Abstract
Myotonic Dystrophy type I (DM1) is caused by an abnormal expansion of CTG triplets in the 3′ UTR of the dystrophia myotonica protein kinase (DMPK) gene, leading to the aggregation of the mutant transcript in nuclear RNA foci. The expanded mutant transcript promotes the sequestration of the MBNL1 splicing factor, resulting in the misregulation of a subset of alternative splicing events. In this study, we identify the DEAD-box RNA helicase p68 (DDX5) in complexes assembled onto in vitro-transcribed CUG repeats. We showed that p68 colocalized with RNA foci in cells expressing the 3′UTR of the DMPK gene containing expanded CTG repeats. We found that p68 increased MBNL1 binding onto pathological repeats and the stem–loop structure regulatory element within the cardiac Troponin T (TNNT2) pre-mRNA, splicing of which is misregulated in DM1. Mutations in the helicase core of p68 prevented both the stimulatory effect of the protein on MBNL1 binding and the colocalization of p68 with CUG repeats, suggesting that remodeling of RNA secondary structure by p68 facilitates MBNL1 binding. We also found that the competence of p68 for regulating TNNT2 exon 5 inclusion depended on the integrity of MBNL1 binding sites. We propose that p68 acts as a modifier of MBNL1 activity on splicing targets and pathogenic RNA.
INTRODUCTION
Myotonic dystrophy type 1 (DM1) is a dominant autosomal neuromuscular disorder, characterized by multisystemic defects affecting muscle, heart, brain and endocrine systems (1). DM1 is one the most frequent form of muscular dystrophy in adults that is caused by an expansion of CTG triplets in the 3′ untranslated region of the dystrophia myotonica protein kinase (DMPK) gene (2–4). The mutant allele is transcribed, correctly spliced and polyadenylated but it is not translated into a protein. Instead, expanded transcripts are retained in the nucleus and accumulate as discrete RNA foci, located at the boundary of nuclear speckles (5–7). However, nuclear/cytoplasmic distribution of mutant DMPK remains controversial as cytoplasmic aggregates have also been detected (8). It is now well established that the expanded CUG repeats are toxic for the cells and play a major role in DM1 pathogenesis. Based on structural studies on short CUG repeats, it has been proposed that expanded CUG repeats fold into an imperfect hairpin structure that interferes with the activities of RNA binding proteins and alters their normal cellular function. The muscleblind-like 1 protein (MBNL1) was identified by its ability to bind to CUG repeats (9). MBNL1 is a splicing factor that interacts with RNA through its zinc-finger domains. Recently it has been shown that MBNL1 binds to single-stranded and structured RNA, which raises the question of how the protein recognizes different RNA targets (10). In the DM1 context, MBNL1 colocalizes with nuclear foci of CUG repeats and its sequestration by the expanded repeats leads to a loss-of-function of the protein [review in refs (11,12)]. Another RNA-binding protein involved in DM1 is CUGBP1. This factor plays an important role in several post-transcriptional processing steps, including translation, RNA stability and alternative splicing. Instead of being sequestered by the repeats, the steady-state level of CUGBP1 is increased in DM1 tissues due to its hyperphosphorylation by protein kinase C, leading to a gain of activity of the protein (13). MBNL1 and CUGBP1 are antagonistic splicing factors that are involved in the reprogramming of splicing events during fetal to post-natal development in muscle (14). The sequestration of MBNL1 and the upregulation of CUGBP1 in DM1 are proposed to trigger the misregulation of alternative splicing of a subset of muscle and brain-specific transcripts, leading to the re-expression of fetal isoforms in adult tissues. Several of these abnormal splicing events correlate with clinical symptoms such as myotonia, insulin resistance and heart conduction defects, which are caused by the misregulation of the muscle-chloride channel, insulin receptor (INSR) and cardiac Troponin T (TNNT2) pre-mRNA, respectively (15–18). In addition to MBNL1 and CUGBP1, other factors such as transcription factors and hnRNP F/H have been proposed to be involved in DM1 (19,20). However, their function in the disease is unclear. Studies using the CUG RNA fly model have identified modifiers of CUG toxicity phenotypes (21), suggesting that factors or signaling pathway other than MBNL1 and CUGBP1 could be involved in DM1 pathogenesis. Recently, the mislocalization of the transcription factor SHARP in DM1 has been associated with alteration of steady-state levels of numerous mRNAs that are important for muscle development (22). Moreover, a recent study has demonstrated that the processing of the pre-miR1 is altered in DM1, reinforcing the idea that other mechanisms are involved in DM1 pathophysiology (23).
The aim of this work was to isolate new factors that bind to CUG repeats. Using an affinity chromatography strategy with an RNA containing 95 pure CUG repeats, we identified the RNA helicase p68 (DDX5). p68 is a prototype of DEAD-box RNA helicase proteins. This family is characterized by a conserved core, consisting of nine conserved motifs including the DEAD signature, which gives rise to the name to these proteins (24). p68 is involved in many aspects of RNA metabolism including transcription, RNA processing, RNA export, translation and mRNA degradation (25,26). Here, we show that p68 colocalizes with RNA foci in different cell types that overexpressed expanded CUG repeats. In vitro experiments suggest that p68 promotes a conformational change of CUG repeats that favors MBNL1 binding or stabilizes the complex through additional interactions. We also found that p68 regulates alternative splicing of TNNT2 exon 5 and we demonstrate that the competence for regulation depends on MBNL1. From these results, we propose that p68 acts as a modifier of MBNL1 activity on splicing targets and pathogenic RNAs.
MATERIALS AND METHODS
Plasmids and constructions
Plasmid CTG 95 was constructed by cloning polymerase chain reaction (PCR) fragment containing CTG repeats in plasmid pSP72 that was digested with PvuII. CTG repeats were obtained by PCR using a sense oligonucleotide containing 7 CTG repeats and an antisense oligonucleotide containing 7 CAG repeats. PCR fragments containing varying lengths of CTG repeats were purified on agarose gel and cloned into the pSP72 plasmid. Several clones were subjected to sequence analysis. According to the orientation of the repeats in the plasmid, different lengths of CUG or CAG repeats were obtained. Plasmid CTG 95 contains 95 CTG repeats. Plasmid CAG 61 contains 61 CAG repeats. Plasmids containing 14 CTG repeats or 16 CAG repeats were chosen for gel retardation assays. Plasmid-containing 62 CCTG repeats were obtained by PCR using a sense oligonucleotide containing 6 CCTG repeats and an antisense oligonucleotide containing 6 CAGG repeats. PCR fragments were cloned into the pSP72 plasmid and proceed as for CTG and CAG plasmids. Plasmids containing the 3′UTR of DMPK gene with 5 or 200 pure CTG repeats were described previously (27). Plasmid-expressing DMPK exons 11–15 containing 960 interrupted CUG repeats in exon 15 were described previously (28). The 3′ UTR of DMPK gene containing 960 interrupted CTG was also cloned into a Tet-on inducible lentiviral construct as previously described (29). Mutations in the helicase core domains II and IV of human p68/DDX5 were made by reverse PCR from pcDNA4 p68/Ha-Myc-His provided by Dr D. Auboeuf. For mutation in domain II (p68 mt2), sense and antisense oligonucleotides are 5′-AGATAGAATGCTTGATATGGGC-3′ and 5′-GCTTCATTAAGGACAAGGTAGG-3′, respectively. For mutation in domain IV (p68 mt4), sense and antisense oligonucleotides are 5′-GCTGTGGAAACCAAAAGAAGA-3′ and 5′-AACAATGGTTTTATTCTCCTTCTC-3′, respectively. The wild-type and mutant 4G cardiac Troponin T (TNNT2) minigenes were described previously (30). Mutant TNNT2, CUG stem (CUGS) and GUF were described previously (31). The TNNT2 RNA used for UV-cross-linking experiments was generated by PCR from the wild-type TNNT2 plasmid using sense and antisense oligonucleotides (5′-ACACATACGATTTAGGTGACACTATAGAACCCAGACTAACCTGT-3′ and 5′-CTGAGGTTCAGGGAGTGG-3′, respectively). Plasmid p68 for expression in Escherichia coli was generated by PCR from pcDNA4 p68/Ha-Myc-His using a sense oligonucleotide (5′-ATCTAGGATCCATGTCGGGTTATTCGA-3′) and an antisense oligonucleotide (5′-AGATCTCGAGTTGGGAATATCCTGT-3′). The PCR product after digestion with NdeI and XhoI was cloned in a variant of pet28 (gift from Dr H. Le Hir) that was digested by NdeI and XhoI. This vector contained in the N-terminal a CBP tag followed by a TEV protease and in the C-terminal a His-tag (32). The deletion of a C-terminal part of p68 giving rise to p68 ΔCt2 was made by reverse PCR using a sense oligonucleotide (5′-CTCGAGCACCACCACCACCACCACTGA-3′) and an antisense oligonucleotide (5′-ATAATTTTCCCTGTCTCTAA-3′) using pet28/p68 as a template. p68ΔCt2 containing mutations in the helicase core domains II and IV were made by reverse PCR from wild-type p68 ΔCt2. Plasmid pGEX-6P1-MBNL1/40 kDa isoform was described previously (33).
