Abstract
Rabies is a progressively fatal and incurable viral encephalitis caused by a lyssavirus infection. Almost all of the 55 000 annual rabies deaths in humans result from infection with dog rabies viruses (RABV). Despite the importance of rabies for human health, little is known about the spread of RABV in dog populations, and patterns of biodiversity have only been studied in limited geographical space. To address these questions on a global scale, we sequenced 62 new isolates and performed an extensive comparative analysis of RABV gene sequence data, representing 192 isolates sampled from 55 countries. From this, we identified six clades of RABV in non-flying mammals, each of which has a distinct geographical distribution, most likely reflecting major physical barriers to gene flow. Indeed, a detailed analysis of phylogeographic structure revealed only limited viral movement among geographical localities. Using Bayesian coalescent methods we also reveal that the sampled lineages of canid RABV derive from a common ancestor that originated within the past 1500 years. Additionally, we found no evidence for either positive selection or widespread population bottlenecks during the global expansion of canid RABV. Overall, our study reveals that the stochastic processes of genetic drift and population subdivision are the most important factors shaping the global phylogeography of canid RABV.
INTRODUCTION
Rabies is one of the most virulent diseases of humans and animals and may have been reported in the Old World before 2300 BC (Steele & Fernandez, 1991; Theodorides, 1986). The development of a vaccine for rabies virus (RABV) by Louis Pasteur in 1886 led to the development of prevention strategies against the disease in the developed world, and this fatal encephalitis is now preventable through the timely administration of post-exposure vaccination and serotherapy (Warrell & Warrell, 2004). Yet, despite these medical advances, more than 50 000 people die of rabies on an annual basis, with 60 % of fatalities occurring in Asia (Knobel et al., 2005).
RABV is a single-stranded, negative-sense lyssavirus (genotype 1; family Rhabdoviridae) with a genome size of approximately 12 kb. The genus Lyssavirus includes a number of important zoonotic bat viruses; phylogenetic analyses have determined both the evolutionary relationships among these lyssaviruses, as well as the existence of seven distinct genotypes, although this number is likely to increase with more intensive sampling (Bourhy et al., 1993; Gould et al., 1998; Kuzmin et al., 2005). As a group, the lyssaviruses are characterized by their ecological association with specific mammalian species, which act as vectors for their transmission, such that a number of phylogenetic lineages co-circulate among a range of mammalian hosts (Davis et al., 2005; Holmes et al., 2002; Kissi et al., 1995).
Of the mammalian RABV, those that circulate in dogs (Canis lupus familiaris) are responsible for more than 99 % of the human cases worldwide (Knobel et al., 2005). However, despite its role as a vector for human disease, the extent and structure of viral biodiversity in this key vector species, as well as the mode and timescale of its evolution, have only been studied on a limited geographical scale. The development of trans-oceanic travel during the 15th century (Leonard et al., 2002; Verginelli et al., 2005) is thought to be responsible for the transmission of rabies to all continents, resulting in the global dissemination of the so-called ‘cosmopolitan’ dog RABV lineage (Badrane & Tordo, 2001; Kissi et al., 1995). Although the hypothesis of RABV dispersal via trans-oceanic travel is often repeated, it has not been subjected to rigorous examination using modern molecular phylogenetics. Furthermore, the evolutionary links between dog viruses and those RABV that circulate in other members of the family Canidae (foxes and raccoon dogs) and in other families such as the Mephitidae (skunk), Procyonidae (raccoon) and Herpestidae (mongoose) were never globally analysed.
To determine the biodiversity of dog RABV, as well as its spatial and temporal dynamics, we sequenced 62 new isolates and analysed a large dataset of RABV including 192 isolates sampled from 55 countries on five continents over a time period of 37 years. To enhance the power of our phylogenetic analysis we investigated evolutionary patterns using sequences of both the complete nucleoprotein (N) (1335 nt) and glycoprotein (G) (1572 nt) genes. Phylogenetic and Bayesian coalescent approaches were applied to reveal both the timescale of RABV evolution in dogs and, for the first time, to explore the global phylogeography of this important human and wildlife pathogen.
METHODS
Phylogenetic analysis
To investigate the global genetic diversity of RABV we analysed a total of 151 sequences (1335 nt) of the N (nucleoprotein) gene for which the time (year) of sampling was available. We also compiled a larger dataset of 190 sequences of the N-terminal region of the N protein (400 nt) for which the sampling date was often unavailable (excluding vaccine strains). In addition, 74 complete G gene sequences (1572 nt) were analysed. In total, 91 N and G gene sequences isolated from non-flying mammals and sampled from 14 countries were newly determined in this study by using methods described previously (Bourhy et al., 1999; Holmes et al., 2002; Kissi et al., 1995) and the primers described in Supplementary Table S1 (available in JGV Online). GenBank accession numbers for the newly acquired sequences are designated EU086128–EU086218 (Supplementary Table S2). Relevant epidemiological information for all those RABV isolates of canid origin analysed in this study is presented in Supplementary Table S2.
For each dataset we inferred maximum-likelihood (ML) phylogenetic trees using the PAUP★ package (Swofford, 2003). In all cases, the best-fit model of nucleotide substitution, determined using MODELTEST (Posada & Crandall, 1998), was the most general GTR+I+Γ4 model. Successive rounds of tree-bisection reconnection branch-swapping were then used to determine the globally optimal phylogeny. To assess the reliability of key nodes on each tree, we used a bootstrap resampling analysis, employing 1000 replicate neighbour-joining trees estimated under the ML substitution model.
