Abstract
Amine end-modified poly(ß-amino ester)s (PBAEs) have generated interest as efficient, biodegradable polymeric carriers for plasmid DNA (pDNA). For cationic, non-degradable polymers, such as polyethylenimine (PEI), the polymer molecular weight (MW) and molecular weight distribution (MWD) significantly affect transfection activity and cytotoxicity. The effect of MW on DNA transfection activity for PBAEs has been less well studied. We applied two strategies to obtain amine end-modified PBAEs varying in MW. In one approach, we synthesized four amine end-modified PBAEs with each at 15 different monomer molar ratios, and observed that polymers of intermediate length mediated optimal DNA transfection in HeLa cells. Biophysical characterization of these feed ratio variants suggested that optimal performance was related to higher DNA complexation efficiency and smaller nanoparticle size, but not to nanoparticle charge. In a second approach, we used preparative size exclusion chromatography (SEC) to obtain well-defined, monodisperse polymer fractions. We observed that the transfection activities of size-fractionated PBAEs generally increased with MW, a trend that was weakly associated with an increase in DNA binding efficiency. Furthermore, this approach allowed for the isolation of polymer fractions with greater transfection potency than the starting material. For researchers working with gene delivery polymers synthesized by step-growth polymerization, our data highlight the potentially broad utility of preparative SEC to isolate monodisperse polymers with improved properties. Overall, these results help to elucidate the influence of polymer MWD on nucleic acid delivery and provide insight toward the rational design of next-generation materials for gene therapy.
1. Introduction
Gene therapy is a promising treatment strategy for many inherited disorders including cystic fibrosis, severe combined immunodeficiency, and hemophilia, in addition to cancer and infectious diseases such as AIDS. Despite recent clinical progress[1], concerns with the use of viral vectors, including immunogenicity, small DNA cargo capacity, and difficulty of large-scale production, have led to continued interest in the development of synthetic carriers[2]. A diverse collection of materials has been studied for potential as synthetic gene delivery agents, including lipids, polymers, polysaccharides, polypeptides, dendrimers, and inorganic nanoparticles[3]. However, sub-optimal delivery efficiency in vivo relative to viral vectors has inhibited their widespread clinical use[4, 5]. Though viruses have been naturally selected to efficiently navigate the multiple intra- and extra-cellular barriers to successful gene transfer, the flexibility of polymer chemistries offers great potential to identify and incorporate functionalities that confer not only effective gene transfection, but also superior biocompatibility, enhanced formulation stability, and low toxicity[6, 7].
Toward this end, a more comprehensive understanding of the structure-property relationship for gene delivery polymers is critical to the elucidation of design principles for future generations of synthetic gene vectors. The molecular weight (MW) and molecular weight distribution (MWD) of cationic polymers are among the factors known to dramatically affect their gene delivery performance. For example, it was reported that higher MW poly(2-(dimethylamino)ethyl methacrylate (PDMAEMA), (Mw > 300 kDa) yielded greater in vitro gene transfection than polymers with lower MW (Mw < 60 kDa)[8]. This trend was confirmed more recently by another group, which found that transfection activity increased with the Mw of PDMAEMA up to at least 915 kDa[9]. Similarly, for various other polymeric carriers including trehalose-based glycopolymers[10], four-branched star vectors[11], and quaternized celluloses[12], higher MW was correlated with increasing gene delivery activity for the range of molecular weights examined. For poly(L-lysine) (PLL), however, polymers of intermediate length (Mw = 54 kDa) produced optimal gene transfection relative to longer (Mw = 225 kDa) or shorter (Mw < 22.4 kDa) variants[13], a phenomenon that has been attributed to an optimal rate of vector unpacking[14].
For polyethylenimine (PEI), there are a variety of studies on the relationship between polymer MW and DNA transfection activity. Using branched PEIs ranging in MW from 0.6 to 70 kDa, one group found that higher MW variants mediated significantly greater in vitro DNA transfection, which they speculated might owe to a greater capacity for endosomal escape [15]. In contrast, another group reported that in vitro transfection activity decreased with increasing MW for three branched PEIs ranging in Mw from 1.8 to 70 kDa [16]. Likewise, in a comparison between a low MW PEI (Mw = 11.9 kDa) with low degrees of branching and a high MW, highly branched PEI (Mw = 1,616 kDa), it was observed the low MW variant had much greater transfection potency and lower toxicity[17]. In another study, 25 kDa branched PEI was fractionated by size, and a particular fraction with Mw of roughly 4–10 kDa displayed optimal performance relative to higher or lower MW fractions[18]. Finally, with linear PEIs ranging in Mw from 1.0–9.0 kDa, it was observed that at low N:P ratios, transfection generally increased with MW, but that at higher N:P ratios, polymers of intermediate length were superior[19].
Poly(β-amino esters) (PBAEs) are a promising class of polymeric gene vectors characterized by their ease of synthesis and biodegradability[20–24]. With their capacity to condense plasmid DNA (pDNA) into nanoparticles on the order of 50–200 nm in diameter, PBAEs have yielded high gene delivery efficiency to a variety of cell types with low toxicity[25, 26]. The use of combinatorial polymer library synthesis coupled with high-throughput screening and characterization has revealed structural motifs associated with highly active gene delivery polymers; in particular, the presence of hydroxyl groups in the side chains and the conjugation of certain primary amines to the chain ends dramatically modulate the efficiency of DNA transfection in different cell types[27–30]. Amine end-capped PBAEs, especially those based on poly(5-amino-1-pentanol-co-1,4-butanediol diacrylate) (C32), have demonstrated potential for a number of clinically relevant applications, including suicide gene therapy for ovarian cancer[31], genetic modification of stem cells for treatment of ischemia[32], and gene transfer to glioblastoma cells[33]. However, the impact of chain length on nucleic acid delivery has not yet been systematically examined for this important group of degradable gene delivery polymers.
