Summary
Quantitative modeling of human brain activity can provide crucial insights about cortical representations [1, 2], and can form the basis for brain decoding devices [3–5]. Recent functional magnetic resonance imaging (fMRI) studies have modeled brain activity elicited by static visual patterns, and have shown that it is possible to reconstruct these images from brain activity measurements [6–8]. However, blood oxygen level dependent (BOLD) signals measured using fMRI are very slow [9], so it has been difficult to model brain activity elicited by dynamic stimuli such as natural movies. Here we present a new motion-energy [10, 11] encoding model that largely overcome this limitation. Our motion-energy model describes fast visual information and slow hemodynamics by separate components. We recorded BOLD signals in occipito-temporal visual cortex of human subjects who passively watched natural movies, and fit the encoding model separately to individual voxels. Visualization of the fit models reveals how early visual areas represent moving stimuli. To demonstrate the power of our approach we also constructed a Bayesian decoder [8], by combining estimated encoding models with a sampled natural movie prior. The decoder provides remarkable reconstructions of natural movies, capturing the spatio-temporal structure of the viewed movie. These results demonstrate that dynamic brain activity measured under naturalistic conditions can be decoded using current fMRI technology.
Results
Many of our visual experiences are dynamic: perception, visual imagery, dreaming and hallucinations all change continuously over time, and these changes are often the most compelling and important aspects of these experiences. Obtaining a quantitative understanding of brain activity underlying these dynamic processes would advance our understanding of visual function. Quantitative models of dynamic mental events could also have important applications as tools for psychiatric diagnosis, and as the foundation of brain machine interface devices [3–5].
Modeling dynamic brain activity is a difficult technical problem. The best tool available currently for non-invasive measurement of brain activity is functional MRI, which has relatively high spatial resolution [12, 13]. However, blood oxygen level dependent (BOLD) signals measured using fMRI are relatively slow [9], especially when compared to the speed of natural vision and many other mental processes. It has therefore been assumed that fMRI data would not be useful for modeling brain activity evoked during natural vision or by other dynamic mental processes.
Here we present a new motion-energy [10, 11] encoding model that largely overcomes this limitation. The model separately describes the neural mechanisms mediating visual motion information and their coupling to much slower hemodynamic mechanisms. In this report we first validate this encoding model by showing that it describes how spatial and temporal information are represented in voxels throughout visual cortex. We then use a Bayesian approach [8] to combine estimated encoding models with a sampled natural movie prior, in order to produce reconstructions of natural movies from BOLD signals.
We recorded BOLD signals from three human subjects while they viewed a series of color natural movies (20 × 20 degrees at 15 Hz). A fixation task was used to control eye position. Two separate data sets were obtained from each subject. The training data set consisted of BOLD signals evoked by 7,200 seconds of color natural movies, where each movie was presented just once. These data were used to fit a separate encoding model for each voxel located in posterior and ventral occipito-temporal visual cortex. The test data set consisted of BOLD signals evoked by 540 seconds of color natural movies, where each movie was repeated ten times. These data were used to assess the accuracy of the encoding model, and as the targets for movie reconstruction. Because the movies used to train and test models were different, this approach provides a fair and objective evaluation of the accuracy of the encoding and decoding models [2, 14].
BOLD signals recorded from each voxel were fit separately using a two-stage process. Natural movie stimuli were first filtered by a bank of neurally-inspired nonlinear units sensitive to local motion-energy [10, 11]. Then L1-regularized linear regression [15, 16] was used to fit a separate hemodynamic coupling term to each nonlinear filter (Fig. 1; also see Supplemental Information). The regularized regression approach used here was optimized to obtain good estimates even for computational models containing thousands of regressors. In this respect our approach differs from the regression procedures used in many other fMRI studies [17, 18].
Figure 1. Schematic diagram of the motion-energy encoding model.
A, Stimuli first pass through a fixed set of nonlinear spatio-temporal motion-energy filters (shown in detail in panel B), and then through a set of hemodynamic response filters fit separately to each voxel. The summed output of the filter bank provides a prediction of BOLD signals. B, The nonlinear motion-energy filter bank consists of several filtering stages. Stimuli are first transformed into the Commission internationale de l'éclairage (CIE) L*A*B* color space and the color channels are stripped off. Luminance signals then pass through a bank of 6,555 spatio-temporal Gabor filters differing in position, orientation, direction, spatial and temporal frequency (see Supplemental Information for details). Motion energy is calculated by squaring and summing Gabor filters in quadrature. Finally, signals pass through a compressive nonlinearity and are temporally down-sampled to the fMRI sampling rate (1 Hz).