Evolutionary and population dynamics
Rates of nucleotide substitution (per site, per year) and the time to most recent common ancestor (TMRCA) for both the complete N and G gene datasets (for which year of sampling was available) were estimated using the Bayesian Markov chain Monte Carlo (MCMC) method available in the BEAST package (Drummond & Rambaut, 2007), again utilizing the GTR+I+Γ4 model of nucleotide substitution. Because preliminary analyses revealed that the population dynamics of RABV supported a model of constant population size through time, we restricted our analysis to this demographic model [although similar evolutionary dynamics, with overlapping 95 % highest posterior density (HPD) values, were estimated under both exponential population growth and Bayesian skyline models; full results available from the authors on request]. We also considered both strict and relaxed (uncorrelated lognormal) molecular clocks; the latter was the best supported under Bayes Factors, as calculated through the TRACER program (http://tree.bio.ed.ac.uk/software/tracer/), although similar results were observed under both clock models. Finally, to remove any estimation error induced by species subdivision, we performed separate analyses on those viruses sampled from terrestrial mammals and those from bats. An equivalent analysis was performed on G sequences for (i) the entire dataset and (ii) terrestrial mammals only. The statistical uncertainty in each parameter estimate was provided by values of the 95 % HPD and all analyses were run for sufficient time to ensure convergence (assessed using the TRACER program).
We used the CODEML program within the PAML package (Yang, 2007) to estimate the mean ratio of non-synonymous (dN) to synonymous (dS) substitutions per site (dN/dS) for RABV from non-flying mammals. This overall dN/dS ratio was calculated using all branches in the N gene ML tree (the one-ratio model). To examine selection pressures in more detail, a separate dN/dS value was estimated for both the external and internal branches of the same phylogeny (the two-ratio model).
Analysis of spatial dynamics
We utilized a parsimony-based approach to determine the geographical structure of dog RABV, based on an ML tree of 130 complete N gene sequences from non-flying mammals (comprising the 126 dated sequences that were used previously and four isolates with no sampling date) rooted by those viruses sampled from bats. This analysis considered global ecoregions, defined as the largest biogeographic divisions on earth, which are likely to influence patterns of viral migration. Hence, ecoregions correspond to homogeneous geographical units of representative habitats and species assemblages (Olson et al., 2001). This approach is justified since the global spread of dogs clearly occurred after their domestication approximately 15 000–17 000 years ago (Leonard et al., 2002; Savolainen et al., 2002; Verginelli et al., 2005) and hence prior to spread of RABV (see Results). Five major ecoregions were distinguished here: Afrotropic, Indomalaya, Neartic, Neotropic and Palearctic. These were subdivided further into smaller subregions representing the geographical proximity of the sampling countries (Olson et al., 2001) (see also http://www.worldwildlife.org/science/ecoregions/biomes.cfm, http://www.nationalgeographic.com/wildworld/terrestrial.html). This classification was chosen as the best approximation to the geographical distribution of the mammalian hosts of RABV. Overall, we collected sequences from 55 different countries, encompassing all 5 major ecoregions.
Each RABV sequence was assigned a character state reflecting its country of origin within a specific ecoregion (Table 1). The number of unambiguous character state changes observed among each region on the ML tree was then recorded and compared with the number expected under the null hypothesis of panmixis, generated by creating 1000 randomized trees. A matrix of differences between the number of character state changes in the observed and expected phylogenies was created to determine the direction of any migration events; the extent of migration (or population subdivision) is defined by the differences between each corresponding pair of expected and observed character states. A negative value indicates the degree of genetic isolation, whereas a positive value signifies migration. All analyses were conducted using PAUP★ (Swofford, 2003).
Table 1. Parsimony analysis of migration frequency and direction among isolates of RABV from terrestrial mammals inferred using the complete N gene.
Positive values (bold type) indicate viral migration between the specified regions (the larger the number, the greater the extent of migration), while negative values are indicative of population subdivision. The table is read across from the ‘origin’ row to the appropriate column (i.e. migration from A to B is 0.64). A–E, Palearctic: A, Europe; B, Afghanistan, Iran, Pakistan; C, Algeria, Egypt, Mauritania, Morocco; D, Kazakhstan, Russia; E, China, Korea. F–H, Indomalaya: F, Cambodia, Laos, Myanmar, Thailand, Vietnam; G, Indian Ocean; H, Indonesia, Philippines. I–N, Afrotropic: I, Mozambique, Namibia, South Africa; J, Benin, Burkina Faso, Democratic Republic of Congo, Guinea, Ivory Coast, Niger; K, Tanzania; L, Ethiopia; M, Oman Sultanate, Saudi Arabia; N, Cameroon, Central African Republic, Chad, Gabon, Nigeria. O, Nearctic: Canada, Greenland. P, Neotropic: Brazil, Colombia, Mexico. Q, Outgroup: India, Sri Lanka.