In this study, we applied two strategies to obtain amine end-modified PBAEs with variation in MW: modulation of monomer stoichiometry and preparative size exclusion chromatography (SEC). Using the first approach, we observed that polymers of intermediate MW mediated optimal DNA transfection activity. For these polymers, we did not observe a significant correlation between polymer MW and toxicity. Optimal performance was associated with higher DNA complexation efficiency and smaller nanoparticle size, but not with nanoparticle ζ-potential. However, using preparative SEC to obtain more monodisperse polymer fractions from a polydisperse starting polymer, we found that the transfection efficiencies of size-fractionated, well-defined PBAEs generally increased with MW. In addition, this approach allowed us to isolate polymer fractions that were more potent than the starting material, which indicates the potentially broad applicability of this separation technique for gene delivery polymers synthesized by step-growth polymerization.
2. Materials and methods
2.1. Materials
1,4-butanediol diacrylate (“C”) and 5-amino-1-pentanol (“32”) were purchased from Alfa Aesar (Ward Hill, MA, USA). 1,3-diaminopropane (“103”), 1,3-pentanediamine (“117”), and 2-methyl-1,5-pentanediamine (“118”) were obtained from Sigma-Aldrich (St. Louis, MO, USA). (PEO)4-bis-amine (“122”) was acquired from Molecular Biosciences (Boulder, CO, USA). All reagents were used without further purification. Plasmid DNA encoding green fluorescent protein (gWiz-GFP) was purchased from Aldevron (Fargo, ND, USA). HeLa cells (ATCC, Manassas, VA, USA) were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Invitrogen). Quant-IT PicoGreen dsDNA reagent was purchased from Invitrogen.
2.2. Polymer synthesis
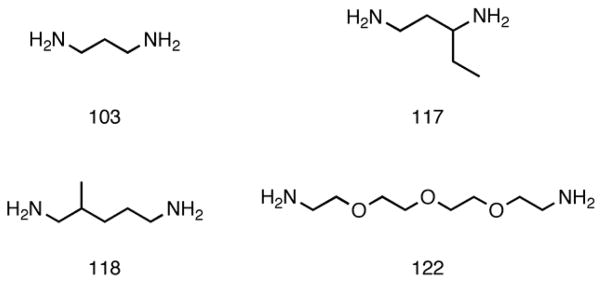
Acrylate-terminated C32 poly(β-amino ester) was synthesized in ~5 g batches by reacting 1,4-butanediol diacrylate (“C”) and 5-amino-1-pentanol (“32”) in bulk at 70°C for 48 h with stirring. To obtain MW variants, 15 monomer molar ratios between 1:1 and 1.3:1 C:32 were chosen for polymerization: 1.0, 1.025, 1.04, 1.05, 1.06, 1.075, 1.1, 1.125, 1.15, 1.175, 1.2, 1.225, 1.25, 1.25, 1.275, and 1.3:1 (C:32). After cooling, 1 g of each batch was end-capped with 2 mmol of each of 4 different amines (103, 117, 118, and 122) by reaction in anhydrous tetrahydrofuran (THF) overnight at RT at a concentration of 100 mg/mL. The following day, the 60 resulting C32 variants were purified by precipitation in anhydrous hexanes (1:3 v/v THF:hexanes) and dried under vacuum for 24 h. Polymers were then dissolved at 100 mg/mL in dimethyl sulfoxide (DMSO) and stored at −20°C until use.
In a typical example, for the 1.1:1 C:32 molar ratio, 3.394 g C (17.12 mmol) was added to 1.605 g 32 (15.55 mmol) in a 20 mL glass vial equipped with stir bar and sealed with a screw cap. It was heated in a reaction block at 70°C for 48 h with stirring. For end-modification of this polymer with the 103 amine, the 5 g batch of C32-Ac was cooled and then dissolved in 10 mL of anhydrous THF; of this solution, 2 mL was transferred to a 20 mL glass vial containing 8 mL of 103 amine at 0.25 M in THF (2 mmol). After stirring overnight at RT, the polymer was isolated by precipitation into hexanes and was then analyzed by SEC.
2.3. Analytical size exclusion chromatography (SEC)
Analytical SEC was performed using a Waters system (Milford, MA) equipped with a 2400 differential refractometer, 515 pump, and 717-plus autosampler. The flow rate was 1 mL/min and the mobile phase was THF. The Styragel columns (Waters) and detector were thermostated at 35ºC. Linear polystyrene standards were used for calibration.
2.4. in vitro GFP plasmid DNA transfection
One day before transfection, 15,000 HeLa cells in 100 μL of medium were seeded into each well of a 96-well polystyrene tissue culture plate. In a typical example, for a 600 ng/well DNA dose, pDNA was diluted to 0.06 mg/mL in 25 mM sodium acetate (NaOAc) buffer at pH 5.2. Polymers were thawed immediately prior to transfection and diluted in NaOAc buffer to a concentration 20, 30, or 40 times that of the DNA concentration, depending on the desired polymer:DNA w/w ratio. To form DNA-polymer nanoparticles, 25 μL of polymer solution was added to 25 μL of DNA in a half-area 96-well plate, mixed by repeated pipetting using a multichannel pipette, and allowed to incubate for 5 min at RT. 30 μL of polymer-DNA complexes were then gently mixed with 195 μL of fresh medium warmed to 37°C. Conditioned medium was removed using a 12-channel aspirating wand and replaced with 150 μL of the complexes diluted in medium. Following a 4 h incubation, complexes were removed with the aid of a multi-channel aspiration wand and replaced with 100 μL of fresh medium. GFP expression was assessed 48 h after transfection by fluorescence-activated cell sorting (FACS).