Affinity capture of protein complexes and MALDI analysis
HeLa cell nuclear extracts were purchased from A. Miller (Cilbiotech, B-7000 Mons, Belgium). Myoblast and myotube nuclear extracts from C2C12 were prepared as described (34). Biotinylated in vitro-transcribed CUG95 repeats bound to streptavidin agarose were incubated with 40% nuclear extracts under splicing conditions as described (35). Proteins eluted from the beads were separated by electrophoresis and detected by Coomassie staining. Fourteen protein bands were excised from the gel and identified using a nanoLC MS/MS-Orbitrap spectrometer (platform of proteomic of South-West Paris, INRA).
Purification of recombinant proteins
Recombinant protein GST-MBNL1 and GST-UAP56 (DDX39) were expressed in E. coli BL21 (DE3) and purified using Glutathione Uniflow resin (BD biosciences Clontech) according to standard procedure. GST-UAP56 was treated by thrombin to eliminate the GST tag according to standard procedure. Recombinant protein p68 ΔCt2 and eIF4A3 were expressed in E. coli BL21 (DE3) and successively purified on calmodulin resin (Stratagene) and on nickel sepharose (GE-healthcare) as previously described (32). Proteins were dialyzed against buffer D (36).
Western blot analysis and antibodies
Western blots were performed according to standard procedure with the following antibodies: anti-MBNL1 monoclonal antibodies [MB1a(4A8), gift from Dr G. Morris (7)], anti-p68 /DDX5 monoclonal antibodies (1/200, SantaCruz, sc 81350), anti-p72/DDX17 polyclonal antibodies (1/1000, Bethyl laboratories A300-509 A), anti-Myc monoclonal antibodies (1/500, sigma), anti-Flag monoclonal antibodies (1/500), anti-Emerin (1/500, SantaCruz, sc25284). The bands were detected with the SuperSignal West Pico detection kit (Pierce) and quantified with a Fuji LAS 3000.
UV-cross-linking, immunoprecipitation and gel retardation experiments
RNAs for gel shift experiments and UV-cross-linking experiments were transcribed using a cap analogue and SP6 or T7 RNA polymerase in the presence of [α-32P]UTP or [α-32P]CTP. Uniformly labeled RNA (10 fmol) were incubated for 15 min at 30°C, in 10 µl with recombinant MBNL1 and p68 as indicated in the figures under splicing conditions with 0.5 mM ATP, 1 mM MgCl2 and 20 mM creatine-phosphate. Samples were UV irradiated for 15 min and treated with 0.3 µg RNase A and 10 U of RNase T1 at 37°C for 30 min. Immunoprecipitation experiments were carried out as described with anti-MBNL1 (clone 3A4, sc47740 SantaCruz) or p68 monoclonal antibodies immobilized on A/G agarose (34). The cross-linked proteins were resolved on a 10% sodium dodecyl sulfate (SDS) polyacrylamide gel and visualized by a PhosphorImager. RNA mobility shift assays were performed as described with recombinant MBNL1 and p68 ΔCt2 as indicated in the figures (37). Recombinant proteins were diluted in Dignam buffer D containing 0.5 mg/ml bovine serum albumin (BSA). The reaction was incubated for 15 min at 30°C and the protein complexes were resolved by electrophoresis on a 4% non-denaturing polyacrylamide gel (39/1), 0.5×Tris/Borate/EDTA buffer (TBE). RNA bands were quantified using a PhosphorImager (Molecular Dynamics).
Cell culture
Cos, HeLa and N1E115 cells were grown at 37°C in Dulbecco's-modified Eagle's medium with 4500 mg/l d-glucose (DMEM) supplemented with 10% fetal bovine serum and antibiotics. C2C12 cells were grown in the same medium supplemented with 20% fetal bovine serum. Human muscle cells were derived from primary human satellite cells and immortalized as described (38). Cells were grown in Ham's F10 medium supplemented with 20% fetal bovine serum and antibiotics. Cultures of human myoblasts were also infected with lentiviral vector expressing the Tet-on inducible 960 CTG construct. Transduction was carried out overnight in the presence of polybrene (4 µg/ml) as previously described (39). Cells were grown in the same medium as above but with 50 µg/ml Gentamicin (Invitrogen). For immunofluorescence cells were seeded in 24-well vessel and grown to ∼90% confluence. Cells were co transfected with 0.4 µg of plasmids expressing the repeats and 0.4 µg of plasmids encoding proteins using lipofectamin 2000 (Invitrogen) according to the manufacturer's recommendations. For plasmid DNA transfection, human muscle cells were seeded in six-well vessel, and grown to ∼90% confluence. Cells were cotransfected with 0.5 µg minigenes and protein expression vectors with lipofectamine LTX plus reagent (Invitrogen, ratio 3 : 1), according to the manufacturer's recommendations. Cells were harvested 24 h later. RNA and proteins were prepared from the same plates using NucleoSpin RNA/protein kit (Macherey-Nagel) and splicing patterns were analyzed by RT–PCR in the presence of [32P]dCTP as previously described (37). PCR products were resolved on 6% denaturing polyacrylamide gels and quantified using a PhosphorImager. siRNAs and primers are listed in Supplementary Table S3. SiRNA duplex was transfected at 50 nM with lipofectamine RNAiMAX (Invitrogen) in growth medium without antibiotics. Twenty-four hours later, the cells were cotransfected with 50 nM siRNA and 0.5 µg of minigene and p68 or MBNL1 expression plasmid using lipofectamine 2000 (Invitrogen). The media was replaced with growth medium lacking antibiotics 3 h later. Cells were harvested 24 h after transfection of the minigene. RNA and proteins were prepared as described above.
RNA–FISH combined with immunofluorescence
FISH was done as described using (CAG)8-Cy3 DNA oligonucleotide probe (40). Glass coverslips containing plated cells were fixed in 4% paraformaldehyde in PBS (pH 7.4) for 20 min and washed three times with PBS. The coverslips were incubated for 5 min in PBS/0.5% Triton X-100 and washed three times with PBS before prehybridization in 30% formamide, 2× SCC for 10 min. The coverslips were hybridized for 2 h at 37°C in 30% formamide, 2× SCC, 2 mM vanadyl ribonucleoside, 10 μg/ml BSA and 0.5 μg (CAG)8-Cy3 DNA oligonucleotide probe (Sigma). The coverslips were washed twice in 2× SCC/30% formamide at 42°C and twice in 1× SCC at room temperature. Following FISH, the coverslips were washed twice in PBS. The coverslips were incubated with primary antibodies DDX5 (1/2000 dilution, Abcam ab21696), c-myc (1/500 dilution, clone 9E10; Santa Cruz), M2-FLAG (1/200 dilution, clone A2220, Sigma) and polyclonal antibodies DDX17 (1/400 dilution, Bethyl laboratories A300-509 A) in PBS/0.1%BSA at room temperature for 60 min. The coverslips were washed four times with PBS/0.1% Tween20 before incubation with a goat anti-rabbit or rabbit anti-mouse secondary antibody conjugated with Alexa-Fluor 488 (1/1000 dilution; Molecular probes, Invitrogen) in PBS/0.1% BSA for 60 min. Then, the coverslips were incubated for 10 min in PBS/0.1% BSA/DAPI (1/10 000 dilution) and rinsed twice in PBS before mounting in Fluorescent Mouting Medium (DAKO). Slides were examined using either a simple fluorescence microscope (Leica) or a Leica DM4000 B confocal microscope, equipped with a Leica 100× HCX Plan Apo CS 1.40 objective, in 1 -μm optical sections.