| Origin | A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 0.64 | −0.41 | 0.28 | −0.87 | −0.79 | −0.29 | −0.44 | −0.34 | −0.50 | −0.17 | −0.07 | −0.15 | 0.40 | −0.15 | −0.42 | −0.39 | |
| B | −0.03 | −0.02 | −0.03 | −0.03 | −0.03 | −0.01 | −0.02 | −0.02 | −0.02 | −0.01 | −0.01 | 1.00 | −0.03 | −0.01 | −0.02 | −0.01 | |
| C | −0.07 | −0.03 | −0.05 | −0.06 | −0.06 | −0.04 | −0.04 | −0.03 | −0.05 | −0.01 | 0 | −0.01 | −0.06 | −0.02 | −0.04 | −0.03 | |
| D | −0.41 | −0.19 | −0.22 | 0.59 | −0.40 | −0.17 | −0.23 | −0.19 | −0.29 | −0.08 | −0.04 | −0.09 | −0.30 | 1.92 | −0.20 | −0.18 | |
| E | −1.35 | −0.56 | −0.63 | −1.17 | −1.27 | −0.43 | 1.28 | −0.53 | −0.81 | −0.21 | −0.10 | −0.23 | −0.94 | −0.24 | −0.66 | −0.53 | |
| F | −0.85 | −0.34 | −0.40 | −0.73 | −0.85 | −0.28 | −0.45 | −0.39 | −0.50 | −0.17 | −0.07 | −0.15 | −0.57 | −0.14 | −0.39 | −0.34 | |
| G | −0.01 | −0.01 | −0.01 | −0.02 | −0.02 | −0.02 | −0.01 | −0.01 | −0.02 | −0.01 | 0 | 0 | −0.01 | −0.01 | −0.01 | −0.01 | |
| H | −0.08 | −0.03 | −0.06 | −0.06 | −0.06 | −0.07 | −0.03 | −0.05 | −0.05 | −0.01 | −0.01 | −0.02 | −0.06 | −0.02 | −0.06 | −0.03 | |
| I | −0.04 | −0.02 | −0.02 | −0.04 | −0.03 | −0.02 | −0.02 | −0.02 | −0.02 | −0.01 | 0 | −0.01 | −0.03 | −0.01 | −0.02 | −0.02 | |
| J | −0.10 | −0.06 | 0.95 | −0.11 | −0.10 | −0.10 | −0.04 | −0.06 | −0.05 | −0.03 | −0.02 | −0.02 | −0.92 | −0.02 | −0.07 | −0.6 | |
| K | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| L | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| M | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −0.01 | 0 | |
| N | −0.20 | −0.08 | −0.10 | −0.18 | −0.20 | −0.20 | −0.07 | −0.10 | 0.92 | 0.87 | 0.96 | −0.02 | −0.04 | −0.03 | −0.09 | −0.08 | |
| O | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| P | 0.95 | −0.03 | 0.96 | −0.05 | −0.05 | −0.06 | −0.03 | −0.04 | −0.02 | −0.03 | −0.01 | 0 | −0.01 | −0.04 | −0.01 | −0.03 | |
| Q | −0.03 | −0.02 | −0.02 | −0.03 | −0.03 | −0.03 | 0.99 | −0.02 | −0.01 | −0.02 | −0.01 | 0 | −0.01 | −0.03 | −0.01 | −0.02 |
RESULTS
Phylogeography of RABV
Consistent with previous analyses based on smaller datasets, the ML trees of complete N genes (25 sequences obtained from bats and 126 sequences from non-flying mammals; 151 sequences in total; Fig. 1), G genes (three sequences from bats, 71 sequences from non-flying mammals; Fig. 2) and partial N genes (27 sequences from bats, 163 sequences from non-flying mammals; Supplementary Fig. S1, available in JGV Online) indicated that viruses generally grouped according to their geographical origin, such that RABV has a clear spatial structure. We explored the causes of this spatial patterning in more detail.
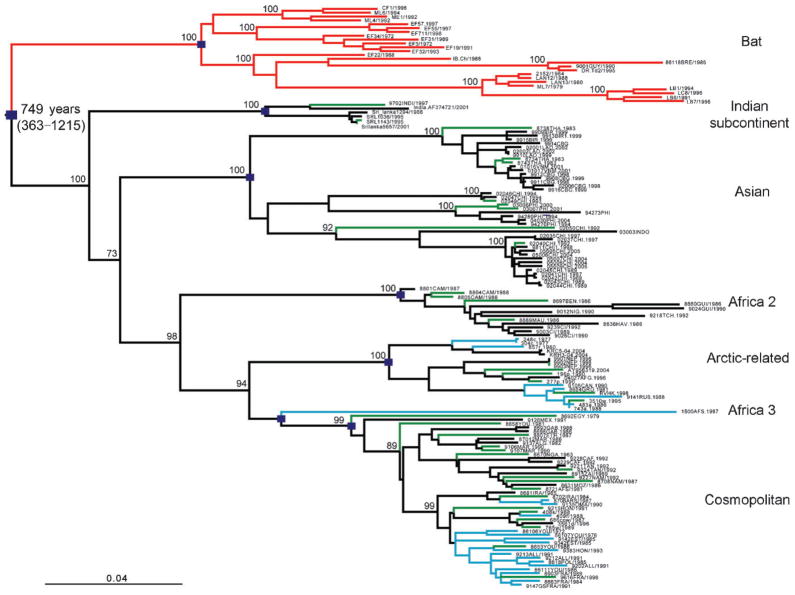
Fig. 1.
ML phylogeny of 151 sequences from the N-coding region of RABV. The estimated TMRCA for this sample of viral lineages, as well as its 95 % HPD values, are indicated. The major clades of RABV are also indicated, denoted by squares at the relevant nodes. Branches are colour-coded by species group: black, dogs; red, bats; blue, alternative reservoir hosts (such as the red fox); green, spill-over hosts (such as humans, bovines, wolf). Horizontal branches are drawn to a scale of nucleotide substitutions per site, with bootstrap support values (>90 % and 73 % for the Indian subcontinent clade) shown for key nodes.
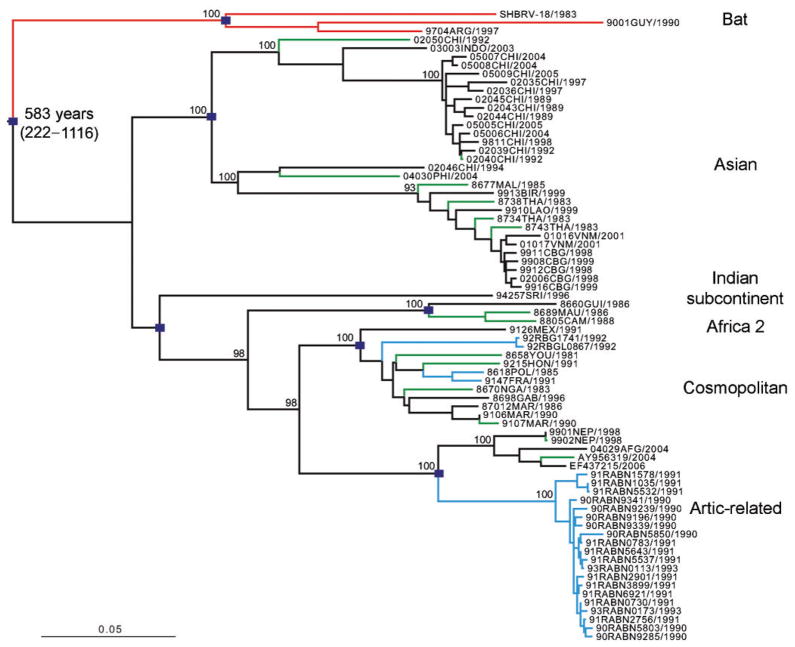
Fig. 2.