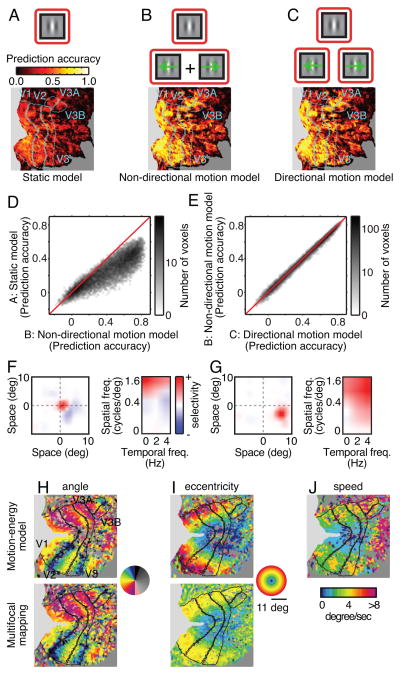
To determine how much motion information is available in BOLD signals we compared prediction accuracy for three different encoding models (Fig. 2A-C): a conventional static model that includes no motion information [8, 19]; a non-directional motion model that represents local motion energy but not direction; and a directional model that represents both local motion energy and direction. Each of these models was fit separately to every voxel recorded in each subject, and the test data were used to assess prediction accuracy for each model. Prediction accuracy was defined as the correlation between predicted and observed BOLD signals. The averaged accuracy across subjects and voxels in early visual areas (V1, V2, V3, V3A and V3B) are 0.24, 0.39 and 0.40 for the static, non-directional and directional encoding models, respectively (Fig. 2D and 2E; see Fig. S1A for subject- and area-wise comparisons). This difference in prediction accuracy is significant (P<0.0001, Wilcoxon signed-rank test). An earlier study showed that the static model tested here recovered much more information from BOLD signals than had been obtained with any previous model [8, 19]. Nevertheless, both motion models developed here provide far more accurate predictions than are obtained with the static model. Note that the difference in prediction accuracy between the directional and non-directional motion models, though significant, is small (Fig. 2E and S1A). This suggests that BOLD signals convey spatially localized but predominantly non-directional motion information. These results show that the motion-energy encoding model predicts BOLD signals evoked by novel natural movies.
Figure 2. The directional motion-energy model capture motion information.
A, (top) The static encoding model includes only Gabor filters that are not sensitive to motion. (bottom) Prediction accuracy of the static model is shown on a flattened map of the cortical surface of one subject (S1). Prediction accuracy is relatively poor. B, The non-directional motion-energy encoding model includes Gabor filters tuned to a range of temporal frequencies, but motion in opponent directions is pooled. Prediction accuracy of this model is better than the static model. C, The directional motion-energy encoding model includes Gabor filters tuned to a range of temporal frequencies and directions. This model provides the most accurate predictions of all models tested. D and E, Voxel-wise comparisons of prediction accuracy between the three models. The directional motion-energy model performs significantly better than the other two models, although the difference between the non-directional and directional motion models is small. See also Figure S1 for subject- and area-wise comparisons. F, The spatial receptive field of one voxel (left), and its spatial and temporal frequency selectivity (right). This receptive field is located near the fovea, and it is high-pass for spatial frequency and low-pass for temporal frequency. This voxel thus prefers static or slow speed motion. G, Receptive field for a second voxel. This receptive field is located lower periphery, and it is band-pass for spatial frequency and high-pass for temporal frequency. This voxel thus prefers higher speed motion than the voxel in F. H, Comparison of retinotopic angle maps estimated using (top) the motion-energy encoding model and (bottom) conventional multi-focal mapping on a flattened cortical map [47]. The angle maps are similar, even though they were estimated using independent data sets and methods. I, Comparison of eccentricity maps estimated as in panel H. The maps are similar except in the far periphery where the multi-focal mapping stimulus was coarse. J, Optimal speed projected on to a flattened map as in panel H. Voxels near the fovea tend to prefer slow speed motion, while those in the periphery tend to prefer high speed motion. See also Figure S1B for subject-wise comparisons.