2.5. FACS analysis
After aspirating conditioned medium, cells were washed with PBS and detached using 25 μL per well of 0.25% trypsin-EDTA (Invitrogen). 50 μL of FACS running buffer, consisting of 98% PBS, 2% FBS, and 1:200 v/v propidium iodide solution (Invitrogen), was added to each well. Cells were mixed thoroughly and then transferred to a 96-well round-bottom plate. GFP expression was measured using FACS on a BD LSR II (Becton Dickinson, San Jose, CA, USA). Propidium iodide (PI) staining was used to exclude dead cells from the analysis. PI staining was also used to determine the viabilities of treated cells relative to non-treated control cells, where the relative viability was calculated as the ratio of live (unstained) treated cells per well to the mean number of live non-treated cells per well. 2D gating was used to separate increased autofluorescence signals from increased GFP signals to more accurately count positively expressing cells. Gating and analysis were performed using FlowJo v8.8 software (TreeStar, Ashland, OR, USA).
2.6. Dynamic light scattering (DLS) measurements
To form complexes using the same concentrations and conditions that were optimal for transfection (40 w/w polymer:DNA), 150 μL of polymer at 2.4 mg/mL in NaOAc buffer was mixed with 150 μL of plasmid DNA (gWiz-GFP) at 0.06 mg/mL in NaOAc buffer. After incubation for 5 min at RT, 1.2 mL of 10% serum-containing medium was added to the mixture, which was then immediately subjected to DLS measurements using a ZetaPALS DLS detector (Brookhaven Instruments Corp., Holtsville, NY, USA, 15-mW laser, incident beam 676 nm). To ensure that all measurements represented the same time point of analysis, the size and ζ-potential data were obtained independently. Correlation functions were collected at a scattering angle of 90°, and particle sizes were calculated using the MAS option of BICs particle sizing software (v. 2.30) using the viscosity and refractive index of water at 25°C. Particle sizes are expressed as effective diameters assuming a log-normal distribution. Electrophoretic mobilities were measured at 25°C using BIC Phase Analysis Light Scattering (PALS) ζ-potential software, and ζ-potentials were calculated using the Smoluchowski model for aqueous suspensions.
2.7. Dye exclusion assay
The dye exclusion assay to determine polymer-DNA complexation efficiency was performed as described previously[29]. Briefly, a working solution of PicoGreen was prepared by diluting 80 μL of stock solution in 15.92 mL NaOAc buffer. In each well of a 96-well plate, 50 μL of polymer at 2.4 mg/mL in NaOAc buffer was added to 50 μL of DNA at 0.06 mg/mL in NaOAc buffer. After 5 min, 100 μL of PicoGreen working solution was added to the complexes. After an additional 5 min incubation, 30 μL was transferred to 200 μL of 10% serum-containing medium in a black 96-well assay plate. The fluorescence was then measured on a Tecan Infinite M1000 plate reader using the FITC filter set (excitation 485 nm, emission 535 nm). The reduction in relative fluorescence (RF), or the relative complexation efficiency, was calculated using the relationship (FDNA− Fsample)/(FDNA − Fblank), where Fsample is the fluorescence of the polymer–DNA–PicoGreen sample, FDNA is the fluorescence of DNA–PicoGreen (no polymer), and Fblank is the fluorescence of a sample with no polymer or DNA (only PicoGreen).
2.8. Preparative SEC
Polymers were fractionated based on size with a Phenogel 5μm MXL gel filtration column (300 mm x 7.8 mm, Phenomenex, P/No. 00H-3087-KO) using THF as the mobile phase at a flow rate of 1 mL/min. The separation was done on a 1200 Series Agilent HPLC system equipped with a UV diode array detector and a 1260 Infinity analytical scale fraction collector. The column compartment was kept at 40°C during fractionation. Polymer fractions were collected at 0.2 min intervals based on the absorption of the polymer at 254 nm. The fractions were transferred into tared vials, dried, weighed, and then dissolved to 100 mg/mL in DMSO. They were stored at −20°C until further use.
3. Results & discussion
3.1. Synthesis and analytical SEC of stoichiometric PBAE variants
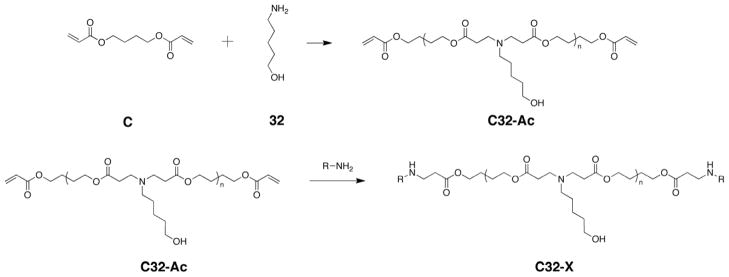
One straightforward method for molecular weight control of polymers synthesized by step-growth addition is to vary the stoichiometric ratio of the starting monomers. According to the Carothers relationship, the weight-average molecular weight Mw depends on the molar ratio of reactants r, the fractional monomer conversion p, and the molecular weight of the polymer repeat unit M0 as follows: Mw = M0(1+p)(1+r)/(1-2pr+r). In this report, the PBAE C32 was synthesized using a range of 15 monomer molar feed ratios between 1:1 and 1.3:1 “C” (1,4-butanediol diacrylate) to “32” (5-amino-1-pentanol); these acrylate-terminated C32 polymers (C32-Ac) were then end-capped with each of four different amine molecules (Fig. 1). These end-capping amines, denoted 103, 117, 118, and 122, were selected because they improved transfection performance relative to unmodified C32 polymer[28].