ML phylogeny of 74 sequences from the complete G-coding region of RABV. The estimated TMRCA for this sample of viral lineages, as well as its 95 % HPD values, are indicated. The major clades of RABV are also indicated, denoted by squares at the relevant nodes. Branches are coloured-coded in the same manner as Fig. 1. Horizontal branches are drawn to scale, with bootstrap support values (>90 %) shown for key nodes.
Notably, the phylogenetic trees of the N and G genes possessed similar topologies, indicating that equivalent clades, with matching geographical distributions, are defined by both trees (Figs 1 and 2). Because of its larger sample size, the N gene tree is particularly informative. Clearly, bat- and dog-associated RABV form distinct phylogenetic groups, with the latter comprising six major clusters (each with strong bootstrap support), identified as the Africa 2, Africa 3, Arctic-related, Asian, Cosmopolitan and Indian subcontinent clades (Fig. 1; Supplementary Fig. S1). In brief, the Cosmopolitan clade included dog, wolf and fox isolates from Europe, the Middle-East, Iran, Kazakhstan and further east to the Republic of Tuva in Russia (Bourhy et al., 1999; David et al., 2007; Kissi et al., 1995; Kuzmin et al., 2004). It also included a number of dog RABV isolates from the Americas (Mexico, Colombia and Brazil) and those previously denoted the Africa 1 lineage, which is distributed in North, Central and South Africa (Kissi et al., 1995), suggesting that they most likely represent secondary migrations from Eurasia. Two other viral clades were specific to Africa. The Africa 2 clade contains dog isolates that have a large geographical range in West Africa, including Mauritania, Guinea, Ivory Coast, Burkina Faso, Cameroon, Benin, Nigeria and Chad (Kissi et al., 1995). In contrast, the Africa 3 group of viruses was associated with carnivores of the family Herpestidae (mainly the yellow mongoose), which is the main vector of rabies in the central plateau of southern Africa (Davis et al., 2007; Nel et al., 2005), although only a single Africa 3 clade virus was available for study here (and none in the G gene tree). The Arctic-related clade included viruses sampled from dog, raccoon dog, arctic fox, red fox, striped skunk and wolf; these circulate as a number of lineages occupying a large area across the Northern hemisphere, ranging from central to eastern Asia (Russia, Nepal, the north of India, Korea) as well as Greenland and North America (Hyun et al., 2005; Kissi et al., 1995; Kuzmin et al., 2004; Mansfield et al., 2006; Park et al., 2005). This clade has also been documented in Iran and Pakistan (Nadin-Davis et al., 2003). We provide the first definitive evidence for a widely distributed Asian clade. This included viruses from South-east Asia, namely Myanmar, Thailand, Laos, Cambodia and Vietnam (Ito et al., 1999; Yamagata et al., 2007), China (Meng et al., 2007; Zhang et al., 2006) and also from Indonesia and The Philippines (Nishizono et al., 2002). Finally, the Indian subcontinent clade of RABV was distributed only within southern India and Sri Lanka (Arai et al., 2001; Nanayakkara et al., 2003). This clade is particularly notable because it occupies a basal position on the non-flying mammal part of the RABV phylogeny based on N sequences (bootstrap support of 73 %), suggesting that it was the first of this group to diverge. Although fewer sequences were available in the G gene tree, the lineage representing the single available Indian subcontinent clade virus is still one of the first to diverge, consistent with the pattern seen in the N gene. Indeed, the divergent position of the Indian subcontinent clade observed in the N gene phylogeny could not be rejected by the G gene data under a Shimodaira–Hasegawa test (P=0.219).
Spatial dynamics of dog RABV
To explore the spatial dynamics of dog-associated RABV in more detail, we determined the extent and pattern of geographical subdivision in our expansive N gene dataset. This revealed a strong population subdivision by geographical region (P≤0.001, compared with the null hypothesis of panmixis), confirming the broad-scale observations from our phylogenetic analysis. Specifically, across 130 sequences from 17 geographical regions, we found only 17 unambiguous changes in geographical location, compared with an average of 33 under the random expectation of panmixis. Similarly, only 14 of 272 pairwise comparisons (5 %) among geographical regions exhibited positive correlations (P>0), indicative of migration between them (Table 1). Further, all positive correlation values were weak, with the strongest evidence for migration between Kazakhstan and Russia to Canada and Greenland, and from China and Korea to The Philippines and Indonesia (Table 1), both probably due to human interventions. It was also striking that the majority of the 14 positive migration events involved contiguous geographical regions (such as those within Africa), with only occasional long distance (trans-continental) migration, particularly involving those viruses present in Latin America. In contrast, 190 of 272 pairwise comparisons (70 %) exhibited negative correlations (P<0), highlighting the strength of population subdivision.
The timescale of RABV evolution
We employed a Bayesian coalescent approach to determine the timescale of RABV evolution. The mean rate of nucleotide substitution estimated for the N gene (assuming an uncorrelated lognormal molecular clock and a constant population size) was 2.3×10−4 substitutions (subs) per site per year (95 % HPD values=1.1–3.6×10−4 subs per site per year). A similar mean rate, with overlapping HPD values, was observed in the G gene (mean=3.9×10−4 subs per site per year; 95 % HPD values=1.2–6.5×10−4 subs per site per year). These rates are in agreement with previous studies of lyssavirus evolution (Badrane & Tordo, 2001; Davis et al., 2005, 2006, 2007; Holmes et al., 2002; Hughes et al., 2004, 2005) and did not differ widely by either demographic or clock model. Although a higher mean rate of evolutionary change was observed in the small sample of N genes from bat viruses (mean=6.3×10−4 subs per site per year; 95 % HPD values=1.8–10.6×10−4 subs per site per year), we noted a major difference in branch lengths between two clades of bat RABV (Fig. 1), with anomalously short internal branches in those viruses associated with Myotis spp. and Eptesicus fuscus bats (Davis et al., 2006). Although this might be indicative of larger-scale differences in substitution rate and might explain the large 95 % HPD values in this case, the small sample size precludes further investigation.