RESULTS
p68/DDX5 and p72/DDX17 form aggregates that colocalize with CUG repeats
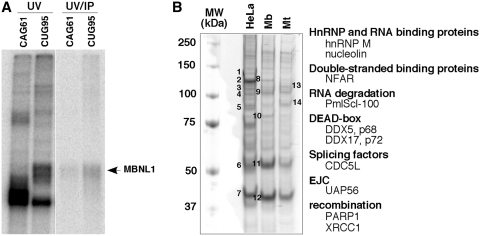
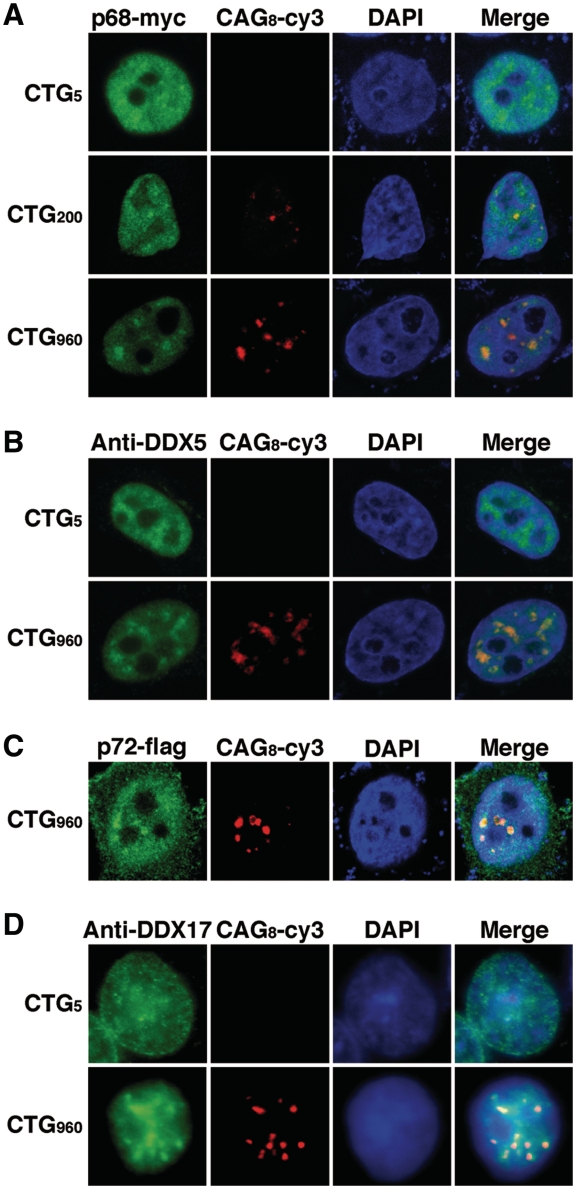
In an attempt to find novel factors involved in DM1 pathophysiology, protein complexes assembled onto biotinylated CUG repeats were purified by affinity chromatography on streptavidin-agarose beads. We verified that MBNL1 from HeLa nuclear extracts became cross-linked to a biotinylated RNA containing 95 pure CUG repeats (Figure 1A). Therefore, streptavidin-agarose beads coupled to the biotinylated (CUG)95 RNA was used to capture protein complexes from HeLa or myogenic nuclear extracts. After several washing steps, proteins were eluted and separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE). Numerous proteins were detected (Figure 1B). Some are common to the three nuclear extracts tested while others seemed to be specific to the cell type or to the differentiation status of muscle cells (Figure 1B). Fourteen bands were selected, from which about 100 proteins were identified by mass-spectrometry (Figure 1B and Supplementary Table S1). We focused on proteins that presented interesting features regarding their properties (double-stranded RNA proteins, splicing factors) or because they are involved in myogenesis (Figure 1B). To test the putative implication of the candidates in DM1, we investigated the localization of the proteins by RNA–FISH/immunofluorescence experiments in Hela cells that had been cotransfected with a plasmid encoding the 3′UTR of the DMPK gene containing either 960 interrupted CTG repeats (28) or non-expanded repeats (5 CTG) as a control. Most of the candidates did not colocalize with RNA foci as exemplified by the RNA helicase UAP56/DDX39 and the splicing factor CDC5L (Supplementary Figure S1 and Supplementary Table S2). In contrast, the RNA helicase p68 (DDX5) colocalized with RNA foci (Figure 2). In the absence of expanded repeats, myc-tagged p68 showed a diffuse localization in the nucleoplasm. However in the presence of expanded repeats, p68-myc formed aggregates that colocalized with the foci as shown by FISH/immunofluorescence using anti-Myc monoclonal antibodies (Figure 2A). The colocalization of exogenous p68 was not cell-line dependent, because aggregates were observed in different cell types such as myogenic (C2C12) and neuronal (NIE115) cell lines (Supplementary Figure S2). We also showed that p68 accumulates with CUG RNA foci in cells that had been cotransfected with a 3′UTR DMPK minigene containing 200 pure CUG repeats (41) (Figure 2A). We next tested whether endogenous p68 colocalizes with CUG repeats. Immunofluorescence with monoclonal p68 antibodies confirmed that endogenous p68 colocalized with expanded CUG repeats (Figure 2B). p72 (DDX17) is a paralog of p68 that is highly similar to p68 (42). p72 was also found in complexes associated with in vitro transcribed 95 CUG repeats. We thus investigated whether p72 colocalized with the 960 CUG repeats. The results showed that p72 tagged with a flag epitope and endogenous p72 formed aggregates that colocalized with RNA foci (Figure 2C and D). In conclusion, our results suggest that the DEAD-box RNA helicases p68 and p72 are novel factors that localize in CUG RNA aggregates that recruit MBNL1.
Figure 1.
Affinity capture of protein complexes from HeLa and myogenic nuclear extracts by an in vitro transcribed RNA containing 95 CUG repeats. (A) MBNL1 is cross-linked to 32P-labeled CUG95 repeats. In vitro transcribed (CUG)95 or (CAG)61 RNAs labelled with [α-32P]CTP were incubated with 40% HeLa nuclear extracts. After UV cross-linking an aliquot was precipitated with anti-MBNL1 antibodies. The cross-linked proteins were separated by SDS-PAGE. (B) Proteins from HeLa nuclear extracts, myoblast (Mb) and myotube (Mt) nuclear extracts from C2C12 were separated onto a 10% polyacrylamide denaturing gel and detected by Coomassie Brilliant Blue staining. Numbers 1 through 14 refer to bands that have been cut out from the gel and identified by mass-spectrometry. A subset of proteins identified by mass spectrometry is indicated.
Figure 2.
p68 and p72 colocalized with CUG RNA foci in HeLa cells. (A) Cells were cotransfected with a plasmid encoding p68-myc and plasmids expressing either 5, 200 or 960 CTG repeats. Expression of p68-myc was detected by immunofluorescence using monoclonal anti-Myc antibodies, followed by rabbit anti-mouse A488 antibodies. Foci were detected by RNA-FISH using Cy3-labeled (CAG)8 probes. Nuclei were stained with DAPI. (B) Endogenous p68 colocalizes with CUG repeats. p68/DDX5 was revealed with a DDX5 monoclonal antibody. (C) Same experiment as in (A) except that cells were cotransfected with a Flag-p72 expression vector. Expression of Flag-p72 was detected by immunofluorescence using anti-Flag antibodies. (D) Endogenous p72 colocalizes with CUG repeats. P72/DDX17 was revealed with DDX17 polyclonal antibodies.
