation since the ability to form a Zn2+-activated binary complex depends on the identity of the base in the gap (Figure 3A). Consistent with this interpretation, 2D NMR experiments on duplex DNA with different nucleotides opposite an abasic site indicate that pyrimidines easily assume an extra-helical position, whereas purines do not (35). The ability to stack within the DNA helix is also a key determinant for DNA polymerase discrimination during non-templated DNA synthesis (14). Thus, when a purine is in the gap the DNA remains in an extended conformation and the nascent base-pair-binding pocket is only partially filled with the templating purine. In contrast, when a pyrimidine is in that position, the primer terminus can position itself in the nascent base-pair-binding pocket to stack with α-helix N in a closed conformation and extrude the unpaired pyrimidine (Figure 6).
It has long been assumed that structures involving extra-helical nucleotides must be feasible as that would explain the ability of X-family polymerases to fill gap lengths longer than a single nucleotide; for such multi-nucleotide gapped substrates, bases in the templating strand downstream from the coding position in the gap must be extruded in order to deter synthesis of deletion intermediates. Crystallographic evidence for such an extra-helical base has been obtained in studies of ternary complex structures of pol λ with 2-nt gapped substrates (36). The templating base hydrogen-bonds with the incoming dNTP, and the downstream unpaired nucleotide is situated outside of the upstream and downstream duplexes, sequestered in a binding pocket formed by residues of α-helix N and the lyase domain. An extra-helical base corresponding to a deletion intermediate was also observed upstream of the active site in pol λ (37). For pol β, an extra-helical templating cytosine base has been observed in a structure containing a primer terminal A-A mismatch where the template A of the mismatch stacks with the primer terminus (38).
Pyrophosphorolysis
The observation of polymerase conformational activation in the absence of an incoming nucleotide was unexpected and appeared to represent an intermediate structure relevant to the reverse reaction, pyrophosphorolysis. This reaction requires several conditions: (i) the primer terminus in the N-site; (ii) substrate PPi; (iii) divalent metal ions; and (iv) conformational activation of the enzyme. The NMR results suggested that Mg2+ should be able to support the reverse reaction for nicked DNA and for 1-nt gapped DNA containing a pyrimidine base in the templating position. Vande Berg et al. (27) previously demonstrated this reaction for a pol β/nicked DNA/Mg2+ complex. Pyrophosphorolysis with a 1-nt pyrimidine gapped DNA at 37°C can readily be observed (Figure 4). Although the NMR data indicates that Zn2+ induced conformational activation for a single nucleotide-gapped DNA substrate with a templating pyrimidine, no pyrophosphorolysis was observed. This may result from the insolubility of Zn2+/PPi at neutral pH, as well as the poorer ability of the electronic configuration of the Zn2+ ion to promote catalysis. We conclude that conformational activation is a necessary, but not sufficient, condition for both the forward and reverse reactions. With adenine in the DNA gap (template position), Mg2+-dependent pyrophosphorolysis was not observed whereas with guanine, weak activity was observed. Alternatively, using identical conditions at room temperature, pyrophosphorolysis is not observed with gapped DNA, consistent with the lack of Mg2+-induced activation at the lower temperature.
E295K pol β
As illustrated in Figure 1, several key charged side-chains that mediate the signal transduction between the nascent base pair and the metal–ion-binding site alter their hydrogen-bonding pattern during conformational activation. Due to the important role of Glu295 in mediating the interaction between Arg283 and Arg258, it was expected that this non-conservative change should inhibit conformational activation. Meeting this expectation, Lang et al. (12) have reported E295K pol β to be inactive. Further insight into the molecular basis for Zn2+-induced conformational activation was obtained in our studies of E295K pol β. NMR experiments with the E295K mutant protein indicate that it is fully capable of binding DNA (Figure 3C), but that binding of the dNTP substrate fails to promote conformational activation. Thus, the methionine residues in the C- and N-subdomains exhibit almost no response to formation of the ternary complex. Consistent with this explanation, we have found that the addition of Zn2+ to the pol β/nicked DNA or 1-nt gapped DNA with a templating thymidine (Figure 3c) also failed to induce conformational activation, indicating that residue Glu295 plays a critical role in both the forward and reverse signaling processes.