Using the same approach, we were able to estimate the timescale of RABV evolution. Specifically, the TMRCA of all the RABV lineages sampled here (both bats and non-flying mammals) was estimated to be approximately 749 years (95 % HPD 363–1215 years), which was similar to the TMRCA estimated for non-flying mammal clades in isolation (761 years with a 95 % HPD 373–1222 years). A similar timescale, with overlapping 95 % HPD values, was observed for the smaller sample of G gene sequences (mean=583 years; 95 % HPD values=222–1116 years), suggesting that these estimates are robust. Hence, there was a relatively rapid lineage radiation during the early evolutionary history of RABV in non-flying mammals. These estimates of age of genetic diversity are also consistent with previous analyses of RABV in Europe, Africa and the Northern hemisphere (Bourhy et al., 1999; Davis et al., 2007) and depict an evolutionary diversification far more recent than the domestication of dogs. In contrast, the genetic diversity within the bat viruses sampled appeared more recently [consistent with previous estimates (Davis et al., 2006; Hughes et al., 2005)], with a mean TMRCA of only 180 years (95 % HPD=69–342 years), although the available dataset is small. Taken together, all of the TMRCAs estimated here suggest that the sampled lineages of RABV originated within the past 1500 years.
Selection pressures on dog-associated RABV
The overall dN/dS of RABV was very low (0.045), indicating that the main evolutionary pressure acting on this virus is strong purifying selection, which is in agreement with previous data (Holmes et al., 2002). This was confirmed by the observation that dN/dS was far higher on external (0.077) compared with internal (0.018) branches, such that most non-synonymous polymorphisms are likely to represent transient deleterious mutations that never achieve fixation (Pybus et al., 2007). This, in turn, suggests that the expansion of the canid clades was not associated with either adaptive evolution or population bottlenecks of sufficient magnitude to result in the fixation of slightly deleterious non-synonymous mutations.
DISCUSSION
The spatial and temporal dynamics of RNA viruses are often reflected by their phylogenetic structure (Biek et al., 2006; Grenfell et al., 2004; Holmes, 2004). As such, detailed phylogenetic analysis of viral populations provides a valuable insight into the pattern and rate of geographical dispersal, especially for viruses that are subject to little natural selection at the epidemiological scale, as is likely to be the case for lyssaviruses (Bourhy et al., 1999; Davis et al., 2005; Holmes et al., 2002; Kissi et al., 1995). The aim of the analysis presented here was to determine the phylogeographic structure of RABV on a global basis and to reconstruct the spatial and temporal dynamics of this virus.
On a broad-scale, our study places the phylogeography of RABV in a global context. Specifically, we show that the current global genetic diversity of RABV from non-flying mammals can be represented by six major and geographically distinct phylogenetic clades, thereby extending previous studies of viral biodiversity. Our analysis also suggests that these clades of terrestrial mammal RABV may have an ancestry that lies with domestic dogs from the south of the Indian subcontinent, as the latter are clearly represented by the most phylogenetically divergent clade in the N gene tree. However, this hypothesis will clearly need to be confirmed with a larger sample of sequences representing a wider range of geographical localities, and with longer sequences to achieve greater phylogenetic support. Further, using coalescent-based methods to estimate times to common ancestry, we were able to show that this evolutionary diversification most likely occurred within the last 1500 years. Consequently, any older canid RABV lineages, proposed to have circulated in the Middle-East more than 2000 years ago (Steele & Fernandez, 1991; Theodorides, 1986), either have not survived to be sampled in the current study, were caused by an independent spill-over from bats that later died out or were due to a different lyssavirus genotype.
Lyssaviruses are zoonotic infections that invariably spill over into non-reservoir hosts (humans, bovines, small ruminants, cats etc). Onward transmission within these dead-end hosts is not sustained, so the successful transmission of RABV in new host species is likely to represent a major adaptive challenge (Kuiken et al., 2006). This is, in part, a reflection of the strong selective constraints that act on RABV, resulting in a high rate of deleterious mutation and hence in relatively low rates of non-synonymous substitution, including at sites that might potentially enhance fitness (Holmes et al., 2002; Kissi et al., 1999). At a larger level, both the N and G gene phylogenies indicate that viruses sampled from other species of the family Canidae, such as foxes and raccoon dogs, as well as hosts belonging to other families within the Carnivora –the Herpestidae in southern Africa and the Mephitidae (skunks) in America – are interspersed within the phylogenetic diversity of dog RABV. While we found no significant evidence for adaptive evolution, our observation strongly suggests that the dog has served as the main vector for inter-species RABV transmission, generating viral lineages that then spread to other taxa. Determining the genetic basis of the traits that govern cross-species transmission clearly represents a major goal for future research on RABV and for emerging viruses in general, although it is important to note that patterns of cross-species transmission may also be in part determined by the ecological factors that shape host contact rates.