To further explore what information can be recovered from these data we estimated the spatial, spatial frequency and temporal frequency tuning of the directional motion-energy encoding model fit to each voxel. The spatial receptive fields of individual voxels are spatially localized (Fig. 2F and 2G, left) and are organized retinotopically (Fig. 2H and 2I), as reported in previous fMRI studies [12, 19–23]. Voxel-based receptive fields also show spatial and temporal frequency tuning (Fig. 2F and 2G, right), as reported in previous fMRI studies [24, 25].
To determine how motion information is represented in human visual cortex we calculated the optimal speed for each voxel by dividing the peak temporal frequency by the peak spatial frequency. Projecting the optimal speed of the voxels onto a flattened map of the cortical surface (Fig. 2J) reveals a significant positive correlation between eccentricity and optimal speed: relatively more peripheral voxels are tuned for relatively higher speeds. This pattern is observed in areas V1, V2 and V3 and for all three subjects (P<0.0001, t-test for correlation coefficient; see Fig. S1B for subject- and area-wise comparisons). To our knowledge this is the first evidence that speed selectivity in human early visual areas depends on eccentricity, though a consistent trend has been reported in human behavioral studies [26–28] and in neurophysiological studies of non-human primates [29, 30]. These results show that the motion-energy encoding model describes tuning for both spatial and temporal information at the level of single voxels.
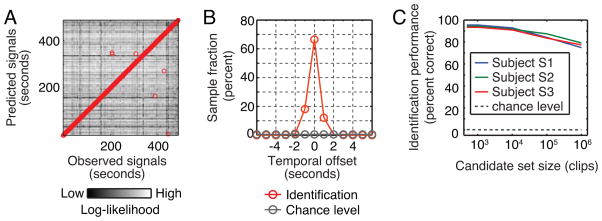
To further characterize the temporal specificity of the estimated motion-energy encoding models we used the test data to estimate movie identification accuracy. Identification accuracy [7, 19] measures how well a model can correctly associate an observed BOLD signal pattern with the specific stimulus that evoked it. Our motion-energy encoding model can identify the specific movie stimulus that evoked an observed BOLD signals 95% (464/486) of the time within ± one volume (one second; Subject S1, Fig 3A and B). This is far above what would be expected by chance (<1%). Identification accuracy (within ± one volume) is greater than 75% for all three subjects even when the set of possible natural movie clips includes one million separate clips chosen at random from the internet (Fig. 3C). This result demonstrates that the motion-energy encoding model is both valid and temporally specific. Furthermore, it suggests that the model might provide good reconstructions of natural movies from brain activity measurements [5].
Figure 3. Identification analysis.
A, Identification accuracy for one subject (S1). The test data in our experiment consisted of 486 volumes (seconds) of BOLD signals evoked by the test movies. The estimated model yielded 486 volumes of BOLD signals predicted for the same movies. The brightness of the point in the mth column and nth row represents the log-likelihood (see Supplemental Information) of the BOLD signals evoked at the mth second given the BOLD signal predicted at the nth second. The highest log-likelihood in each column is designated by a red circle and thus indicates the choice of the identification algorithm. B, Temporal offset between the correct timing and the timing identified by the algorithm, for the same subject shown in panel A. The algorithm was correct to within ± one volume (second) 95% of the time (464/486); chance performance is less than 1% (3/486; i.e., three volumes centered at the correct timing). C, Scaling of identification accuracy with set size. To understand how identification accuracy scales with size of stimulus set we enlarged the identification stimulus set to include additional stimuli drawn from a natural movie database (but not actually used in the experiment). For all three subjects identification accuracy (within ± one volume) is greater than 75% even when set of potential movies includes one million clips. This is far above chance (gray dashed line).