Other implications for polymerase fidelity
It remains to be seen if metal-induced activation is specific to gap-filling DNA polymerases that must inherently interact with duplex DNA upstream and downstream of the growing primer terminus. The domain organization of pol β includes an 8-kDa domain that anchors the enzyme to the downstream duplex (39). When the primer 3′-terminus is near, the polymerase domain interacts with the upstream duplex DNA to complete gap-filling DNA synthesis. In contrast, replicative DNA polymerases interact primarily with upstream duplex DNA and only to a lesser extent with downstream single-stranded DNA. The low deletion-frameshift fidelity of polymerases belonging to the X-family is consistent with the suggestion that these polymerases modulate template/primer positioning during DNA synthesis. DNA polymerase β readily utilizes the downstream template base when the coding nucleotide base is absent (as in an abasic site) (40).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Results, Supplementary Figures S1–S5, Supplementary Tables S1 and S2, and Supplementary References [15–16 and 41–42].
FUNDING
Intramural research program of the National Institute of Environmental Health Sciences, National Institutes of Health, under projects (Z01-ES050147 to R.E.L.) and (Z01-ES050158 to S.H.W.), in association with National Institutes of Health (Grant 1U19CA105010). Funding for open access charge: Research Project Numbers (Z01-ES050147 to R.E.L. and Z01-ES050158 to S.H.W.). National Institute of Environmental Health Sciences.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Steitz TA, Smerdon SJ, Jager J, Joyce CM. A unified polymerase mechanism for nonhomologous DNA and RNA polymerases. Science. 1994;266:2022–2025. doi: 10.1126/science.7528445. [DOI] [PubMed] [Google Scholar]
- 2.Batra VK, Beard WA, Shock DD, Krahn JM, Pedersen LC, Wilson SH. Magnesium induced assembly of a complete DNA polymerase catalytic complex. Structure. 2006;14:757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin P, Pedersen LC, Batra VK, Beard WA, Wilson SH, Pedersen LG. Energy analysis of chemistry for correct insertion by DNA polymerase β. Proc. Natl Acad. Sci. USA. 2006;103:13294–13299. doi: 10.1073/pnas.0606006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sirover MA, Loeb LA. Metal activation of DNA synthesis. Biochem. Biophys. Res. Commun. 1976;70:812–817. doi: 10.1016/0006-291x(76)90664-1. [DOI] [PubMed] [Google Scholar]
- 5.Batra VK, Beard WA, Shock DD, Pedersen LC, Wilson SH. Structures of DNA polymerase β with active site mismatches suggest a transient abasic site intermediate during misincorporation. Mol. Cell. 2008;30:315–324. doi: 10.1016/j.molcel.2008.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snow ET, Xu LS, Kinney PL. Effects of nickel ions on polymerase-activity and fidelity during DNA-replication in vitro. Chemico-Biol. Interactions. 1993;88:155–173. doi: 10.1016/0009-2797(93)90089-h. [DOI] [PubMed] [Google Scholar]
- 7.Tabor S, Richardson CC. Effect of manganese ions on the incorporation of dideoxynucleotides by bacteriophage-T7 DNA-polymerase and Escherichia-coli DNA-polymerase-I. Proc. Natl Acad. Sci. USA. 1989;86:4076–4080. doi: 10.1073/pnas.86.11.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beard WA, Shock DD, Vande Berg BJ, Wilson SH. Efficiency of correct nucleotide insertion governs DNA polymerase fidelity. J. Biol. Chem. 2002;277:47393–47398. doi: 10.1074/jbc.M210036200. [DOI] [PubMed] [Google Scholar]
- 9.Kunkel TA, Bebenek R. DNA replication fidelity. Annu. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 10.Beard WA, Shock DD, Yang X-P, DeLauder SF, Wilson SH. Loss of DNA polymerase β stacking interactions with templating purines, but not pyrimidines, alters catalytic efficiency and fidelity. J. Biol. Chem. 2002;277:8235–8242. doi: 10.1074/jbc.M107286200. [DOI] [PubMed] [Google Scholar]