Our phylogenetic analysis of migration patterns is notable in that it reveals a strong population subdivision in RABV on a global scale, in contrast to the more fluid dynamics seen when the virus spreads through a specific geographical region (Biek et al., 2007; Real & Biek, 2007). The geographical spread of RABV in non-flying mammals at a global level (and over a period of less than 1500 years) has therefore occurred at such a low rate that its phylogenetic structure is dominated by population subdivision rather than gene flow (Criscione & Blouin, 2005). Hence, despite the relatively recent timescale of RABV evolution, its current biodiversity is characterized by a series of spatially distinct clusters that experience relatively little contact among them. It is likely that this lack of admixture reflects the influence of major geographical barriers to gene flow, as previously demonstrated for RABV in Europe (Bourhy et al., 1999). Indeed, the importance of physical isolation is supported by the phylogeographical patterns observed here, which suggest that both the Himalayan mountains and the Sahara Desert have acted as barriers to gene flow; the former explaining, in part, the spatial partitioning within the Asian clade, and the latter, the different phylogenetic groups seen in Africa. Conversely, a lack of major physical barriers, thereby enabling gene flow, may explain why those viruses from the Arctic-related clade occupy such a wide geographical range. Alternatively, it may be that after initial colonization there is little viral spread to adjoining regions, perhaps because immigrating viruses have a low probability of establishment in areas where other RABV already circulate (Biek et al., 2007; Real & Biek, 2007). However, whether such exclusion barriers to gene flow can explain broad-scale phylogeographic patterns is uncertain.
There are now several examples illustrating how the long distance transmission of RABV is facilitated by human-mediated animal movements (Fevre et al., 2006), including the translocation of infected raccoons from Florida to Virginia for hunting (Jenkins & Winkler, 1987) and the importation of dog rabies in Flores Islands in Indonesia in 1997 (Windiyaningsih et al., 2004), both of which resulted in the rapid spread of RABV. Indeed, the movement of rabid domestic dogs is clearly still a major threat for rabies-free areas (Bourhy et al., 2005). However, the strong population subdivision observed here suggests that, other than large-scale and often inter-continental translocations, humans were not normally responsible for the dispersal of rabid animals and hence RABV. Further, the lack of admixture among clades supports the idea that, over longer timescales, the persistence of RABV in its enzootic stage does not depend upon regular immigration of infected individuals (Biek et al., 2007). Rather, it is more likely that the dispersal of RABV reflects the gradual spatial spread of virus within animals that themselves move relatively small distances, as previously demonstrated in Europe with red foxes and raccoon dogs (Bourhy et al., 1999, 2005) and in North American raccoons (Biek et al., 2007). The only exceptions found in our study, which most likely reflect human intervention, are the migration of virus from Russia to Canada and Greenland and from China to the Philippines and Indonesia.
The phylogenetic pattern depicted here – of distinct, geographically based clades with few intermediate lineages – is in contrast with recent studies of raccoon RABV in North America. In this case, phylogenies of isolates sampled over a period of 30 years were characterized by high rates of branching near the root of the tree, indicative of both spatial and demographical expansion (Biek et al., 2007). There are two explanations for the long-term phylogeographical pattern revealed here: that fitness differences among lineages have enabled some to out-compete others, resulting in a selective purging of lineages, or that intermediate lineages have died out because of stochastic processes alone. Although it is possible that the fixation of advantageous mutations that enable RABV to adapt to new host species has occurred but cannot be detected by current methods, the very low dN/dS values observed throughout RABV evolution, as well as their bias towards external branches, suggests that purifying, rather than positive selection, dominates evolutionary dynamics. Therefore, we propose that random processes have had a more profound effect on long-term phylogeographical patterns in RABV. Specifically, over extended time periods, many of those lineages that appear in short-term demographical expansions are lost randomly by genetic drift, leaving the spatially disjunct phylogeographical clades observed here. This stochastic picture of RABV phylogeography, analogous to an allopatric model of speciation, is also compatible with simple population genetic theory. In a haploid population, the mean time to common ancestry, 2Net (where t is the generation time between transmission events and Ne the effective population size), is likely to be ~2 months in the case of canid RABV (Fekadu, 1991). In both this study and that of Biek et al. (2007), Net ranges from 102 to 103, leading to mean TMRCAs of a few hundred years, indicating that both studies are in agreement regarding the timescale of RABV evolution, as depicted here. Hence, the dual processes of random genetic drift and geographical isolation alone are likely to be sufficient to explain the long-term phylogeographical patterns of dog-associated RABV.
Supplementary Material
Acknowledgments
This work was supported by the ‘Programme de Recherches Avancées de Coopérations Franco-Chinoises’ (PRA B01-06): ‘Epidémiologie moléculaire de la rage en Chine et standardisation des techniques de détection du virus rabique’, by the European Union through the FP6/VIZIER program, and by NIH grant GM080533-01. We are also grateful to the genomic platform of Institut Pasteur, Paris, France and particularly to M. Tichit and C. Bouchier for their help with sequencing. We also thank Andres Paez Fernandez for providing RABV isolates from Colombia and Alan Calaor from the Research Institute for Tropical Medicine, Metro Manila, Philippines, for his help.
Footnotes
GenBank/EMBL/DDBJ accession numbers for the newly acquired N and G gene sequences are designated EU086128–EU086218.
A supplementary figure and two supplementary tables are available with the online version of this paper.