We used a Bayesian approach [8] to reconstruct movies from the evoked BOLD signals (see also Fig. S2). We estimated the posterior probability by combining a likelihood function (given by the estimated motion-energy model; see Supplemental Information) and a sampled natural movie prior. The sampled natural movie prior consists of ~18 million seconds of natural movies sampled at random from the internet. These clips were assigned uniform prior probability (and consequently, all other clips were assigned zero prior probability; note also that none of the clips in the prior were used in the experiment). Furthermore, to make decoding tractable reconstructions were based on one second clips (15 frames), using BOLD signals with a delay of four seconds. In effect, this procedure enforces an assumption that the spatio-temporal stimulus that elicited each measured BOLD signal must be one of the movie clips in the sampled prior.
Fig. 4 shows typical reconstructions of natural movies obtained using the motion-energy encoding model and the Bayesian decoding approach (see Movie S1 for the corresponding movies). The posterior probability was estimated across the entire sampled natural movie prior separately for each BOLD signal in the test data. The peak of this posterior distribution is the conventional maximum a posteriori (MAP) reconstruction [8] for each BOLD signal (see second row in Fig. 4). When the sampled natural movie prior contains clips that are similar to the viewed clip then the MAP reconstructions are good (e.g., the close-up of a human speaker shown in Fig. 4A). However, when the prior contains no clips similar to the viewed clip then the reconstructions are poor (e.g., Fig. 4B). This likely reflects both the limited size of the sampled natural movie prior and noise in the fMRI measurements. One way to achieve more robust reconstructions without enlarging the prior is to interpolate over the sparse samples in the prior. We therefore created an averaged high posterior (AHP) reconstruction, by averaging the 100 clips in the sampled natural movie prior that had the highest posterior probability (see also Fig. S2; Note that the AHP reconstruction can be viewed as a Bayesian version of bagging [31]). The AHP reconstruction captures the spatio-temporal structure within a viewed clip even when it is completely unique (e.g., the spreading of an inkblot from the center of the visual field shown in Fig. 4B).
Figure 4. Reconstructions of natural movies from BOLD signals.
A, First row: Three frames from a natural movie used in the experiment, taken one second apart. Second through sixth rows: frames from the five clips with the highest posterior probability. The maximum a posteriori (MAP) reconstruction is shown in row two. Seventh row: The averaged high posterior (AHP) reconstruction. The MAP provides good reconstruction of the second and third frames, while AHP provide more robust reconstructions across frames. B and C, Additional examples of reconstructions, format same as in panel A. D, Reconstruction accuracy (correlation in motion-energy; see Supplemental Information) for all three subjects. Error bars indicate ± 1 s.e.m. across one-second clips. Both the MAP and AHP reconstructions are significant, though the AHP reconstructions are significantly better than the MAP reconstructions. Dashed lines show chance performance (P=0.01). See also Figure S2.
To quantify reconstruction quality we calculated the correlation between the motion-energy content of the original movies and their reconstructions (see Supplemental Information). A correlation of 1.0 indicates perfect reconstruction of the spatio-temporal energy in the original movies and a correlation of 0.0 indicates that the movies and their reconstruction are spatio-temporally uncorrelated. The results for both MAP and AHP reconstructions are shown in Fig. 4D. In both cases reconstruction accuracy is significantly higher than chance (P<0.0001, Wilcoxon rank-sum test; see Supplemental Information). Furthermore, AHP reconstructions are significantly better than MAP reconstructions (P<0.0001, Wilcoxon signed-rank test). Although still crude (motion-energy correlation ~ 0.3), these results validate our general approach to reconstruction and demonstrate that the AHP estimate improves reconstruction over the MAP estimate.
Discussion
In this study we developed a new encoding model that predicts BOLD signals in early visual areas with unprecedented accuracy. By using this model in a Bayesian framework we provide the first reconstructions of natural movies from human brain activity. This is a critical step toward the creation of brain reading devices that can reconstruct dynamic perceptual experiences. Our solution to this problem rests on two key innovations. The first is a new motion-energy encoding model that is optimized for use with fMRI, and that aims to reflect the separate contributions of the underlying neuronal population and hemodynamic coupling (Fig. 1). This encoding model recovers fine temporal information from relatively slow BOLD signals. The second is a sampled natural movie prior that is embedded within a Bayesian decoding framework. This approach provides a simple method for reconstructing spatio-temporal stimuli from the sparsely sampled and slow BOLD signals.