p68 is a modifier of MBNL1 binding on CUG repeats
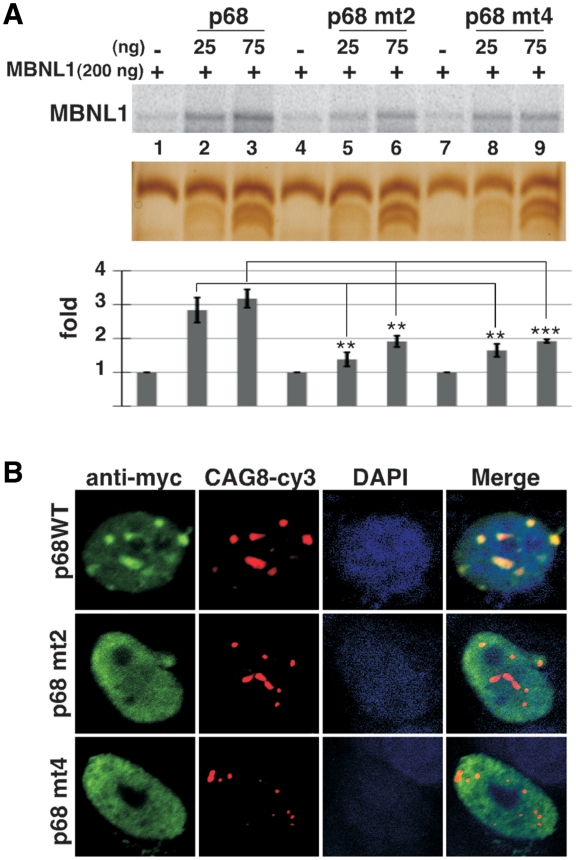
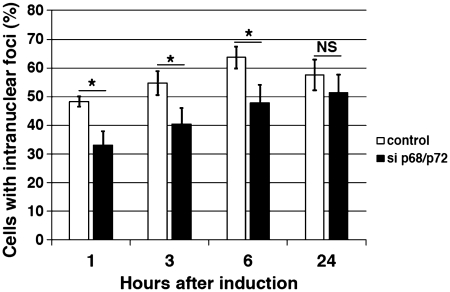
Structural analyses have shown that MBNL1 binds to CUG repeats via its Watson-Crick interface, suggesting that the long A-form stem of the CUG repeats must be partially unfolded (43). One function ascribed to DEAD-box RNA helicases is to separate strands of short RNA duplexes to promote the remodeling of RNA–protein complexes (25). As RNA chaperones, these proteins facilitate the correct folding of structured-RNA molecules (44). Thus, we hypothesized that RNA helicase p68 and/or p72 could locally disrupt the G–C pairs of the CUG helix allowing the recruitment or the stabilization of MBNL1 to the repeats. To test this hypothesis, recombinant protein MBNL1 was cross-linked to in vitro transcribed 32P RNA containing 95 CUG repeats in the absence or presence of recombinant p68 protein under splicing conditions. We focused on p68 because recombinant p72 was insoluble in our hands. To increase the solubility of the recombinant p68 in E. coli, 100 amino-acids of p68 were deleted from the C-terminus, keeping NLS and NES signals intact (45). This truncated p68 protein (p68ΔCt2) was still able to colocalize with the RNA foci of expanded CUG repeats and to regulate splicing (see below) as did the full-length p68 (Supplementary Figure S3). As previously observed in other studies (9), MBNL1 was cross-linked to CUG repeats even in the absence of p68 (Figure 3A). However, in the presence of increasing concentrations of recombinant p68, the amount of MBNL1 cross-linked to the repeats was significantly increased (Figure 3A and supplementary Figure S4). Immunoprecipation with monoclonal antibodies against MBNL1 confirmed that the cross-linked band was indeed MBNL1 (Figure 3B). As already reported, p68 was not efficiently cross-linked to RNA using UV-cross-linking (46). However, we confirmed the interaction of p68 with CUG repeats by gel retardation assays (Supplementary Figure S5). It has been shown that MBNL1 was recruited to RNA foci in cells expressing CAG repeats (28). Thus, we tested whether p68 had an effect on MBNL1 binding on these repeats. Results shown in Figure 3C indicated that p68 was able to increase the binding of MBNL1 on CAG repeats in a similar range to those obtained on CUG repeats. Myotonic dystrophy type 2 (DM2) is the second form of myotonic dystrophy (47). It has been shown that expanded CCUG repeats in DM2 also form RNA foci that recruit MBNL1 (48). Interestingly, the stimulating effect of p68 was less efficient on CCUG repeats than on the other repeats (Figure 3C). This suggests that the effect of p68 on MBNL1 binding might depend on particular structural features embedded in the repeats. We next tested whether the stimulating effect on MBNL1 binding was specific to p68. UV-cross-linking experiments were performed in the presence of UAP56/DDX39, one of the candidates found in complexes assembled onto (CUG)95 repeats and another DEAD-box protein, eIF4A3. The stimulation was markedly reduced in the presence of either UAP56 or eIF4A3, suggesting that p68 exhibits specificity toward the repeats despite the fact that the catalytic core of the DEAD-box helicase family is highly conserved (Figure 3D). To further characterize the requirement of p68 for MBNL1 binding, we tested whether mutations that affected ATPase/helicase activity and RNA binding altered the stimulatory effect of p68 on MBNL1 binding. Mutations D248N of the DEAD-box motif and mutation F346A in motif IV, which have been shown to be important for these activities in other DEAD-box proteins (24,49) reduced by 2-fold the ability of p68 to stimulate MBNL1 binding (Figure 4A). Consistent with this in vitro analysis, mutations in both the DEAD motif and RNA binding motif IV abolish colocalization of p68 to RNA foci (Figure 4B). Altogether, these results suggest that p68 acts as a specific factor to facilitate MBNL1 binding onto CUG repeats. Previous experiments have proposed that MBNL1 is a primary determinant of RNA foci formation (50). To examine whether knockdown of both p68 and p72 by siRNA has an effect on RNA foci formation, human muscle cells were transduced with a 960 CTG construct under the control of a Tet-on inducible promoter. RNA foci were detected as early as 1 h following the addition of doxycycline to the culture medium (Figure 5). In contrast, the number of nuclei showing RNA foci was significantly reduced in induced cells treated by siRNA against p68 and p72, particularly at the early time points (1, 3 and 6 h) following doxycycline addition. No difference could be observed at 24 h between induced-cells that were treated or non-treated by siRNA against both p68 and p72 (Figure 5). Altogether, our results suggest that p68 and p72 play a role in the formation of RNA foci by facilitating the binding of MBNL1 and that the interaction of these proteins with RNA foci is transient.
Figure 3.
p68 increases the binding of MBNL1 to 95 CUG repeats. (A) Plasmid (CUG)95 was in vitro transcribed in the presence of [α-32P]UTP. Labeled (CUG)95 RNA was incubated with a constant amount of recombinant MBNL1 protein (200 ng) and increasing amounts of recombinant p68ΔCt2 under splicing conditions with ATP. Proteins cross-linked to labeled RNA were separated on a 10% SDS–PAGE. Bottom image shows a silver stain of a gel run in parallel. Note that p68 ΔCt2 migrates as two bands that are recognized by anti-p68 antibodies (data not shown). (B) The cross-linked proteins shown in (A) were immunoprecipitated with anti-MBNL1 antibodies and separated by SDS-PAGE. (C) (CUG)95, (CAG)61 and (CCUG)62 RNAs were labeled with [α-32P]CTP and used for UV-cross-linking experiments. (D) The increase of cross-linked MBNL1 to CUG repeats is specific to p68. Labeled (CUG)95 was incubated with 200 ng of recombinant MBNL1 protein and 25 or 75 ng of recombinant p68ΔCt2, UAP56 or eIF4A3 proteins. Quantifications result from three independent experiments, with error bars indicating standard deviation. *P < 0.05, **P < 0.01, ***P < 0.001.
Figure 4.
(A) Mutations in the helicase core domain of p68 modify its ability to increase MBNL1 binding. UV-cross-linking experiments with labeled (CUG)95 were performed with 200 ng of recombinant MBNL1 and 25 or 75 ng of wild-type recombinant p68ΔCt2 or p68ΔCt2 mutated in domain II (p68 mt2, mutation D248N), or in domain IV (p68 mt4, mutation F346A). Quantifications result from three independent experiments, with error bars indicating standard deviation. *P < 0.05, **P < 0.01, ***P < 0.001. (B) Mutations in the helicase core domain of p68 strongly affect colocalization of the protein with CUG repeats. RNA/FISH experiments were performed as described in Figure 2 with wild-type p68-myc expressing vector or p68-myc expressing vectors mutated in domain II (DEAD) or IV.
Figure 5.