References
- Arai YT, Takahashi H, Kameoka Y, Shiino T, Wimalaratne O, Lodmell DL. Characterization of Sri Lanka rabies virus isolates using nucleotide sequence analysis of nucleoprotein gene. Acta Virol. 2001;45:327–333. [PubMed] [Google Scholar]
- Badrane H, Tordo N. Host switching in Lyssavirus history from the Chiroptera to the Carnivora orders. J Virol. 2001;75:8096–8104. doi: 10.1128/JVI.75.17.8096-8104.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek R, Drummond AJ, Poss M. A virus reveals population structure and recent demographic history of its carnivore host. Science. 2006;311:538–541. doi: 10.1126/science.1121360. [DOI] [PubMed] [Google Scholar]
- Biek R, Henderson JC, Waller LA, Rupprecht CE, Real LA. A high-resolution genetic signature of demographic and spatial expansion in epizootic rabies virus. Proc Natl Acad Sci U S A. 2007;104:7993–7998. doi: 10.1073/pnas.0700741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhy H, Kissi B, Tordo N. Molecular diversity of the Lyssavirus genus. Virology. 1993;194:70–81. doi: 10.1006/viro.1993.1236. [DOI] [PubMed] [Google Scholar]
- Bourhy H, Kissi B, Audry L, Smreczak M, Sadkowska-Todys M, Kulonen K, Tordo N, Zmudzinski JF, Holmes EC. Ecology and evolution of rabies virus in Europe. J Gen Virol. 1999;80:2545–2557. doi: 10.1099/0022-1317-80-10-2545. [DOI] [PubMed] [Google Scholar]
- Bourhy H, Dacheux L, Strady C, Mailles A. Rabies in Europe in 2005. Euro Surveill. 2005;10:213–216. [PubMed] [Google Scholar]
- Criscione CD, Blouin MS. Effective sizes of macroparasite populations: a conceptual model. Trends Parasitol. 2005;21:212–217. doi: 10.1016/j.pt.2005.03.002. [DOI] [PubMed] [Google Scholar]
- David D, Hughes GJ, Yakobson BA, Davidson I, Un H, Aylan O, Kuzmin IV, Rupprecht CE. Identification of novel canine rabies virus clades in the Middle East and North Africa. J Gen Virol. 2007;88:967–980. doi: 10.1099/vir.0.82352-0. [DOI] [PubMed] [Google Scholar]
- Davis PL, Holmes EC, Larrous F, Van der Poel WH, Tjornehoj K, Alonso WJ, Bourhy H. Phylogeography, population dynamics, and molecular evolution of European bat lyssaviruses. J Virol. 2005;79:10487–10497. doi: 10.1128/JVI.79.16.10487-10497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis PL, Bourhy H, Holmes EC. The evolutionary history and dynamics of bat rabies virus. Infect Genet Evol. 2006;6:464–473. doi: 10.1016/j.meegid.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Davis PL, Rambaut A, Bourhy H, Holmes EC. The evolutionary dynamics of canid and mongoose rabies virus in southern Africa. Arch Virol. 2007;152:1251–1258. doi: 10.1007/s00705-007-0962-9. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekadu M. Canine rabies. In: Baer GM, editor. The Natural History of Rabies. 2. Boca Raton, USA: CRC Press; 1991. pp. 367–378. [Google Scholar]
- Févre EM, Bronsvoort BM, Hamilton KA, Cleaveland S. Animal movements and the spread of infectious diseases. Trends Microbiol. 2006;14:125–131. doi: 10.1016/j.tim.2006.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould AR, Hyatt AD, Lunt R, Kattenbelt JA, Hengstberger S, Blacksell SD. Characterisation of a novel lyssavirus isolated from Pteropid bats in Australia. Virus Res. 1998;54:165–187. doi: 10.1016/s0168-1702(98)00025-2. [DOI] [PubMed] [Google Scholar]
- Grenfell BT, Pybus OG, Gog JR, Wood JL, Daly JM, Mumford JA, Holmes EC. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- Holmes EC. The phylogeography of human viruses. Mol Ecol. 2004;13:745–756. doi: 10.1046/j.1365-294x.2003.02051.x. [DOI] [PubMed] [Google Scholar]
- Holmes EC, Woelk CH, Kassis R, Bourhy H. Genetic constraints and the adaptive evolution of rabies virus in nature. Virology. 2002;292:247–257. doi: 10.1006/viro.2001.1271. [DOI] [PubMed] [Google Scholar]
- Hughes GJ, Paez A, Boshell J, Rupprecht CE. A phylogenetic reconstruction of the epidemiological history of canine rabies virus variants in Colombia. Infect Genet Evol. 2004;4:45–51. doi: 10.1016/j.meegid.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Hughes GJ, Orciari LA, Rupprecht CE. Evolutionary timescale of rabies virus adaptation to North American bats inferred from the substitution rate of the nucleoprotein gene. J Gen Virol. 2005;86:1467–1474. doi: 10.1099/vir.0.80710-0. [DOI] [PubMed] [Google Scholar]
- Hyun BH, Lee KK, Kim IJ, Lee KW, Park HJ, Lee OS, An SH, Lee JB. Molecular epidemiology of rabies virus isolates from South Korea. Virus Res. 2005;114:113–125. doi: 10.1016/j.virusres.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Ito N, Sugiyama M, Oraveerakul K, Piyaviriyakul P, Lumlertdacha B, Arai YT, Tamura Y, Mori Y, Minamoto N. Molecular epidemiology of rabies in Thailand. Microbiol Immunol. 1999;43:551–559. doi: 10.1111/j.1348-0421.1999.tb02441.x. [DOI] [PubMed] [Google Scholar]
- Jenkins SR, Winkler WG. Descriptive epidemiology from an epizootic of raccoon rabies in the Middle Atlantic States, 1982–1983. Am J Epidemiol. 1987;126:429–437. doi: 10.1093/oxfordjournals.aje.a114674. [DOI] [PubMed] [Google Scholar]
- Kissi B, Tordo N, Bourhy H. Genetic polymorphism in the rabies virus nucleoprotein gene. Virology. 1995;209:526–537. doi: 10.1006/viro.1995.1285. [DOI] [PubMed] [Google Scholar]
- Kissi B, Badrane H, Audry L, Lavenu A, Tordo N, Brahimi M, Bourhy H. Dynamics of rabies virus quasispecies during serial passages in heterologous hosts. J Gen Virol. 1999;80:2041–2050. doi: 10.1099/0022-1317-80-8-2041. [DOI] [PubMed] [Google Scholar]
- Knobel DL, Cleaveland S, Coleman PG, Févre EM, Meltzer MI, Miranda ME, Shaw A, Zinsstag J, Meslin FX. Reevaluating the burden of rabies in Africa and Asia. Bull World Health Organ. 2005;83:360–368. [PMC free article] [PubMed] [Google Scholar]