Our results provided the first evidence that there is a positive correlation between eccentricity and speed tuning in human early visual areas. This provides a functional explanation for previous behavioral studies indicating that speed sensitivity depends on eccentricity [26–28]. This systematic variation in speed tuning across the visual field may be an adaptation to the non-uniform distribution of speed signals induced by selective foveation in natural scenes [32]. From the perspective of decoding, this result suggests that we might further optimize reconstruction by including eccentricity-dependent speed tuning in the prior.
We found that a motion-energy model that incorporates directional motion signals was only slightly better than a model that does not include direction. We believe that this likely reflects limitations in the spatial resolution of fMRI recordings. Indeed, a recent study reported that hemodynamic signals were sufficient to visualize a columnar organization of motion direction in macaque area V2 [33]. Future fMRI experiments at higher spatial or temporal resolution [34, 35] might therefore be able to recover clearer directional signals in human visual cortex.
In preliminary work for this study we explored several encoding models that incorporated color information explicitly. However, we found that color information did not improve the accuracy of predictions or identification beyond what could be achieved with models that include only luminance information. We believe that this reflects the fact that luminance and color borders are often correlated in natural scenes [36, 37]; but see [38]. (Note that when iso-luminant, mono-chromatic stimuli are used, color can be reconstructed from evoked BOLD signals [39].) The correlation between luminance and color information in natural scenes has an interesting side effect: our reconstructions tend to recover color borders (e.g., borders between hair vs. face or face vs. body), even though the encoding model makes no use of color information. This is a positive aspect of the sampled natural movie prior and provides additional cues to aid in recognition of reconstructed scenes (see also [40]).
We found that the quality of reconstruction could be improved by simply averaging around the maxiumum of the posterior movies. This suggests that reconstructions might be further improved if the number of samples in the prior is much larger than the one used here. Likelihood estimation (and thus reconstruction) would also improve if additional knowledge about the neural representation of movies were used to construct better encoding models (e.g., [41]).
In a landmark study Thirion et al. [6] first reconstructed static imaginary patterns from BOLD signals in early visual areas. Other studies have decoded subjective mental states, such as the contents of visual working memory [42], or whether subjects are attending to one or another orientation or direction [3, 43]. The modeling framework presented here provides the first reconstructions of dynamic perceptual experiences from BOLD signals. Therefore, this modeling framework might also permit reconstruction of dynamic mental content such as continuous natural visual imagery. In contrast to earlier studies that reconstruct visual patterns defined by checkerboard contrast [6, 7], our framework could potentially be used to decode involuntary subjective mental states (e.g., dreaming or hallucination), though it would be difficult to determine whether the decoded content was accurate. One recent study showed that BOLD signals elicited by visual imagery are more prominent in ventral-temporal visual areas than in early visual areas [44]. This finding suggests that a hybrid encoding model that combines the structural motion-energy model developed here with a semantic model of the form developed in previous studies [8, 45, 46] could provide even better reconstructions of subjective mental experiences.
Experimental Procedures
Stimuli
Visual stimuli consisted of color natural movies drawn from the Apple QuickTime HD gallery (http://www.apple.com/quicktime/guide/hd/) and YouTube (http://www.youtube.com/; see the list of movies in Supplemental Information). The original high-definition movies were cropped to a square and then spatially down-sampled to 512 by 512 pixels. Movies were then clipped to 10–20 seconds in length, and the stimulus sequence was created by randomly drawing movies from the entire set. Movies were displayed using a VisuaStim LCD goggles system (20x20 degrees, 15 Hz). A colored fixation spot (4 pixels or 0.16 degree square) was presented on top of the movie. The color of the fixation spot changed three times per second to ensure that it was visible regardless of the color of the movie.
MRI parameters
The experimental protocol was approved by the Committee for the Protection of Human Subjects at University of California at Berkeley. Functional scans were conducted using a 4 Tesla Varian INOVA scanner (Varian, Inc., Palo Alto, CA) with a quadrature transmit/receive surface coil (Midwest RF, LLC, Hartland, WI). Scans were obtained using T2*-weighted gradient-echo EPI: TR = 1 second, TE = 28 ms, Flip angle = 56 degrees, voxel size = 2.0 × 2.0 × 2.5 mm3, and FOV = 128 × 128 mm2. The slice prescription consisted of 18 coronal slices beginning at the posterior pole and covering the posterior portion of occipital cortex.