P68 and p72 facilitate RNA foci formation. Human muscle cells expressing 960-interrupted CTG repeats under the control of a Tet-on inducible promoter was used to attend the early steps of RNA foci formation. The number of nuclei containing foci was determined at different times after doxycycline addition (1, 3, 6 and 24 h) in non-treated or treated cells by siRNA to inhibit p68/p72. SiRNA duplexes were transfected 48 h before CUG repeats induction by doxycycline. Quantification results from three independent experiments. Approximately 300 cells were counted for each condition, with error bars indicating standard deviation *P < 0.05, **P < 0.01, ***P < 0.001.
p68 regulates alternative splicing of TNNT2 that is misregulated in DM1
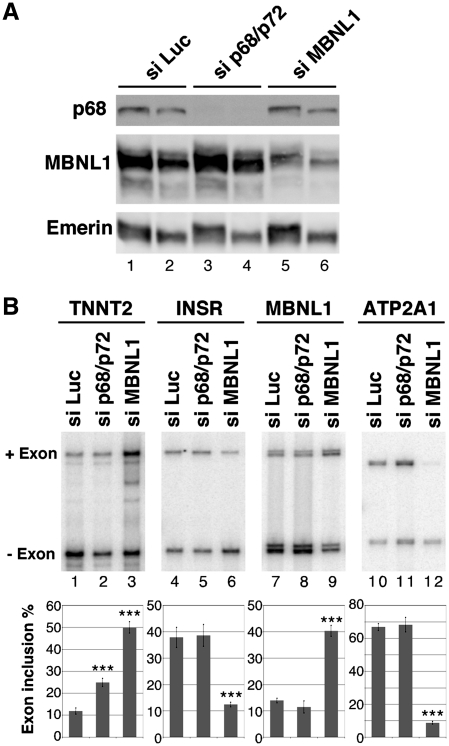
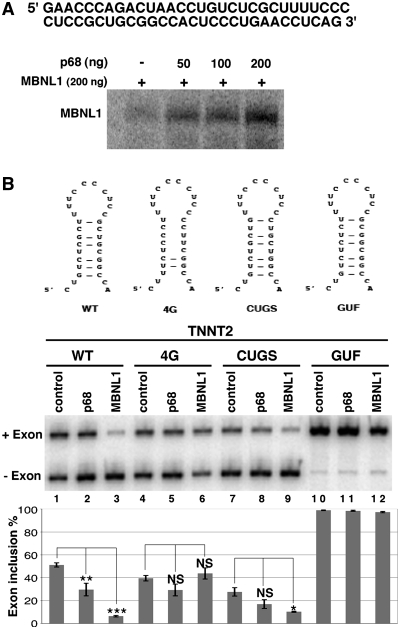
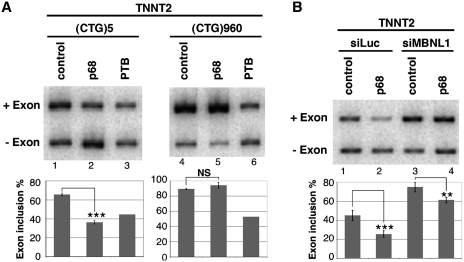
One of the characteristic features of DM1 is the misregulation of alternative splicing of a subset of transcripts that resulted in the re-expression of fetal isoforms in adult tissues. It was shown that some of these events are responsive to MBNL1. In this study, we have found that p68 modifies MBNL1 binding on CUG repeats. Therefore, we wondered whether p68 played a role in the regulation of alternative splicing events that are regulated by MBNL1 and misregulated in DM1. To test this hypothesis we first inactivated both p68 and p72 by siRNA interference using a siRNA that targeted both proteins. Western blot analysis showed that about 95% of p68 was depleted from the cells (Figure 6A). The level of p72 depletion was also around 95% (Supplementary Figure S6). The knockdown of both p68 and p72-activated splicing of endogenous TNNT2 alternative exon 5 as a MBNL1 knockdown did, but it had no effect on alternative splicing of INSR exon 11 or on either sarco/endoplasmic reticulum Ca2+-ATPase (ATP2A1) exon 22 or MBNL1 exon 7 (Figure 6B). Thus, these results suggest that p68 and/or p72 regulates a subset of alternative splicing events. It has been found that p68 and p72 could display distinct functions. Notably, it has been shown that p68 and p72 have different abilities to activate splicing of the CD44 alternative exons (51). To test whether the increase of TNNT2 exon 5 inclusion upon inactivation of p68 and p72 was specific to one of these two proteins, we performed individual p68 and p72 knockdown experiments. All of the siRNAs tested that targeted different regions of p68 or p72 were able to increase TNNT2 exon 5 inclusion (Supplementary Figure S6). This suggests that both p68 and p72 are regulators of splicing TNNT2 exon 5. Recently, it has been proposed that MBNL1 binding sites identified in TNNT2 are embedded in a stem–loop structure containing pyrimidine-pyrimidine mismatches that is similar to the CUG repeats, suggesting that MBNL1 recognizes these structures through a common mechanism (31). Thus, we hypothesized that RNA helicases p68 and p72 might facilitate MBNL1 binding in TNNT2 pre-mRNA as it does for the CUG repeats. To test this proposal, we performed UV-cross-linking experiments with an RNA encompassing the regulatory stem–loop element. Figure 7A showed that indeed p68 increased the binding of MBNL1 onto the regulatory element. Then, we asked whether mutations that reduce MBNL1 binding would impair regulation of TNNT2 exon 5 by p68. To answer this question, we used several TNNT2 minigenes (obtained from the groups of T. Cooper and A. Berglund), in which several mutations had been introduced to disrupt MBNL1 binding and/or the stem–loop RNA structure (Figure 7B) (30,31). Human myoblasts were cotransfected with wild-type and mutant TNNT2 minigenes that include exon 4–6 and with vectors overexpressing p68 or MBNL1. In agreement with siRNA experiments, overexpression of p68 repressed exon 5 inclusion as did MBNL1 (Figure 7B). As previously reported by Ho et al. and Warf et al., the 4G minigene in which mutations disrupted the RNA structure and MBNL1 binding did not respond to MBNL1 overexpression (Figure 7B). Interestingly, the 4G mutant was no longer able to respond to p68. In contrast, the CUGS mutant that still responded to MBNL1 also remained responsive to p68 regulation (Figure 7B). Finally, the GUF mutant that resulted in complete inclusion of exon 5 and became insensitive to MBNL1 regulation was also insensitive to p68 regulation (Figure 7B). These results suggested that the competence for regulating alternative splicing of TNNT2 by p68 depended on the presence of MBNL1 binding sites and the ability of MBNL1 to regulate this transcript. Therefore, we can anticipate that the inactivation of MBNL1 would result in the inability of p68 to regulate TNNT2 exon 5. To validate this hypothesis, we first cotransfected human myoblasts with the 3′DMPK UTR containing 5 or 960 CTG repeats and the TNNT2 minigene in the presence or absence of a vector coding for p68 (Figure 8A). We used an expression vector encoding PTB that is a repressor of exon 5 inclusion as a control (52). As previously observed in other studies and consistent with sequestration of MBNL1 by the repeats, expressing CUG repeats resulted in an increase of TNNT2 exon 5 inclusion (12) (Figure 8A). Interestingly, p68 was not able to regulate inclusion of TNNT2 exon 5 when 960 CUG repeats were expressed. In contrast, p68 was still able to regulate TNNT2 exon 5 in control experiments where only five CUG repeats were expressed. This finding was not due to a general effect of the repeats on splicing, because repression of exon 5 inclusions by PTB was the same regardless of the expressed CUG repeats length. We then tested the ability of p68 to regulate TNNT2 exon 5 inclusions after inactivation of MBNL1 by siRNA (Figure 8B). In agreement with previous studies, knockdown of MBNL1 increased splicing of TNNT2 exon 5 (Figure 8B). As already shown (Figure 8B), cotransfecting cells with a plasmid coding p68 resulted in a 2-fold decrease of TNNT2 exon 5 splicing. In contrast, splicing of TNNT2 exon 5 became less sensitive to p68 when cells were deprived of MBNL1. Consistent with these results, splicing of TNNT2 exon 5 became unresponsive to the knockdown of both p68 and p72 in DM1 myoblasts (Supplementary Figure S7). Altogether, these results demonstrate that p68 regulates the splicing of TNNT2 exon 5 alternative exon and suggest that p68-mediated regulation depends on MBNL1 binding.
Figure 6.
p68 regulates splicing of TNNT2 alternative exon 5. (A) Western blot analysis of human myoblasts treated with a control luciferase siRNA, (siLuc), sip68/p72 or siMBNL1. About 15 µg (lanes 1, 3 and 5) or 7.5 µg (lanes 2, 4 and 6) of proteins were loaded per lane. The blot was probed with antibodies against p68, MBNL1 and emerin as a control. (B) RT–PCR analysis of endogenous transcripts in human myoblast cells transfected with a control siRNA (siLuc), siRNA against p68/p72 or siRNA against MBNL1. The histogram shows the quantification of exon inclusion. Results are derived from six independent experiments for TNNT2 and four independent experiments for INSR, MBNL1 and ATP2A1 with error bars indicating standard deviation. *P < 0.05, **P < 0.01, ***P < 0.001.
Figure 7.