- Kuiken T, Holmes EC, McCauley J, Rimmelzwaan GF, Williams CS, Grenfell BT. Host species barriers to influenza virus infections. Science. 2006;312:394–397. doi: 10.1126/science.1122818. [DOI] [PubMed] [Google Scholar]
- Kuzmin IV, Botvinkin AD, McElhinney LM, Smith JS, Orciari LA, Hughes GJ, Fooks AR, Rupprecht CE. Molecular epidemiology of terrestrial rabies in the former Soviet Union. J Wildl Dis. 2004;40:617–631. doi: 10.7589/0090-3558-40.4.617. [DOI] [PubMed] [Google Scholar]
- Kuzmin IV, Hughes GJ, Botvinkin AD, Orciari LA, Rupprecht CE. Phylogenetic relationships of Irkut and West Caucasian bat viruses within the Lyssavirus genus and suggested quantitative criteria based on the N gene sequence for lyssavirus genotype definition. Virus Res. 2005;111:28–43. doi: 10.1016/j.virusres.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Leonard JA, Wayne RK, Wheeler J, Valadez R, Guillen S, Vila C. Ancient DNA evidence for Old World origin of New World dogs. Science. 2002;298:1613–1616. doi: 10.1126/science.1076980. [DOI] [PubMed] [Google Scholar]
- Mansfield KL, Racloz V, McElhinney LM, Marston DA, Johnson N, Rønsholt L, Christensen LS, Neuvonen E, Botvinkin AD, et al. Molecular epidemiological study of Arctic rabies virus isolates from Greenland and comparison with isolates from throughout the Arctic and Baltic regions. Virus Res. 2006;116:1–10. doi: 10.1016/j.virusres.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Meng SL, Yan JX, Xu GL, Nadin-Davis SA, Ming PG, Liu SY, Wu J, Ming HT, Zhu FC, et al. A molecular epidemiological study targeting the glycoprotein gene of rabies virus isolates from China. Virus Res. 2007;124:125–138. doi: 10.1016/j.virusres.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Nadin-Davis SA, Simani S, Armstrong J, Fayaz A, Wandeler AI. Molecular and antigenic characterization of rabies viruses from Iran identifies variants with distinct epidemiological origins. Epidemiol Infect. 2003;131:777–790. doi: 10.1017/s095026880300863x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanayakkara S, Smith JS, Rupprecht CE. Rabies in Sri Lanka: splendid isolation. Emerg Infect Dis. 2003;9:368–371. doi: 10.3201/eid0903.020545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nel LH, Sabeta CT, von Teichman B, Jaftha JB, Rupprecht CE, Bingham J. Mongoose rabies in southern Africa: a reevaluation based on molecular epidemiology. Virus Res. 2005;109:165–173. doi: 10.1016/j.virusres.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Nishizono A, Mannen K, Elio-Villa LP, Tanaka S, Li KS, Mifune K, Arca BF, Cabanban A, Martinez B, et al. Genetic analysis of rabies virus isolates in the Philippines. Microbiol Immunol. 2002;46:413–417. doi: 10.1111/j.1348-0421.2002.tb02714.x. [DOI] [PubMed] [Google Scholar]
- Olson DM, Dinerstein E, Wikramanayake ED, Burgess ND, Powell GVN, Underwood EC, D’Amico JA, Itoua I, Strand HE, et al. Terrestrial ecoregions of the world: a new map of life on earth. Bioscience. 2001;51:933–938. [Google Scholar]
- Park YJ, Shin MK, Kwon HM. Genetic characterization of rabies virus isolates in Korea. Virus Genes. 2005;30:341–347. doi: 10.1007/s11262-005-6777-4. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Pybus OG, Rambaut A, Belshaw R, Freckleton RP, Drummond AJ, Holmes EC. Phylogenetic evidence for deleterious mutation load in RNA viruses and its contribution to viral evolution. Mol Biol Evol. 2007;24:845–852. doi: 10.1093/molbev/msm001. [DOI] [PubMed] [Google Scholar]
- Real LA, Biek R. Spatial dynamics and genetics of infectious diseases on heterogeneous landscapes. J R Soc Interface. 2007;4:935–948. doi: 10.1098/rsif.2007.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen P, Zhang YP, Luo J, Lundeberg J, Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science. 2002;298:1610–1613. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- Steele JH, Fernandez PJ. History of rabies and global aspects. In: Baer GM, editor. The Natural History of Rabies. 2. Boca Raton, USA: CRC Press; 1991. pp. 1–26. [Google Scholar]
- Swofford DL. PAUP 4.0 User’s Manual: Phylogenetic Analysis Using Parsimony. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- Theodorides J. Histoire de la Rage. Paris: Masson; 1986. p. 289. [Google Scholar]
- Verginelli F, Capelli C, Coia V, Musiani M, Falchetti M, Ottini L, Palmirotta R, Tagliacozzo A, De Grossi Mazzorin I, Mariani-Costantini R. Mitochondrial DNA from prehistoric canids highlights relationships between dogs and South-East European wolves. Mol Biol Evol. 2005;22:2541–2551. doi: 10.1093/molbev/msi248. [DOI] [PubMed] [Google Scholar]
- Warrell MJ, Warrell DA. Rabies and other lyssavirus diseases. Lancet. 2004;363:959–969. doi: 10.1016/S0140-6736(04)15792-9. [DOI] [PubMed] [Google Scholar]
- Windiyaningsih C, Wilde H, Meslin FX, Suroso T, Widarso HS. The rabies epidemic on Flores Islands, Indonesia (1998–2003) J Med Assoc Thai. 2004;87:1389–1393. [PubMed] [Google Scholar]
- Yamagata J, Ahmed K, Khawplod P, Mannen K, Xuyen DK, Loi HH, Dung NV, Nishizono A. Molecular epidemiology of rabies in Vietnam. Microbiol Immunol. 2007;51:833–840. doi: 10.1111/j.1348-0421.2007.tb03979.x. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang YZ, Xiong CL, Zou Y, Wang DM, Jiang RJ, Xiao QY, Hao ZY, Zhang LZ, Yu YX, Fu ZF. Molecular characterization of rabies virus isolates in China during 2004. Virus Res. 2006;121:179–188. doi: 10.1016/j.virusres.2006.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.