Data collection
Functional MRI scans were made from three human subjects, S1 (author S.N., age 30), S2 (author T.N., age 34) and S3 (author A.V., age 23). All subjects were healthy and had normal or corrected-to-normal vision. The training data were collected in 12 separate 10 minute blocks (7200 seconds total). The training movies were shown only once each. The test data were collected in 9 separate 10 minute blocks (5400 seconds total) that consists of 9 minute movies repeated 10 times each. To minimize effects from potential adaptation and long-term drift in the test data, the 9 minute movies were divided into 1 minute chunks and these were randomly permuted across blocks. Each test block was thus constructed by concatenating 10 separate one minute movies. All data were collected across multiple sessions for each subject, and each session contained multiple training and test blocks. The training and test data sets used different movies.
Supplementary Material
Highlights.
A new motion-energy model can describe BOLD signals evoked by natural movies.
The model reveals how motion information is represented in early visual areas.
Speed tuning in human early visual areas depends on eccentricity.
The model provides reconstructions of natural movies from evoked BOLD signals.
Acknowledgments
We thank B. Inglis for assistance with MRI, and K. Kay and K. Hansen for assistance with retinotopic mapping. We also thank M. Oliver, R. Prenger, D. Stansbury, A. Huth and J. Gao for their help in various aspects of this research. This work was supported by NIH and NEI.
Footnotes
The authors declare no conflict of interest.
Additional methods can be found in Supplemental Information online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wu MC, David SV, Gallant JL. Complete functional characterization of sensory neurons by system identification. Annu Rev Neurosci. 2006;29:477–505. doi: 10.1146/annurev.neuro.29.051605.113024. [DOI] [PubMed] [Google Scholar]
- 2.Naselaris T, Kay KN, Nishimoto S, Gallant JL. Encoding and decoding in fMRI. Neuroimage. 2011;56:400–410. doi: 10.1016/j.neuroimage.2010.07.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamitani Y, Tong F. Decoding the visual and subjective contents of the human brain. Nat Neurosci. 2005;8:679–685. doi: 10.1038/nn1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haynes JD, Rees G. Decoding mental states from brain activity in humans. Nat Rev Neurosci. 2006;7:523–534. doi: 10.1038/nrn1931. [DOI] [PubMed] [Google Scholar]
- 5.Kay KN, Gallant JL. I can see what you see. Nat Neurosci. 2009;12:245. doi: 10.1038/nn0309-245. [DOI] [PubMed] [Google Scholar]
- 6.Thirion B, Duchesnay E, Hubbard E, Dubois J, Poline JB, Lebihan D, Dehaene S. Inverse retinotopy: inferring the visual content of images from brain activation patterns. Neuroimage. 2006;33:1104–1116. doi: 10.1016/j.neuroimage.2006.06.062. [DOI] [PubMed] [Google Scholar]
- 7.Miyawaki Y, Uchida H, Yamashita O, Sato MA, Morito Y, Tanabe HC, Sadato N, Kamitani Y. Visual image reconstruction from human brain activity using a combination of multiscale local image decoders. Neuron. 2008;60:915–929. doi: 10.1016/j.neuron.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Naselaris T, Prenger RJ, Kay KN, Oliver M, Gallant JL. Bayesian reconstruction of natural images from human brain activity. Neuron. 2009;63:902–915. doi: 10.1016/j.neuron.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friston KJ, Jezzard P, Turner R. Analysis of functional MRI time-series. Human Brain Mapping. 1994;1:153–171. [Google Scholar]
- 10.Adelson EH, Bergen JR. Spatiotemporal energy models for the perception of motion. J Opt Soc Am A. 1985;2:284–299. doi: 10.1364/josaa.2.000284. [DOI] [PubMed] [Google Scholar]
- 11.Watson AB, Ahumada Jr. Model of human visual-motion sensing. J Opt Soc Am A. 1985;2:322–341. doi: 10.1364/josaa.2.000322. [DOI] [PubMed] [Google Scholar]