Fig. 1.
As expected, SEC analysis revealed that the molecular weight of these end-modified PBAEs decreased as the feed ratios deviated from stoichiometric unity (Fig. 2). The polymers ranged in Mw from a maximum of 16.4 kDa for C32-118 synthesized at a feed ratio of 1:1 C:32 to a minimum of 2.4 kDa for C32-103 synthesized at 1.3:1 C:32. Because the end-modified polymers were synthesized from the same 15 batches of intermediate C32-Ac polymer, the differences in MW observed between end-modified PBAEs at the same monomer feed ratio may reflect a limited extent of polymer cross-linking or degradation.
Fig. 2.
3.2. Plasmid DNA transfection and cytotoxicity
To investigate the relationship between the molecular weight of end-modified PBAEs and in vitro transfection activity, we formed complexes using the synthesized polymers and GFP-encoding plasmid DNA (pDNA), and then incubated these nanoparticles with cultured HeLa cells in serum-containing growth medium for 4 h. Two days after transfection, we used fluorescence activated cell sorting (FACS) to quantify the proportion of HeLa cells expressing GFP.
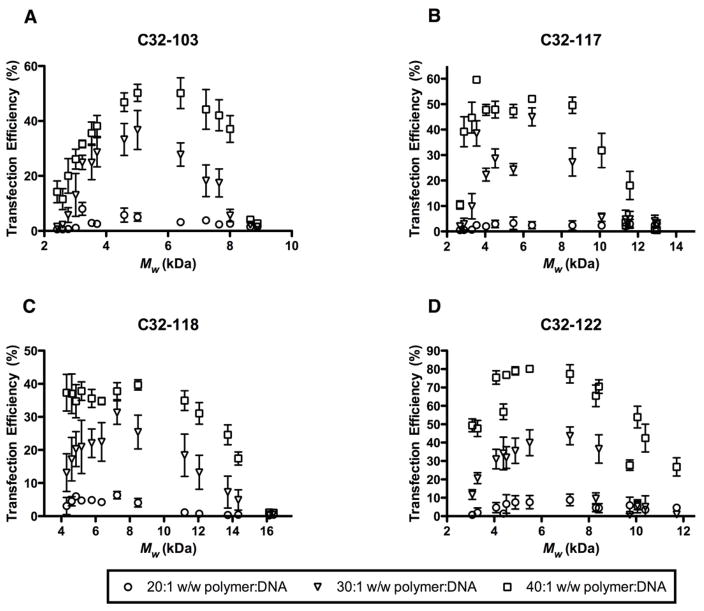
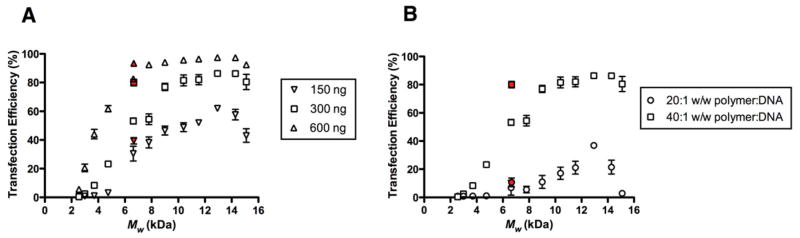
When the transfection efficiencies of the various end-modified PBAEs were correlated with Mw, we observed that for a given weight ratio of polymer:DNA, polymers of intermediate length (Mw ~ 5–8 kDa) generally outperformed polymers with higher or lower Mw (Fig. 3). For example, at a polymer:DNA w/w ratio of 40:1 (equivalent to N:P ratio of ~44:1), C32-122 with Mw = 5.5 kDa (synthesized at a C:32 molar feed ratio of 1.125:1) successfully transfected ~80% of HeLa cells, in contrast to ~26% for the highest MW variant (Mw = 11.7 kDa) and ~49% for the lowest MW variant (Mw = 3.0 kDa). At a lower polymer:DNA w/w ratio of 30:1 (N:P ~ 33), this trend of optimal transfection activity for polymers of intermediate MW persisted. However, at a w/w ratio of 20:1 or below (N:P ~ 22), none of the MW variants yielded significant transfection.
Fig. 3.