RNA stem–loop mutations that abolish MBNL1 binding impair the regulation by p68. (A) p68 stimulates binding of MBNL1 onto the stem–loop regulatory element of TNNT2. Labeled stem–loop regulatory RNA was incubated with a constant amount of recombinant MBNL1 protein (200 ng) and increasing amounts of recombinant p68ΔCt2. Proteins cross-linked to labeled RNA were separated on a 10% SDS–PAGE. The TNNT2 RNA used for cross-linking experiments is indicated. (B) RT–PCR analysis of splicing of TNNT2 alternative exon 5 in wild-type and mutant minigenes. Schematic representation of wild-type and mutant stem–loop structure within TNNT2 intron 4 [adapted from ref. (31)] is shown. Human myoblast cells were cotransfected with wild-type or mutant TNNT2 minigenes and expression vector coding for p68 (lanes 2, 5, 8 and 11) or MBNL1 (lanes 3, 6, 9 and 12). The histogram shows the quantification of exon 5 inclusion. Results are derived from at least three experiments, except for lanes 4, 6, 7, 9, 10 and 12, which are derived from two experiments. *P < 0.05, **P < 0.01, ***P < 0.001. NS: non-specific.
Figure 8.
Sequestration of MBNL1 by CUG repeats or MBNL1 knockdown by siRNA makes splicing of TNNT2 exon 5 non-responsive to p68. (A) RT–PCR analysis of TNNT2 minigene in human myoblast cells cotransfected with the 3′ UTR DMPK minigene containing 5 (lanes 1–3) or 960 (lanes 4 to 6) CTG repeats and a p68 expression vector (lanes 2 and 5) or as a control a PTB expression vector (lanes 3 and 6). The histogram shows the quantification of exon 5 inclusion. Results are from at least two experiments except lanes 3 and 6, for which a single experiment was performed. (B) RT–PCR analysis of TNNT2 minigene in human myoblast cells transfected with a control siRNA (lanes 1 and 2) or siRNA against MBNL1 (lanes 3 and 4) and a p68 expression vector (lanes 2 and 4). The histogram shows the quantification of exon 5 inclusion. Results are from four experiments with error bars indicating standard deviation. *P < 0.05, **P < 0.01, ***P < 0.001.
DISCUSSION
DEAD-box proteins are a well-characterized class of RNA helicases that play essential roles in cellular processes involving RNA (25,26). Their basic functions are to remodel RNA and RNA–protein complexes. In this study, we identified DEAD-box RNA helicases p68 and p72 in complexes that assembled onto pure CUG repeats. We showed that these proteins colocalized with RNA foci formed by the 3′UTR of DMPK mRNA-containing 200 or 960 CUG repeats. In particular, we found that p68 increases the binding of MBNL1 onto in vitro transcribed CUG repeats. Moreover, we showed that TNNT2-mediated splicing regulation by p68 is dependent on MBNL1 binding sites. On the basis of our results we suggest that p68 acts as a modifier of MBNL1 activity by facilitating the binding of MBNL1 onto the repeats and splicing target.
Affinity chromatography is a powerful method that has been used to decode the composition of protein complexes that assemble onto RNA. Using pure CUG repeats as bait, about 100 proteins were identified by mass-spectrometry. Many of them are hnRNP and are involved in several aspect of RNA metabolism (Figure 1 and Supplementary Table 1). Surprisingly, MBNL1 was not found in MS spectra. A similar observation was made by Kim et al. (53), who also did not detect MBNL1 in complexes purified using 46CUG repeats. However, in accordance with Miller et al. (9), we showed that MBNL1 is one of the major proteins from nuclear extracts that becomes cross-linked to the 95 CUG repeats (Figure 1A). In addition, MBNL1 was detected in the elution fraction from the affinity chromatography by western blot analysis (data not shown). Thus, the reason for the absence of MBNL1 from MS-spectra is not completely understood. One possible explanation is that, unfortunately, the band containing MBNL1 was not excised from the gel for identification by mass-spectrometry. Another possibility is that in our experimental conditions other proteins compete with MBNL1 for binding to the repeats. Among the proteins that we identified, only the DEAD-box proteins p68 and p72 form aggregates that colocalize with expanded CUG repeats. Interestingly, a study utilizing coexpression frequency shows that MBNL1 is coexpressed with p68 and p72, suggesting that these proteins could belong to the same signaling pathway (54). Additionally, another report shows that p68 is found in complexes associated with MBNL1 in myoblasts (55). The colocalization of p68 and p72 with RNA foci was observed with overexpressed RNA containing interrupted 960 CUG repeats or pure 200 CUG repeats and was independent of the cell lines. We also found that endogenous p68 and p72 colocalized with the repeats, as did exogenous proteins, suggesting that p68 and p72 are indeed recruited to RNA foci. However, in contrast to MBNL1 our results also show that only a fraction of p68 and p72 aggregates with RNA foci. p68 and p72 proteins are involved in multiple biological pathways and play a crucial role during development (25,26). It has been shown that the disruption of either p68 or p72 genes in mice results in a very severe phenotype that leads to early lethality (56). In addition, the same study found that the processing of several microRNAs involved in cell survival is affected in p68 and p72 homozygous mice (56). Thus, according to these findings, a sequestration mechanism of p68 and p72 that leads to a deficiency of protein function similar to that of MBNL1 seems unlikely. In fact, no colocalization of p68 with RNA foci could be observed in muscle tissues from DM1 patients or mice expressing large CTG expansions (data not shown). As recently reported, we never observed recruitment of p68 in RNA foci of DM1 myoblasts (55). However, taking advantage of an inducible system in which the expression of expanded CUG repeats is under the control of an inducible promoter, we were able to show that the knockdown of both p68 and p72 has an effect on RNA foci formation (Figure 5). The effect on the number of newly formed foci was transient following doxycycline addition suggesting that p68 and p72 act at an early stage of RNA aggregation and then are released from the RNA foci. Interestingly a recent model has been proposed by Junghans to conciliate some conflicting results of DM1 pathobiology. This model poises the existence of insoluble and soluble RNA foci with distinct binding properties (57). Thus, one possibility would be that p68 and p72 intervene in the transition between these two forms of RNA foci.
One general task of DEAD-box RNA helicase is to modify RNA structure and/or RNA protein complexes (25,26). A recent crystal analysis of two zinc-finger domains of MBNL1 with a short CGCUGU RNA shows that MBNL1 targets GC steps, indicating that G–C base pairing within the helix has to be distorted (43). On the basis of our results, we propose that the recruitment of p68 to the CUG repeats would promote structural changes of the RNA that would facilitate and/or stabilize the binding of the splicing factor MBNL1 to the repeats. Consistent with that proposal, mutations in the helicase core domain of p68 strongly decreased both the stimulation of p68 on MBNL1 binding and the colocalization of p68 with CUG repeats (Figure 4). Although we focused on the role of p68, data in other publications suggest that p68 and p72 can exist as heterodimer in the cell (58). Indeed, our results show that p68 and p72 have the same function, suggesting that at least a fraction of p68 and p72 acts functionally as heterodimers (Figure 2 and Supplementary Figure S6). We also observed that the stimulatory effect operates on CAG repeats and on the regulatory intronic sequence of TNNT2 pre-mRNA (Figures 3C and 7A). Interestingly, several biochemical and structural studies predicted that pathogenic CUG repeats and TNNT2 RNA adopt similar RNA secondary structure (31,59). For CAG repeats, recent structural analysis also suggests that CAG and CUG duplexes share structural similarities (60). Thus, we can hypothesize that these common structural elements compose the basis for p68 recognition and subsequently for the increase of MBNL1 on the RNA. In contrast, p68 only slightly increases the binding of MBNL1 onto CCUG repeats. Interestingly, MBNL1 binds to CCUG RNA with an affinity 2-fold stronger than CUG RNA (31). However, it has been shown that CCUG has a decreased stability compared to CUG (31). Thus, it is tempting to speculate that the requirement for p68 might depend on the stability of the RNA structure. We can suggest that the less an RNA is structured, the less p68 will be needed to distort the structure. Our results are consistent with a recent study that shows that MBNL1 is able to bind to both single-stranded and double-stranded RNAs (10). The mechanism by which p68 favors the binding of MBNL1 to the CUG repeats remains unknown. Time course experiments show that there is a huge increase of MBNL1 binding in the first few minutes following incubation with p68 (Supplementary Figure S4). It is known that the DEAD-box RNA helicases as RNA chaperones, modify the folding of RNA molecules. Thus, we can speculate that p68 accelerates the transition from misfolded RNA conformation to correct folding RNA structure, suitable for MBNL1 binding. Several studies point to the importance of U–U mismatches for MBNL1 binding (59,61). Thus, we propose that the mismatches could provide an anchoring site for p68 from which local strand separation could be initiated which would allow the interaction of MBNL1 to the repeats. It also has been shown that MBNL1 forms a ring-like structure with CUG repeats. From this finding we can suggest that in vivo p68 accelerates formation of this structure that is proposed to be the basis for MBNL1 sequestration (59).