- 12.Engel SA, Rumelhart DE, Wandell BA, Lee AT, Glover GH, Chichilnisky EJ, Shadlen MN. fMRI of human visual cortex. Nature. 1994;369:525. doi: 10.1038/369525a0. [DOI] [PubMed] [Google Scholar]
- 13.Logothetis NK. What we can do and what we cannot do with fMRI. Nature. 2008;453:869–878. doi: 10.1038/nature06976. [DOI] [PubMed] [Google Scholar]
- 14.Kriegeskorte N, Simmons WK, Bellgowan PS, Baker CI. Circular analysis in systems neuroscience: the dangers of double dipping. Nat Neurosci. 2009;12:535–540. doi: 10.1038/nn.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Osher S. Coordinate descent optimization for l minimization with application to compressed sensing; a greedy algorithm. Inverse Problems and Imaging. 2009;3:487–503. [Google Scholar]
- 16.Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society Series B. 1996;58:267–288. [Google Scholar]
- 17.Friston KJ, Frith CD, Turner R, Frackowiak RS. Characterizing evoked hemodynamics with fMRI. Neuroimage. 1995;2:157–165. doi: 10.1006/nimg.1995.1018. [DOI] [PubMed] [Google Scholar]
- 18.Boynton GM, Engel SA, Glover GH, Heeger DJ. Linear systems analysis of functional magnetic resonance imaging in human V1. J Neurosci. 1996;16:4207–4221. doi: 10.1523/JNEUROSCI.16-13-04207.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kay KN, Naselaris T, Prenger RJ, Gallant JL. Identifying natural images from human brain activity. Nature. 2008;452:352–355. doi: 10.1038/nature06713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sereno MI, Dale AM, Reppas JB, Kwong KK, Belliveau JW, Brady TJ, Rosen BR, Tootell RBH. Borders of multiple visual areas in humans revealed by functional magnetic resonance imaging. Science. 1995;268:889–893. doi: 10.1126/science.7754376. [DOI] [PubMed] [Google Scholar]
- 21.DeYoe EA, Carman GJ, Bandettini P, Glickman S, Wieser J, Cox R, Miller D, Neitz J. Mapping striate and extrastriate visual areas in human cerebral cortex. Proc Natl Acad Sci U S A. 1996;93:2382–2386. doi: 10.1073/pnas.93.6.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wandell BA, Dumoulin SO, Brewer AA. Visual field maps in human cortex. Neuron. 2007;56:366–383. doi: 10.1016/j.neuron.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 23.Dumoulin SO, Wandell BA. Population receptive field estimates in human visual cortex. Neuroimage. 2008;39:647–660. doi: 10.1016/j.neuroimage.2007.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh KD, Smith AT, Greenlee MW. Spatiotemporal frequency and direction sensitivities of human visual areas measured using fMRI. Neuroimage. 2000;12:550–564. doi: 10.1006/nimg.2000.0642. [DOI] [PubMed] [Google Scholar]
- 25.Henriksson L, Nurminen L, Hyvarinen A, Vanni S. Spatial frequency tuning in human retinotopic visual areas. J Vis. 2008;8(5):1–13. doi: 10.1167/8.10.5. [DOI] [PubMed] [Google Scholar]
- 26.Kelly DH. Retinal inhomogeneity. I. Spatiotemporal contrast sensitivity. J Opt Soc Am A. 1984;1:107–113. doi: 10.1364/josaa.1.000107. [DOI] [PubMed] [Google Scholar]
- 27.McKee SP, Nakayama K. The detection of motion in the peripheral visual field. Vision Res. 1984;24:25–32. doi: 10.1016/0042-6989(84)90140-8. [DOI] [PubMed] [Google Scholar]
- 28.Orban GA, Van Calenbergh F, De Bruyn B, Maes H. Velocity discrimination in central and peripheral visual field. J Opt Soc Am A. 1985;2:1836–1847. doi: 10.1364/josaa.2.001836. [DOI] [PubMed] [Google Scholar]
- 29.Orban GA, Kennedy H, Bullier J. Velocity sensitivity and direction selectivity of neurons in areas V1 and V2 of the monkey: influence of eccentricity. J Neurophysiol. 1986;56:462–480. doi: 10.1152/jn.1986.56.2.462. [DOI] [PubMed] [Google Scholar]
- 30.Yu HH, Verma R, Yang Y, Tibballs HA, Lui LL, Reser DH, Rosa MG. Spatial and temporal frequency tuning in striate cortex: functional uniformity and specializations related to receptive field eccentricity. Eur J Neurosci. 2010;31:1043–1062. doi: 10.1111/j.1460-9568.2010.07118.x. [DOI] [PubMed] [Google Scholar]