- 11.Beard WA, Osheroff WP, Prasad R, Sawaya MR, Jaju M, Wood TG, Kraut J, Kunkel TA, Wilson SH. Enzyme-DNA interactions required for efficient nucleotide incorporation and discrimination in human DNA polymerase β. J. Biol. Chem. 1996;271:12141–12144. doi: 10.1074/jbc.271.21.12141. [DOI] [PubMed] [Google Scholar]
- 12.Lang T, Dalal S, Chikova A, DiMaio D, Sweasy JB. The E295K DNA polymerase beta gastric cancer-associated variant interferes with base excision repair and induces cellular transformation. Mol. Cell. Biol. 2007;27:5587–5596. doi: 10.1128/MCB.01883-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J. A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase β. Biochemistry. 1996;35:12762–12777. doi: 10.1021/bi9529566. [DOI] [PubMed] [Google Scholar]
- 14.Beard WA, Shock DD, Batra VK, Pedersen LC, Wilson SH. DNA polymerase β substrate specificity: side chain modulation of the ‘A-rule’. J. Biol. Chem. 2009;284:31680–31689. doi: 10.1074/jbc.M109.029843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bose-Basu B, DeRose EF, Kirby TW, Mueller GA, Beard WA, Wilson SH, London RE. Dynamic characterization of a DNA repair enzyme: NMR studies of [methyl-13C]methionine-labeled DNA polymerase β. Biochemistry. 2004;43:8911–8922. doi: 10.1021/bi049641n. [DOI] [PubMed] [Google Scholar]
- 16.Kirby TW, DeRose EF, Beard WA, Wilson SH, London RE. A thymine isostere in the templating position disrupts assembly of the closed DNA polymerase β ternary complex. Biochemistry. 2005;44:15230–15237. doi: 10.1021/bi0511742. [DOI] [PubMed] [Google Scholar]
- 17.Meyer PR, Matsuura SE, So AG, Scott WA. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl Acad. Sci. USA. 1998;95:13471–13476. doi: 10.1073/pnas.95.23.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanes JW, Johnson KA. A novel mechanism of selectivity against AZT by the human mitochondrial DNA polymerase. Nucleic Acids Res. 2007;35:6973–6983. doi: 10.1093/nar/gkm695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koczor CA, Lewis W. Nucleoside reverse transcriptase inhibitor toxicity and mitochondrial DNA. Exp. Opin. Drug Metab. Toxicol. 2010;6:1493–1504. doi: 10.1517/17425255.2010.526602. [DOI] [PubMed] [Google Scholar]
- 20.John BK, Plant D, Hurd RE. Improved proton-detected heteronuclear correlation using gradient-enhanced Z and ZZ filters. J. Magn. Reson. 1993;101:113–117. [Google Scholar]
- 21.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 22.Johnson BA, Blevins RA. NMRView: a computer-program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 23.Zhu G, Bax A. Improved linear prediction of damped NMR signals using modified forward backward linear prediction. J. Magn. Reson. 1992;100:202–207. [Google Scholar]
- 24.Sawaya MR, Prasad P, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 25.Iwanaga A, Ouchida M, Miyazaki K, Hori K, Mukai T. Functional mutation of DNA polymerase β found in human gastric cancer - inability of the base excision repair in vitro. Mutat. Res. 1999;435:121–128. doi: 10.1016/s0921-8777(99)00036-1. [DOI] [PubMed] [Google Scholar]
- 26.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase β. Chem. Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 27.Vande Berg BJ, Beard WA, Wilson SH. DNA structure and aspartate 276 influence nucleotide binding to human DNA polymerase β: implication for the identity of the rate-limiting conformational change. J. Biol. Chem. 2001;276:3408–3416. doi: 10.1074/jbc.M002884200. [DOI] [PubMed] [Google Scholar]