In this study, we provide evidence that the alternative splicing of TNNT2, which is misregulated in DM1 is regulated by p68. We also found that alternative splicing of MBNL1 exon 7, ATP2A1 exon 22 or INSR exon 11, which are regulated by MBNL1 and misregulated in DM1, are not sensitive to p68 and/or p72, suggesting that these proteins regulate specific classes of transcripts. This finding raises an intriguing question. Why does alternative splicing of some transcripts regulated by MBNL1 require p68? The answer is not yet understood, but could be related to the architecture of the MBNL1 binding sites. Indeed, a recent study of Cass and coworkers suggests that different modes of MBNL1 recognition must operate for single-stranded and structured RNA targets (10). In the case of TNNT2, MBNL1 binds to a short stem–loop RNA structure, and we show that p68 increases the binding of MBNL1 onto this RNA structure. In contrast, at least for the INSR and MBNL1 transcripts no potential RNA secondary structure has been identified within the MBNL1 response element (62,63). Thus, we can propose that p68 would be required only for splicing RNA targets in which MBNL1 binding sites are embedded into a stem–loop structure. The modification of RNA stem–loop structure is a common theme for regulating alternative splicing by p68. However, the mechanism by which p68 mediates its effect is different according to the different targets. In the case of alternative splicing of Tau exon 10 and IDX alternative exon of c-H-ras pre-mRNA, it has been shown that p68 promotes the access of U1snRNP to the 5′ splice site (64) or block the binding of hnRNP H (65), respectively. Our findings provide a new mode for how p68 can regulate alternative splicing by increasing the activity of a splicing factor. We also found that preventing the binding of MBNL1 onto the RNA, either by sequestration of the protein by expanded CUG repeats or by mutations in the MBNL1 binding sites, made TNNT2 exon 5 insensitive to p68 regulation. This suggests that the repression of TNNT2 exon 5 by p68 is dependent on MBNL1 binding to the regulatory element, and it supports the idea that p68 would not act as a splicing factor per se, but rather, would be a coregulator or a coadaptator of MBNL1 binding. Thus, we favor a model according to which p68 modifies the stem–loop RNA structure of TNNT2 as it does on the toxic CUG repeats, which allows the stabilization of MBNL1.
In conclusion, our results have identified a modifier of MBNL1 activity. We suggest that through a common mechanism, the DEAD-box RNA helicase p68 modifies the RNA secondary structures of splicing targets and pathological RNAs to facilitate or stabilize MBNL1 binding that may contribute to the pathogenesis of DM1.
SUPPLEMENTARY DATA
Supplementary data is available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–7, Supplementary Reference [66].
FUNDING
Centre National de la Recherche Scientifique (CNRS); Association Française contre les Myopathies (AFM); Paris-Sud University, the AFM and Fondation des Treilles fellowships (to F.X.L.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to T. Cooper (Baylor University) for DMPK CTG 960 plasmid, TNNT2 and mutant TNNT2 4G minigenes; M. Mahadevan (University of Virginia) for DMPK CTG 5 and CTG 200 plasmids; A. Berglund (University of Oregon) for mutant TNNT2 CUGS and GUF plasmids; S. Kato (University of Tokyo), F. Fuller-Pace (University of Dundee) and D. Auboeuf (Centre Léon Bérard, Lyon) for p68 and p72 plasmids; G. Morris (Centre for Inherited Neuromuscular Disease; Robert Jones and Agnes Hunt Orthopaedic Hospital) for the MBNL1 (MB1a) monoclonal antibodies; H. Le Hir (Ecole Normale Supérieure, Paris) for recombinant eIF4A3 protein; O. Cordin and J. Banroques (Institut de Biologie Physico-chimique, Paris) for recombinant UAP56 protein. N. Gourrier and C. Branlant (Laboratoire AREMS, Nancy) for GST-MBNL1 plasmid; N. Sergeant for N1E115 cells. They acknowledge Kyle Tanner and Katriona Laurent-Price for their constant interest and for carefully reading the manuscript. They also thank all the members of the French DM1 network for helpful discussions.
REFERENCES
- 1.Harper PS. Myotonic Dystrophy. London: W. B. Saunders; 2001. [Google Scholar]
- 2.Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 3.Mahadevan M, Tsilfidis C, Sabourin L, Shutler G, Amemiya C, Jansen G, Neville C, Narang M, Barcelo J, O'Hoy K, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3' untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 4.Fu YH, Pizzuti A, Fenwick RG, Jr, King J, Rajnarayan S, Dunne PW, Dubel J, Nasser GA, Ashizawa T, de Jong P, et al. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992;255:1256–1258. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- 5.Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell. Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis BM, McCurrach ME, Taneja KL, Singer RH, Housman DE. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl Acad. Sci. USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holt I, Mittal S, Furling D, Butler-Browne GS, Brook JD, Morris GE. Defective mRNA in myotonic dystrophy accumulates at the periphery of nuclear splicing speckles. Genes Cells. 2007;12:1035–1048. doi: 10.1111/j.1365-2443.2007.01112.x. [DOI] [PubMed] [Google Scholar]
- 8.Schoser B, Timchenko L. Myotonic dystrophies 1 and 2: complex diseases with complex mechanisms. Curr. Genomics. 2010;11:77–90. doi: 10.2174/138920210790886844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cass DM, Hotchko R, Barber P, Jones K, Gates DP, Berglund JA. The four Zn fingers of MBNL1 provide a flexible platform for recognition of its RNA binding elements. BMC Mol. Biol. 2011;12:20. doi: 10.1186/1471-2199-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Rourke JR, Swanson MS. Mechanisms of RNA-mediated disease. J. Biol. Chem. 2009;284:7419–7423. doi: 10.1074/jbc.R800025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu. Rev. Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 13.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 15.Charlet BN, Savkur RS, Singh G, Philips AV, Grice EA, Cooper TA. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 16.Mankodi A, Takahashi MP, Jiang H, Beck CL, Bowers WJ, Moxley RT, Cannon SC, Thornton CA. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 17.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 18.Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 19.Ebralidze A, Wang Y, Petkova V, Ebralidse K, Junghans RP. RNA leaching of transcription factors disrupts transcription in myotonic dystrophy. Science. 2004;303:383–387. doi: 10.1126/science.1088679. [DOI] [PubMed] [Google Scholar]
- 20.Paul S, Dansithong W, Kim D, Rossi J, Webster NJ, Comai L, Reddy S. Interaction of muscleblind, CUG-BP1 and hnRNP H proteins in DM1-associated aberrant IR splicing. EMBO J. 2006;25:4271–4283. doi: 10.1038/sj.emboj.7601296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Lopez A, Monferrer L, Garcia-Alcover I, Vicente-Crespo M, Alvarez-Abril MC, Artero RD. Genetic and chemical modifiers of a CUG toxicity model in Drosophila. PLoS One. 2008;3:e1595. doi: 10.1371/journal.pone.0001595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dansithong W, Jog SP, Paul S, Mohammadzadeh R, Tring S, Kwok Y, Fry RC, Marjoram P, Comai L, Reddy S. RNA steady-state defects in myotonic dystrophy are linked to nuclear exclusion of SHARP. EMBO Rep. 2011;12:735–742. doi: 10.1038/embor.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rau F, Freyermuth F, Fugier C, Villemin JP, Fischer MC, Jost B, Dembele D, Gourdon G, Nicole A, Duboc D, et al. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat. Struct. Mol. Biol. 2011;18:840–850. doi: 10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
- 24.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 25.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janknecht R. Multi-talented DEAD-box proteins and potential tumor promoters: p68 RNA helicase (DDX5) and its paralog, p72 RNA helicase (DDX17) Am. J. Transl. Res. 2010;2:223–234. [PMC free article] [PubMed] [Google Scholar]
- 27.Amack JD, Paguio AP, Mahadevan MS. Cis and trans effects of the myotonic dystrophy (DM) mutation in a cell culture model. Hum. Mol. Genet. 1999;8:1975–1984. doi: 10.1093/hmg/8.11.1975. [DOI] [PubMed] [Google Scholar]