- 31.Domingos P. Why does bagging work? A Bayesian account and its implications. In: Heckerman D, Mannila H, Pregibon D, Uthurusamy R, editors. Proceedings of the Third International Conference on Knowledge Discovery and Data Mining. 1997. pp. 155–158. [Google Scholar]
- 32.Eckert MP, Buchsbaum G. Efficient coding of natural time varying images in the early visual system. Philos Trans R Soc Lond B Biol Sci. 1993;339:385–395. doi: 10.1098/rstb.1993.0038. [DOI] [PubMed] [Google Scholar]
- 33.Lu HD, Chen G, Tanigawa H, Roe AW. A motion direction map in macaque v2. Neuron. 2010;68:1002–1013. doi: 10.1016/j.neuron.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moeller S, Yacoub E, Olman CA, Auerbach E, Strupp J, Harel N, Ugurbil K. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn Reson Med. 2010;63:1144–1153. doi: 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feinberg DA, Moeller S, Smith SM, Auerbach E, Ramanna S, Glasser MF, Miller KL, Ugurbil K, Yacoub E. Multiplexed echo planar imaging for sub-second whole brain FMRI and fast diffusion imaging. PLoS One. 2010;5:e15710. doi: 10.1371/journal.pone.0015710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fine I, MacLeod DI, Boynton GM. Surface segmentation based on the luminance and color statistics of natural scenes. J Opt Soc Am A Opt Image Sci Vis. 2003;20:1283–1291. doi: 10.1364/josaa.20.001283. [DOI] [PubMed] [Google Scholar]
- 37.Zhou C, Mel BW. Cue combination and color edge detection in natural scenes. J Vis. 2008;8(4):1–25. doi: 10.1167/8.4.4. [DOI] [PubMed] [Google Scholar]
- 38.Hansen T, Gegenfurtner KR. Independence of color and luminance edges in natural scenes. Vis Neurosci. 2009;26:35–49. doi: 10.1017/S0952523808080796. [DOI] [PubMed] [Google Scholar]
- 39.Brouwer GJ, Heeger DJ. Decoding and reconstructing color from responses in human visual cortex. J Neurosci. 2009;29:13992–14003. doi: 10.1523/JNEUROSCI.3577-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliva A, Schyns PG. Diagnostic colors mediate scene recognition. Cogn Psychol. 2000;41:176–210. doi: 10.1006/cogp.1999.0728. [DOI] [PubMed] [Google Scholar]
- 41.Bartels A, Zeki S, Logothetis NK. Natural vision reveals regional specialization to local motion and to contrast-invariant, global flow in the human brain. Cereb Cortex. 2008;18:705–717. doi: 10.1093/cercor/bhm107. [DOI] [PubMed] [Google Scholar]
- 42.Harrison SA, Tong F. Decoding reveals the contents of visual working memory in early visual areas. Nature. 2009;458:632–635. doi: 10.1038/nature07832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamitani Y, Tong F. Decoding seen and attended motion directions from activity in the human visual cortex. Curr Biol. 2006;16:1096–1102. doi: 10.1016/j.cub.2006.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddy L, Tsuchiya N, Serre T. Reading the mind's eye: decoding category information during mental imagery. Neuroimage. 2010;50:818–825. doi: 10.1016/j.neuroimage.2009.11.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell TM, Shinkareva SV, Carlson A, Chang KM, Malave VL, Mason RA, Just MA. Predicting human brain activity associated with the meanings of nouns. Science. 2008;320:1191–1195. doi: 10.1126/science.1152876. [DOI] [PubMed] [Google Scholar]
- 46.Li LJ, Socher R, Fei-Fei L. Towards total scene understanding: Classification, annotation and segmentation in an automatic framework. IEEE Conference on Computer Vision and Pattern Recognition; 2009. pp. 2036–2043. [Google Scholar]
- 47.Hansen KA, David SV, Gallant JL. Parametric reverse correlation reveals spatial linearity of retinotopic human V1 BOLD response. Neuroimage. 2004;23:233–241. doi: 10.1016/j.neuroimage.2004.05.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.