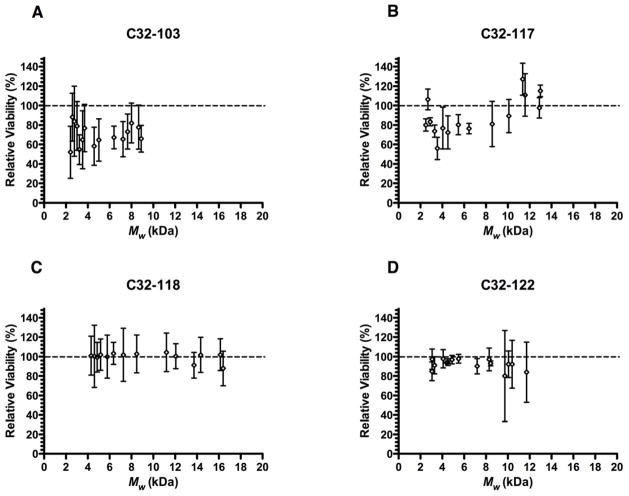
Because of the frequent reports of a correlation between toxicity and polymer MW for many polymers, including PEI, we examined this relationship by staining transfected cells with propidium iodide immediately prior to FACS. When the viabilities of transfected cells relative to non-treated control cells were plotted against polymer Mw at the highest polymer:DNA w/w ratio used (40:1), we did not observe any significant association between the length of end-modified PBAEs and their toxicity during transfection (Fig. 4). Although there was some toxicity associated with C32-103 and C32-117, for each of these polymers, it did not increase with increasing MW. Importantly, for the most effective end-modified PBAE, C32-122, the relative viabilities of transfected cells were not significantly reduced. These results suggest that PBAEs have low toxicity over a wide range of MW, and therefore show promise as non-toxic transfection materials.
Fig. 4.
3.3. Biophysical characterization of polymer/DNA nanoparticles
For various other polymeric gene delivery systems, the relationship between MW and gene delivery has been associated with variation in nanoparticle size and charge. To assess the hypothesis that the dependence of transfection efficiency on the MW of end-modified PBAEs reflects biophysical characteristics of the polymer/DNA nanoparticles, we used dynamic light scattering (DLS) to measure the sizes and ζ-potentials of complexes formed from our stoichiometric variants. Polymers and plasmid DNA were mixed at the optimal weight ratio for transfection, 40:1 w/w polymer:DNA (N:P ~ 44:1). To replicate transfection conditions, complexes were prepared by repeated pipetting in 25 mM sodium acetate buffer at pH 5.2, incubated for 5 min at room temperature to allow for self-assembly, and then subjected to DLS immediately following dilution in 10% serum-containing medium.
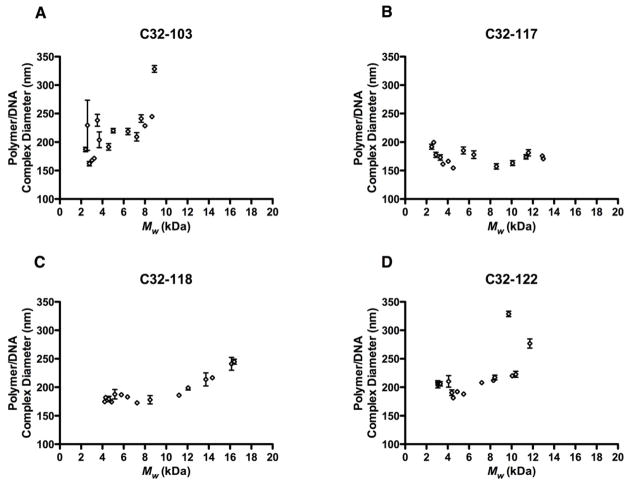
We observed that for three of the four end-modified PBAEs, C32-118, C32-122, and to a limited extent, C32-103, polymer/DNA nanoparticle diameters were relatively uniform with respect to Mw up to ~6-8 kDa, but above this threshold, particle size increased with increasing Mw (Fig. 5). The most effective end-modified PBAEs generally yielded nanoparticles less than 200 nm in size. In contrast to all of the other polymers tested, for C32-117, there was no clear association between polymer length and nanoparticle size, with all polymer variants resulting in complexes between 150-200 nm in diameter. Although for a given end-modified PBAE, smaller nanoparticles generally mediated more efficient transfection (Figure S1), the smallest complexes were never the most effective, which suggests the significance of factors other than particle size.
Fig. 5.
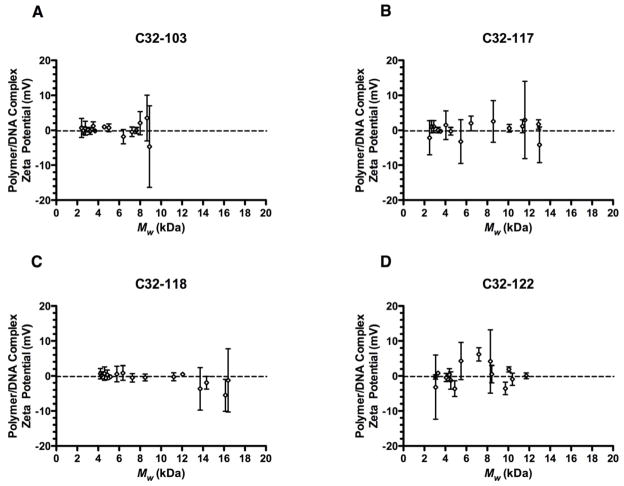
Because we wanted a direct comparison with our in vitro transfection results, we chose to perform DLS analysis, including the charge measurements, with complexes that were diluted in serum-containing medium. Under these conditions, we observed no significant variation in the nanoparticle ζ-potential, with most polymers producing complexes that were near-neutral in serum-containing medium regardless of MW (Fig. 6). We speculated that either the lack of a buffering agent or the presence of negatively-charged serum proteins could generate spurious data or otherwise mask an underlying trend. However, when we conducted charge measurements of C32-122/DNA nanoparticles in 25 mM sodium acetate buffer at pH 5.2, all particles were similarly characterized by near-neutral ζ-potentials within a narrow range of −1 to +1 mV (Figure S2). Therefore, for these end-modified PBAEs, the superior transfection activity of polymers of intermediate length depends to a certain degree on smaller nanoparticle size, but is independent of nanoparticle charge.