- 28.Barshop BA, Adamson DT, Vellom DC, Rosen F, Epstein BL, Seegmiller JE. Luminescent immobilized enzyme test systems for inorganic pyrophosphate: assays using firefly luciferase and nicotinamide-mononucleotide adenylyl transferase or adenosine-5′-triphosphate sulfurylase. Anal. Biochem. 1991;197:266–272. doi: 10.1016/0003-2697(91)90387-9. [DOI] [PubMed] [Google Scholar]
- 29.Dunlap CA, Tsai M-D. Use of 2-aminopurine and tryptophan fluorescence as probes in kinetic analyses of DNA polymerase β. Biochemistry. 2002;41:11226–11235. doi: 10.1021/bi025837g. [DOI] [PubMed] [Google Scholar]
- 30.Dahlberg ME, Benkovic SJ. Kinetic mechanism of DNA polymerase I (Klenow Fragment): identification of a second conformational change and evaluation of the internal equilibrium constant. Biochemistry. 1991;30:4835–4843. doi: 10.1021/bi00234a002. [DOI] [PubMed] [Google Scholar]
- 31.Patel SS, Wong I, Johnson KA. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 32.Arndt JW, Gong W, Zhong X, Showalter AK, Liu J, Dunlap CA, Lin Z, Paxson C, Tsai M-D, Chan MK. Insight into the catalytic mechanism of DNA polymerase β: structures of intermediate complexes. Biochemistry. 2001;40:5368–5375. doi: 10.1021/bi002176j. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-Diaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC. A closed conformation for the pol λ catalytic cycle. Nat. Struct. Mol. Biol. 2005;12:97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- 34.Sarafianos SG, Clark AD, Jr, Das K, Tuske S, Birktoft JJ, Ilankumaran P, Ramesha AR, Sayer JM, Jerina DM, Boyer PL, et al. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. EMBO J. 2002;21:6614–6624. doi: 10.1093/emboj/cdf637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuniasse P, Fazakerley GV, Guschlbauer W, Kaplan BE, Sowers LC. The abasic site as a challenge to DNA polymerase: a nuclear magnetic resonance study of G, C and T opposite a model abasic site. J. Mol. Biol. 1990;213:303–314. doi: 10.1016/S0022-2836(05)80192-5. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Diaz M, Bebenek K, Larrea AA, Havener JM, Perera L, Krahn JM, Pedersen LC, Ramsden DA, Kunkel TA. Template strand scrunching during DNA gap repair synthesis by human polymerase λ. Nat. Struct. Mol. Biol. 2009;16:967–972. doi: 10.1038/nsmb.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Diaz M, Bebenek K, Krahn JM, Pedersen LC, Kunkel TA. Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase. Cell. 2006;124:331–342. doi: 10.1016/j.cell.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 38.Batra VK, Beard WA, Shock DD, Pedersen LC, Wilson SH. Nucleotide-induced DNA polymerase active site motions accomodating a mutagenic DNA intermediate. Structure. 2005;13:1225–1233. doi: 10.1016/j.str.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Prasad R, Beard WA, Wilson SH. Studies of gapped DNA substrate binding by mammalian DNA polymerase ß: dependence on 5′-phosphate group. J. Biol. Chem. 1994;269:18096–18101. [PubMed] [Google Scholar]
- 40.Efrati E, Tocco G, Eritja R, Wilson SH, Goodman MF. Abasic translesion synthesis by DNA polymerase β violates the “A-rule”: novel types of nucleotide incorporation by human DNA polymerase ß at an abasic lesion in different sequence contexts. J. Biol. Chem. 1997;272:2559–2569. doi: 10.1074/jbc.272.4.2559. [DOI] [PubMed] [Google Scholar]
- 41.Mueller GA, DeRose EF, Kirby TW, London RE. NMR assignment of polymerase beta labeled with H-2, C-13, and N-15 in complex with substrate DNA. Biomol. NMR Assignments. 2007;1:33–35. doi: 10.1007/s12104-007-9007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogers MT, Woodbrey JC. A proton magnetic resonace study of hindered internal rotation in some substituted N,N-dimethylamides. J. Phys. Chem. 1962;66:540–546. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.