Abstract
Mucins glycoproteins contribute to lung pathophysiology in asthma. The protein backbone of mucin glycoproteins are encoded by specific MUC genes, which exhibit a high degree of polymorphisms that generate a variable number of tandem repeat (VNTR) domains. MUC7 typically encodes for 6 VNTR, each with 23 amino acids. In a Northern-European cohort a polymorphism encoding MUC7*5 (5-VNTR) is in 100% linkage disequilibrium with the single nucleotide polymorphism rs9982010 and associated with a decreased risk of being asthmatic and having better lung function. African-Americans have 5 to 10-fold increase in the incidence of asthma relative to Caucasians, believed to be partially associated with higher genetic susceptibility. Occurrence of the rs9982010 and MUC7 allelic frequencies was evaluated in inner-city African-Americans to test their association with being asthmatic. A logistic regression analysis showed that having the MUC7*5-VNTR allele decreased the likelihood of being asthmatic (OR=0.173 (CI: 0.041–0.737) and p-value of <0.018) and not in a strong linkage disequilibrium with the rs9982010 (r2=0.03; OR=66; CI: 5.913–736.72). A novel MUC7*4-VNTR polymorphism, identified in an African-American non-asthmatic individual, was linked to a structural rearrangement of the VNTR domain. These data extend the association of MUC7*5 allelic polymorphisms and asthma to inner-city African-Americans.
Keywords: mucin genes, asthma, genetic polymorphism, African-American, inner-city
INTRODUCTION
Asthma is a complex, multifactorial disease reflecting genetic and environmental components. Asthma is now regarded as having multiple different subtypes rather than being a single disease entity 1. The morbidity and mortality associated with asthma are disproportionately high among minority pediatric populations, particularly those who reside in densely populated inner-city areas 2–6. African Americans are hospitalized for asthma three times more often than other Americans, and African Americans living in inner-cities are two to six times more likely to die from asthma 7. Inner-city and social risk factors likely explain some of the disparities in incidence; however genotype is an important determinant of host immune responses and contributes to the overall predisposition of individuals to asthma. The contributions of genetic background to the development of asthma in minority populations are understudied.
Mucin glycoproteins (mucins) are the major macromolecular components of the lung mucus layer, which protects the respiratory tract epithelium against infectious agents, allergens and environmental toxins. Mucins are overproduced in asthma, as well as other lung diseases, and contribute to airway pathophysiology and thus to disease morbidity and mortality 8–12. MUC genes encode the protein backbone of mucins and most of the MUC genes have polymorphisms that encode a variable number of tandem repeats (VNTR) 13 [reviewed in 12] that can result in differences in the length of the protein backbone. Genetic analyses of some MUC genes have been carried out in patients with atopy and or asthma. A longer VNTR length in the MUC2 gene is associated with a cohort of atopic, non-asthmatic patients, but no associated differences with asthma and VNTR domains of MUC1, MUC4, MUC5AC, or MUC5B genes have been found 14.
However, a study using a Northern European cohort showed an association between the risk of being diagnosed with asthma and a polymorphism in MUC7 15. Typically, MUC7 encodes 6 non-perfect VNTR (MUC7*6) of 23 amino acids; with the less common MUC7 polymorphic variant containing 5 VNTR (MUC7*5) 16. A study by Kirkbride et al. (2001) in a Northern European cohort identified an association of the MUC7*5 allele with a decreased risk of an asthma diagnosis 15. A subsequent study identified the rs998210 single nucleotide polymorphism (SNP) in 100% linkage disequilibrium (LD) with the MUC7*5 polymorphism in the same Northern European population 17. Since African Americans have a higher prevalence of asthma, we hypothesized that they would also have a lower prevalence of the apparently protective MUC7*5 allele. We therefore investigated the MUC7 VNTR domain and the occurrence of the rs998210 single nucleotide polymorphism (SNP) in the MUC7 gene to determine its association with asthma in inner-city African American children. These 2 polymorphisms were focused on as they have are the only MUC7 polymorphisms that have been associated with asthma to date.
MATERIALS AND METHODS
Human subjects
The Asthma Severity Modifying Polymorphisms (AsthMaP) Project provided asthmatic patient samples for our study. AsthMaP is a study of gene-environment interactions in inner-city pediatric asthma patients treated at Children’s National Medical Center (CNMC), Washington, DC. The non-asthmatic controls were adolescents selected from an ongoing genetic study at CNMC on metabolic syndrome in inner-city adolescents.