Fig. 6.
3.4. Relative DNA binding efficiency
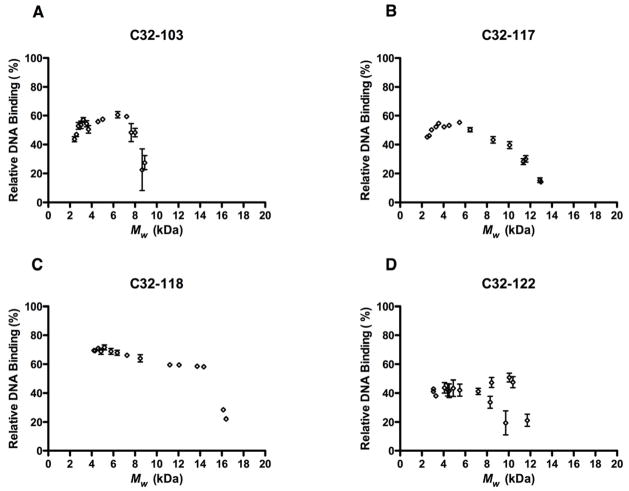
To assess whether the presence of an optimal polymer MW for transfection reflects enhanced condensation and loading of free plasmid DNA into nanoparticles, we quantified the DNA binding efficiencies of these end-modified PBAE variants by measuring the reduction in relative fluorescence due to the protection of entrapped DNA from intercalation by the PicoGreen dye[3434]. Using the same conditions as those for transfection as well as for the DLS measurements above, we found that for at least two of the four end-modified PBAEs, C32-103 and C32-117, polymers of intermediate MW entrapped plasmid DNA with greater efficiency than higher or lower MW variants (Fig. 7). For these polymers, the variants with the greatest gene delivery activity in HeLa cells also complexed DNA the most efficiently (Fig. S3). For C32-118 and C32-122, however, complexation efficiency generally decreased with polymer MW, and the variants most effective at transfection did not coincide with those that bound DNA with the highest efficiency.
Fig. 7.
From these data, we hypothesize that at least for C32-103 and C32-117, the relatively high DNA binding efficiencies of polymers of intermediate MW contribute to their enhanced transfection. Though useful in establishing trends when comparing variants of a particular end-modified polymer, the complexation efficiency values obtained from this dye exclusion assay in general appeared to have limited predictive power with respect to transfection activity; for instance, C32-122 variants loaded DNA the least efficiently relative to other end-capped C32 polymers, but actually transfected HeLa cells the most efficiently. These results suggest that for a given gene delivery polymer, DNA complexation efficiency is one important factor among others, including chemical composition and nanoparticle size, responsible for the variation in transfection activity due to polymer MW.
3.5. Preparative SEC
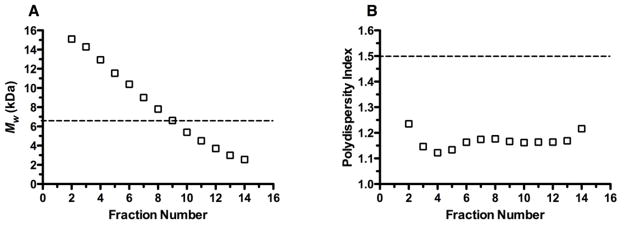
Although they vary in MW, the stoichiometric PBAE variants synthesized above through step-growth polymerization are characterized by broad MWDs. Besides their high polydispersity indices (PDIs), variation of the diacrylate:amine ratio in the polymerization reaction could alter the degree to which polymers are amine end-capped. To obtain polymers varying in MW, but characterized by a narrower MWD, we re-synthesized a particular feed ratio variant of C32-122 (C:32 = 1.1:1; Mw = 6.69 kDa, PDI = 1.50) and subjected it to preparative SEC using an HPLC system with an automated liquid fraction collector (Fig. S4). Analytical SEC of ~13 of the 20 fractions collected (representing ~75% of the total polymer mass loaded) revealed that Mw decreased smoothly with elution time (Fig. 8A). Furthermore, this method allowed for the isolation of fractions with lower PDI (<1.2) than the crude polymer (Fig. 8B).
Fig. 8.
We performed a pDNA transfection experiment using these fractionated C32-122 polymers and assessed the overall transfection efficiency in HeLa cells after 48 h. When polymer length was correlated with transfection efficiency at high DNA doses and polymer:DNA w/w ratio, we observed that transfection efficiency generally increased with polymer Mw (Fig. 9). At a lower DNA dose, or at a lower polymer:DNA w/w ratio, we found that some high MW fractions exhibited dramatically greater potency than fractions with low MW. These high MW fractions were also more potent than the crude, unfractionated polymer. For example, at a DNA dose of 150 ng per well with a polymer:DNA w/w ratio of 40:1, C32-122 with a Mw of 12.9 kDa transfected ~62% of cells, in contrast to <10% for polymers below 5 kDa, and ~39% for the crude polymer. These transfection data underscore the importance of using freshly synthesized materials when working with degradable gene-delivery polymers, since the MW threshold for optimal performance can be rather narrow, and the consequence of a small degree of degradation accordingly steep. Furthermore, the isolation of monodisperse polymers improved consistency of physical properties across the samples, and enhanced delivery at higher MW.
Fig. 9.