- 28.Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA. Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J. Cell Sci. 2005;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- 29.Chaouch S, Mouly V, Goyenvalle A, Vulin A, Mamchaoui K, Negroni E, Di Santo J, Butler-Browne G, Torrente Y, Garcia L, et al. Immortalized skin fibroblasts expressing conditional MyoD as a renewable and reliable source of converted human muscle cells to assess therapeutic strategies for muscular dystrophies: validation of an exon-skipping approach to restore dystrophin in Duchenne muscular dystrophy cells. Hum. Gene Ther. 2009;20:784–790. doi: 10.1089/hum.2008.163. [DOI] [PubMed] [Google Scholar]
- 30.Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 33.Tran H, Gourrier N, Lemercier C, Dhaenens CM, Vautrin A, Fernandez-Gomez FJ, Arandel L, Carpentier C, Obriot H, Eddarkaoui S, et al. Analysis of exonic-regions involved in nuclear localization, splicing activity and dimerization of muscleblind-like-1 isoforms. J. Biol. Chem. 2011;286:16435–16446. doi: 10.1074/jbc.M110.194928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauliere J, Sureau A, Expert-Bezancon A, Marie J. The polypyrimidine tract binding protein (PTB) represses splicing of exon 6B from the beta-tropomyosin pre-mRNA by directly interfering with the binding of the U2AF65 subunit. Mol. Cell Biol. 2006;26:8755–8769. doi: 10.1128/MCB.00893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Expert-Bezancon A, Le Caer JP, Marie J. Heterogeneous nuclear ribonucleoprotein (hnRNP) K is a component of an intronic splicing enhancer complex that activates the splicing of the alternative exon 6A from chicken beta-tropomyosin pre-mRNA. J. Biol. Chem. 2002;277:16614–16623. doi: 10.1074/jbc.M201083200. [DOI] [PubMed] [Google Scholar]
- 36.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sureau A, Sauliere J, Expert-Bezancon A, Marie J. CELF and PTB proteins modulate the inclusion of the beta-tropomyosin exon 6B during myogenic differentiation. Exp. Cell Res. 2011;317:94–106. doi: 10.1016/j.yexcr.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Zhu CH, Mouly V, Cooper RN, Mamchaoui K, Bigot A, Shay JW, Di Santo JP, Butler-Browne GS, Wright WE. Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: consequences in aging muscle and therapeutic strategies for muscular dystrophies. Aging Cell. 2007;6:515–523. doi: 10.1111/j.1474-9726.2007.00306.x. [DOI] [PubMed] [Google Scholar]
- 39.Francois V, Klein AF, Beley C, Jollet A, Lemercier C, Garcia L, Furling D. Selective silencing of mutated mRNAs in DM1 by using modified hU7-snRNAs. Nat. Struct. Mol. Biol. 2011;18:85–87. doi: 10.1038/nsmb.1958. [DOI] [PubMed] [Google Scholar]
- 40.Taneja KL. Localization of trinucleotide repeat sequences in myotonic dystrophy cells using a single fluorochrome-labeled PNA probe. Biotechniques. 1998;24:472–476. doi: 10.2144/98243rr02. [DOI] [PubMed] [Google Scholar]
- 41.Amack JD, Mahadevan MS. The myotonic dystrophy expanded CUG repeat tract is necessary but not sufficient to disrupt C2C12 myoblast differentiation. Hum. Mol. Genet. 2001;10:1879–1887. doi: 10.1093/hmg/10.18.1879. [DOI] [PubMed] [Google Scholar]
- 42.Lamm GM, Nicol SM, Fuller-Pace FV, Lamond AI. p72: a human nuclear DEAD box protein highly related to p68. Nucleic Acids Res. 1996;24:3739–3747. doi: 10.1093/nar/24.19.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teplova M, Patel DJ. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat. Struct. Mol. Biol. 2008;15:1343–1351. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woodson SA. Taming free energy landscapes with RNA chaperones. RNA Biol. 2010;7:677–686. doi: 10.4161/rna.7.6.13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H, Gao X, Huang Y, Yang J, Liu ZR. P68 RNA helicase is a nucleocytoplasmic shuttling protein. Cell Res. 2009;19:1388–1400. doi: 10.1038/cr.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu ZR, Sargueil B, Smith CW. Detection of a novel ATP-dependent cross-linked protein at the 5' splice site-U1 small nuclear RNA duplex by methylene blue-mediated photo-cross-linking. Mol. Cell Biol. 1998;18:6910–6920. doi: 10.1128/mcb.18.12.6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 48.Day JW, Ranum LP. RNA pathogenesis of the myotonic dystrophies. Neuromuscul Disord. 2005;15:5–16. doi: 10.1016/j.nmd.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 49.Banroques J, Cordin O, Doere M, Linder P, Tanner NK. A conserved phenylalanine of motif IV in superfamily 2 helicases is required for cooperative, ATP-dependent binding of RNA substrates in DEAD-box proteins. Mol. Cell Biol. 2008;28:3359–3371. doi: 10.1128/MCB.01555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dansithong W, Paul S, Comai L, Reddy S. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J. Biol. Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- 51.Honig A, Auboeuf D, Parker MM, O'Malley BW, Berget SM. Regulation of alternative splicing by the ATP-dependent DEAD-box RNA helicase p72. Mol. Cell Biol. 2002;22:5698–5707. doi: 10.1128/MCB.22.16.5698-5707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Charlet BN, Logan P, Singh G, Cooper TA. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 53.Kim DH, Langlois MA, Lee KB, Riggs AD, Puymirat J, Rossi JJ. HnRNP H inhibits nuclear export of mRNA containing expanded CUG repeats and a distal branch point sequence. Nucleic Acids Res. 2005;33:3866–3874. doi: 10.1093/nar/gki698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson BJ, Giguere V. Identification of novel pathway partners of p68 and p72 RNA helicases through Oncomine meta-analysis. BMC Genomics. 2007;8:419. doi: 10.1186/1471-2164-8-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paul S, Dansithong W, Jog SP, Holt I, Mittal S, Brook D, Morris GE, Comai L, Reddy S. Expanded CUG repeats dysregulate RNA splicing by altering the stoichiometry of the muscleblind 1 complex. J. Biol. Chem. 2011;286:38427–38438. doi: 10.1074/jbc.M111.255224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat. Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 57.Junghans RP. Dystrophia myotonia: why focus on foci? Eur. J. Hum. Genet. 2009;17:543–553. doi: 10.1038/ejhg.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogilvie VC, Wilson BJ, Nicol SM, Morrice NA, Saunders LR, Barber GN, Fuller-Pace FV. The highly related DEAD box RNA helicases p68 and p72 exist as heterodimers in cells. Nucleic Acids Res. 2003;31:1470–1480. doi: 10.1093/nar/gkg236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan Y, Compton SA, Sobczak K, Stenberg MG, Thornton CA, Griffith JD, Swanson MS. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Atomic resolution structure of CAG RNA repeats: structural insights and implications for the trinucleotide repeat expansion diseases. Nucleic Acids Res. 2010;38:8370–8376. doi: 10.1093/nar/gkq700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warf MB, Diegel JV, von Hippel PH, Berglund JA. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc. Natl Acad. Sci. USA. 2009;106:9203–9208. doi: 10.1073/pnas.0900342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grammatikakis I, Goo YH, Echeverria GV, Cooper TA. Identification of MBNL1 and MBNL3 domains required for splicing activation and repression. Nucleic Acids Res. 2011;39:2769–2780. doi: 10.1093/nar/gkq1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gates DP, Coonrod LA, Berglund JA. Auto-regulated splicing of the muscleblind-like 1 (MBNL1) pre-mRNA. J. Biol. Chem. 2011;286:34224–34233. doi: 10.1074/jbc.M111.236547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kar A, Fushimi K, Zhou X, Ray P, Shi C, Chen X, Liu Z, Chen S, Wu JY. RNA helicase p68 (DDX5) regulates tau exon 10 splicing by modulating a stem-loop structure at the 5′ splice site. Mol. Cell Biol. 2011;31:1812–1821. doi: 10.1128/MCB.01149-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Camats M, Guil S, Kokolo M, Bach-Elias M. P68 RNA helicase (DDX5) alters activity of cis- and trans-acting factors of the alternative splicing of H-Ras. PLoS One. 2008;3:e2926. doi: 10.1371/journal.pone.0002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uhlmann-Schiffler H, Rossler OG, Stahl H. The mRNA of DEAD box protein p72 is alternatively translated into an 82-kDa RNA helicase. J. Biol. Chem. 2002;277:1066–1075. doi: 10.1074/jbc.M107535200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.