DNA Isolation
Whole blood or buccal swab samples were collected from asthmatic individuals and non-asthmatic controls under an IRB approved protocol. Genomic DNA was isolated by standard protocols.
MUC7 VNTR Polymorphism Genotyping
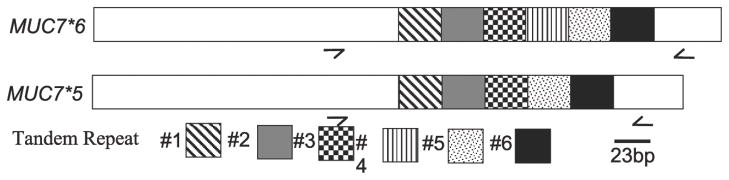
The approach utilized by Rousseau, et al. 17 was used to evaluate the VNTR polymorphisms in the genomic MUC7 gene for each subject. Briefly, PCR amplification of genomic DNA was carried out using primers designed to span the entire VNTR domain. The location of primers is indicated by arrows in Figure 1. The sense primer 5′-cagaatgccaccaccatatcttcaa-3′ and the antisense primer 5′-ggtgcaagagtagttggggaagaat-3′ are located at 400–425 and 959–984 nt, respectively, on the genomic MUC7 DNA in exon 3 (chr 4q13-q21, accession number L13283).
Figure 1.
Schematic of MUC7 cDNA. Each of the tandem repeats, 69 bp in length, are identified. The arrows either side of the TR domain indicated location of primers used for genotyping.
DNA Sequencing
PCR products were electrophoresed on a 2% ethidium bromide agarose gel and visualized on a Chemidoc Imager (BioRad, Hercules, CA). Bands identified for DNA sequencing were excised and extracted using QiaQuick gel extraction kit (Qiagen, Valencia, CA), ligated into pCRII-TOPO (Invitrogen, Carlsbad, CA) and was sequenced (Davis Sequencing, Davis, CA).
SNP Analysis
TaqMan® SNP Genotyping Assays Analysis of the rs998210 SNP was carried out using specifically designed kits (Applied Biosystems, Foster City, CA) on an ABI 7900HT TaqMan machine. The SNP Genotyping Assay targeted to MUC7 determined the C/T transition, located at Chr.4 71380925.
Statistical Analysis
The frequency of each MUC7 polymorphism was imported into a contingency table (Table 2). A chi-square test was then used to statistically evaluate whether there was an association between the MUC7 polymorphisms and asthma. To evaluate the associated risk of an asthma diagnosis and the MUC7 allelic polymorphisms in an African American cohort, logistic regression models were used to generate relative odds ratios and 95% confidence intervals (Stata V10, StataCorp, College Station, TX). These analysis were repeated to determine the association of the MUC7*5 polymorphism and the rs998210 SNP in the same population. Hardy-Weinberg equilibrium was tested for each SNP using a 1 degree of freedom chi square test.
Table 2.
Frequencies of the MUC7 polymorphic alleles.
| Control | Athsma | ||||
|---|---|---|---|---|---|
| Tandem Repeat | *6 | *5 | *4 | *6 | *5 |
| Number of people | 67 | 6 | 1 | 165 | 3 |
| Allelic Frequency | 0.91 | 0.08 | 0.01 | 0.99 | 0.01 |
Due to the limited availability of a control population a post-hoc power analysis was performed to determine the likelihood of finding a significant difference.
RESULTS
Demographics
The sample population was limited by the enrollment of the pediatric population in the AsthMaP study and other non-airway related studies. Our population contained 84 inner-city children in the AsthMaP study and 37 non-asthmatic controls. The age and gender of the cohort are reported in Table 1. Overall, the sample population was 46% male and 57% asthmatic.
Table 1.
Demographic data on the human subject population.
| Total Population | Male | Female | Age range yrs. (Mean) | |
|---|---|---|---|---|
| Control | 37 | 13 | 24 | 14–20 (18) |
| Asthma | 84 | 45 | 39 | 4–18 (10) |
The post-hoc power analysis comparing the TR polymorphism in asthmatics and non-asthmatics, samples sizes of 84 and 37 respectively, showed that we had a 66% power to detect a significant difference (p=<0.05).
Allelic variation in the number of VNTR domains
The two previously reported MUC7 VNTR polymorphisms -- MUC7*6 and MUC7*5 --were identified in our study. They were observed as either homozygous 6/6 (Figure2, Lane 1: 559 bp) or heterozygous 6/5 (Figure2, Lane 2: 490 bp) allelic pairings.