When we conducted DLS measurements of the DNA nanoparticles formed using these size-fractionated polymers in serum-containing media, we did not observe significant variation in particle diameter; however, the variation in relative DNA binding efficiency among the size-fractionated polymers appeared to correlate to a certain extent with the observed transfection activities (Fig. S5). In contrast to our data with the polymer feed ratio variants, we did observe an association between polymer MW and relative cell viability following transfection, which was most prominent at the highest dose and polymer:DNA weight ratio tested (Fig. S6). Given that under these conditions some polymer fractions transfected nearly 100% of cells, the transfection and viability data taken together imply that a careful selection of polymer MW, DNA dose, and weight ratio (in addition to other known variables such as cell density) could permit highly efficient gene delivery with low cytotoxicity.
As we suggested above, we hypothesize that the observed differences in trends between the C32-122 feed ratio variants and the size-fractionated C32-122 polymer likely result from the high polydispersities of the feed ratio variants as well as possible heterogeneity in the extent of amine end-modification. For researchers working with gene delivery polymers synthesized by step-growth polymerization, our data highlight the potentially broad utility of preparative SEC for the isolation of well-defined, monodisperse fractions with higher transfection potency than the starting material. We expect that this approach should also help to reduce variability between batches of polymers.
Because both data sets suggested a role for DNA complexation efficiency in modulating the relationship between polymer MW and transfection activity, we hypothesize that the best gene delivery polymers owe their performance at least in part to an optimal balance of the rates of complex formation during particle assembly and unpacking within the cell. Additional studies are required to address this hypothesis in greater detail.
4. Conclusions
Working with amine end-modified PBAEs, a promising class of degradable gene delivery polymers, we investigated the effect of polymer MW on the transfection activity, toxicity, and biophysical properties of the resulting polymer-DNA nanoparticles. Using variation of monomer stoichiometry, we observed that polymers of intermediate length mediated optimal DNA transfection in HeLa cells. Characterization of these feed ratio variants suggested that optimal performance was related to higher polymer-DNA complexation efficiency and smaller nanoparticle size, but not to nanoparticle charge. In contrast, using preparative SEC to obtain well-defined, monodisperse polymer fractions, we observed that the transfection activities of size-fractionated PBAEs generally increased with MW, a trend that was weakly associated with more efficient DNA binding. Our ability to isolate polymer fractions with higher transfection potency than the starting material indicates the potentially broad utility of this approach. These results further our understanding of the influence of polymer MWD on nucleic acid delivery, a critical aspect of the structure-property relationship for gene delivery materials.
Supplementary Material
Acknowledgments
The authors thank Dr. Christopher Levins, Dr. Ana Jaklenec, Patrick Fenton, Glenn Paradis, and Lenny Rigione for assistance with instrumentation, and Dr. Janet Zoldan for kindly providing HeLa cells. This work was supported by Alnylam Pharmaceuticals and the National Institutes of Health (NIH) Grants R01-EB000244-27 and 5-R01-CA132091-04. A.A.E. acknowledges graduate research fellowship support from the National Science Foundation. D.J.S. acknowledges postdoctoral support from NIH NRSA award F32-EB011867.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sheridan C. Gene therapy finds its niche. Nat Biotechnol. 2011;29:459. doi: 10.1038/nbt.1769. [DOI] [PubMed] [Google Scholar]
- 2.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4:346–58. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 3.Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chem Rev. 2009;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 4.Glover DJ, Lipps HJ, Jans DA. Towards safe, non-viral therapeutic gene expression in humans. Nat Rev Genet. 2005;6:299–310. doi: 10.1038/nrg1577. [DOI] [PubMed] [Google Scholar]
- 5.Edelstein M. J Gene Med. Wiley; 2011. Gene therapy clinical trials worldwide. [DOI] [PubMed] [Google Scholar]
- 6.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4:581–93. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 7.Wong SY, Pelet JM, Putnam D. Polymer systems for gene delivery--past, present, and future. Progress in Polymer Science. 2007;32:799–837. [Google Scholar]
- 8.van de Wetering P, Cherng JY, Talsma H, Crommelin DJ, Hennink WE. 2-(dimethylamino)ethyl methacrylate based (co)polymers as gene transfer agents. J Control Release. 1998;53:145–53. doi: 10.1016/s0168-3659(97)00248-4. [DOI] [PubMed] [Google Scholar]
- 9.Layman JM, Ramirez SM, Green MD, Long TE. Influence of polycation molecular weight on poly(2-dimethylaminoethyl methacrylate)-mediated DNA delivery in vitro. Biomacromolecules. 2009;10:1244–52. doi: 10.1021/bm9000124. [DOI] [PubMed] [Google Scholar]
- 10.Srinivasachari S, Liu Y, Prevette LE, Reineke TM. Effects of trehalose click polymer length on pDNA complex stability and delivery efficacy. Biomaterials. 2007;28:2885–98. doi: 10.1016/j.biomaterials.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 11.Nemoto Y, Borovkov A, Zhou Y-M, Takewa Y, Tatsumi E, Nakayama Y. Impact of molecular weight in four-branched star vectors with narrow molecular weight distribution on gene delivery efficiency. Bioconjug Chem. 2009;20:2293–9. doi: 10.1021/bc900283h. [DOI] [PubMed] [Google Scholar]
- 12.Song Y, Wang H, Zeng X, Sun Y, Zhang X, Zhou J, et al. Effect of molecular weight and degree of substitution of quaternized cellulose on the efficiency of gene transfection. Bioconjug Chem. 2010;21:1271–9. doi: 10.1021/bc100068f. [DOI] [PubMed] [Google Scholar]
- 13.Wolfert MA, Dash PR, Nazarova O, Oupicky D, Seymour LW, Smart S, et al. Polyelectrolyte vectors for gene delivery: influence of cationic polymer on biophysical properties of complexes formed with DNA. Bioconjug Chem. 1999;10:993–1004. doi: 10.1021/bc990025r. [DOI] [PubMed] [Google Scholar]
- 14.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67:598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 15.Godbey WT, Wu KK, Mikos AG. Size matters: Molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. J Biomed Mater Res. 1999;45:268–75. doi: 10.1002/(sici)1097-4636(19990605)45:3<268::aid-jbm15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 16.Morimoto K, Nishikawa M, Kawakami S, Nakano T, Hattori Y, Fumoto S, et al. Molecular weight-dependent gene transfection activity of unmodified and galactosylated polyethyleneimine on hepatoma cells and mouse liver. Mol Ther. 2003;7:254–61. doi: 10.1016/s1525-0016(02)00053-9. [DOI] [PubMed] [Google Scholar]
- 17.Fischer D, Bieber T, Li Y, Elsässer HP, Kissel T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm Res. 1999;16:1273–9. doi: 10.1023/a:1014861900478. [DOI] [PubMed] [Google Scholar]
- 18.Werth S, Urban-Klein B, Dai L, Höbel S, Grzelinski M, Bakowsky U, et al. A low molecular weight fraction of polyethylenimine (PEI) displays increased transfection efficiency of DNA and siRNA in fresh or lyophilized complexes. Journal of Controlled Release. 2006;112:257–70. doi: 10.1016/j.jconrel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Breunig M, Lungwitz U, Liebl R, Fontanari C, Klar J, Kurtz A, et al. Gene delivery with low molecular weight linear polyethylenimines. J Gene Med. 2005;7:1287–98. doi: 10.1002/jgm.775. [DOI] [PubMed] [Google Scholar]
- 20.Lynn DM, Langer R. Degradable poly(β-amino esters): synthesis, characterization, and self-assembly with plasmid DNA. J Am Chem Soc. 2000;122:10761–8. [Google Scholar]
- 21.Lynn D, Anderson D, Putnam D, Langer R. Accelerated discovery of synthetic transfection vectors: parallel synthesis and screening of a degradable polymer library. J Am Chem Soc. 2001;123:8155–6. doi: 10.1021/ja016288p. [DOI] [PubMed] [Google Scholar]
- 22.Akinc A, Lynn D, Anderson D, Langer R. Parallel synthesis and biophysical characterization of a degradable polymer library for gene delivery. J Am Chem Soc. 2003;125:5316–23. doi: 10.1021/ja034429c. [DOI] [PubMed] [Google Scholar]
- 23.Anderson D, Lynn D, Langer R. Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angew Chem Int Ed. 2003;42:3153–8. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 24.Anderson D, Peng W, Akinc A, Hossain N, Kohn A, Padera R, et al. A polymer library approach to suicide gene therapy for cancer. Proc Natl Acad Sci USA. 2004;101:16028–33. doi: 10.1073/pnas.0407218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luten J, van Nostrum CF, De Smedt SC, Hennink WE. Biodegradable polymers as non-viral carriers for plasmid DNA delivery. J Control Release. 2008;126:97–110. doi: 10.1016/j.jconrel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 26.Green JJ, Langer R, Anderson DG. A combinatorial polymer library approach yields insight into nonviral gene delivery. Acc Chem Res. 2008;41:749–59. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson D, Akinc A, Hossain N, Langer R. Structure/property studies of polymeric gene delivery using a library of poly(β-amino esters) Mol Ther. 2005;11:426–34. doi: 10.1016/j.ymthe.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 28.Zugates G, Peng W, Zumbuehl A, Jhunjhunwala S, Huang Y, Langer R, et al. Rapid optimization of gene delivery by parallel end-modification of poly(3-amino ester)s. Mol Ther. 2007;15:1306–12. doi: 10.1038/sj.mt.6300132. [DOI] [PubMed] [Google Scholar]
- 29.Green JJ, Zugates GT, Tedford NC, Huang Y, Griffith LG, Lauffenburger DA, et al. Combinatorial modification of degradable polymers enables transfection of human cells comparable to adenovirus. Adv Mater. 2007;19:2836–42. [Google Scholar]
- 30.Sunshine J, Green JJ, Mahon KP, Yang F, Eltoukhy AA, Nguyen DN, et al. Small-molecule end-groups of linear polymer determine cell-type gene-delivery efficacy. Adv Mater. 2009;21:4947–51. doi: 10.1002/adma.200901718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang YH, Zugates GT, Peng W, Holtz D, Dunton C, Green JJ, et al. Nanoparticle-delivered suicide gene therapy effectively reduces ovarian tumor burden in mice. Cancer Res. 2009;69:6184–91. doi: 10.1158/0008-5472.CAN-09-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang F, Cho SW, Son SM, Bogatyrev SR, Singh D, Green J, et al. Genetic engineering of human stem cells for enhanced angiogenesis using biodegradable polymeric nanoparticles. Proc Natl Acad Sci USA. 2010;107:3317–22. doi: 10.1073/pnas.0905432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tzeng SY, Guerrero-Cazares H, Martinez EE, Sunshine JC, Quinones-Hinojosa A, Green JJ. Non-viral gene delivery nanoparticles based on poly(β-amino esters) for treatment of glioblastoma. Biomaterials. 2011;32:5402–10. doi: 10.1016/j.biomaterials.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Welz C, Fahr A. Spectroscopic methods for characterization of nonviral gene delivery systems from a pharmaceutical point of view. Applied Spectroscopy Reviews. 2001;36:333–97. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.