Figure 2.
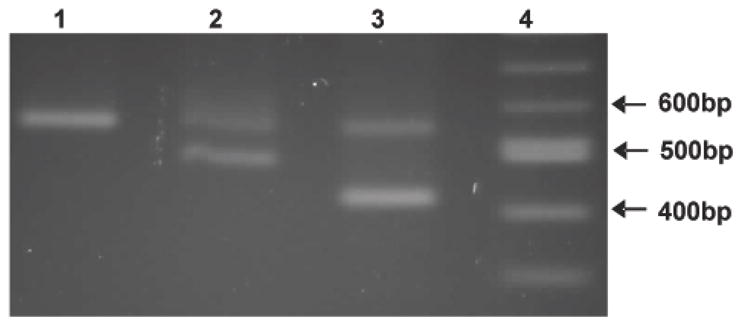
Electrophoresis of representative PCR products showing allelic variation in the MUC7 tandem repeat domain. Amplicons were separated on a 2% agarose gel. [predicted sizes are: 6* -559bp; 5* - 490bp; 4* - 421bp]. Lane 1, 100bp ladder; Lane 2, 6*/4* alleles; Lane 3, 6*/5* alleles; Lane 4, 6*/6* alleles.
We also identified, in a control subject, what looked like a novel polymorphism predicted to encode four VNTR (Figure2, Lane 3: 421 bp), as the amplicon corresponded to 69 bp (size of a single TR) less than MUC7*5. To assess this, DNA from the PCR product (Figure 2) was sequenced and shown to encode 4 VNTR. The first encoded VNTR repeat domain contained two SNPs that altered the genotype and resulted in changes in the amino acid sequence (Figure 3). The SNPs resulted in a P169T and a T176S change, indicated in Figure 3 as TR1Δ2 with highlighted changes. Unlike MUC7*5, MUC7*4 showed a rearrangement of the order of its VNTR domains, as TR1Δ2 was followed by TR2, TR1 and TR2 (Figure 3). This previously unidentified MUC7 VNTR is now designated MUC7*4.
Figure 3.

Schematic of MUC7*4 cDNA/protein showing the changes in the VNTR domain. The order of the repeats is: TR1▲2, TR2, TR1 and TR2. DNA and amino acid sequence of TR1 compared to the altered TR1 (TR1▲2), in the MUC7*4 allelic polymorphism.
Frequency analysis of the MUC7*6 and MUC7*5 allelic polymorphism
DNA samples from non-asthmatic and asthmatic patients (Table 1) were analyzed for the frequency of each polymorphism. The frequencies identified in the asthmatic population were: MUC7*6 allele, 0.99 and MUC7*5 allele, 0.01. In the control population the frequency results were: MUC7*6 allele, 0.91; MUC7*5 allele; 0.08 and MUC7*4 allele, 0.01 (Table 2).
We evaluated the expression of MUC7 allelic polymorphisms and the associated risk of being diagnosed with asthma both in the AsthMaP and control cohorts. A logistic regression analysis of the association of MUC7*5 allelic polymorphism and not being diagnosed with asthma gave an odds ratio (OR) of 0.173 with a confidence interval (CI) of 0.041 – 0.737 and a p-value of 0.018 (Table 3). These data were supported by a chi2 analysis showing a significant association of MUC7*5 (p = 0.01) with not being diagnosed with asthma as a child.
Table 3.
Logistic regression analysis of the MUC7 VNTR polymorphisms and its association with being asthmatic.
| Genotype | Cases | Controls | Odds Ratio | p-Value | 95% CI |
|---|---|---|---|---|---|
| 6/6 | 81 | 28 | 1.0 | ||
| 6/5 | 3 | 6 | 0.173 | 0.018 | 0.041–0.737 |
SNP analysis of rs998210
The previously identified SNP rs9982010 in the second intron of the MUC7 gene, was earlier shown to be in 100% LD with the MUC7*5 allelic polymorphism in a Northern European cohort 17. This SNP was also analyzed in our African American cohort. Our data showed that with the T to C conversion there is a low LD between the C/T SNP and the MUC7*5 allelic polymorphism. These data show that the African Americans population only showed an LD measured by an r2 of 0.03. These findings are not in concordance with the results in the Northern European cohort where a 100% LD with the polymorphic allele is observed. Both the MUC7*5 and the SNP were in Hardy Weinberg equilibrium (MUC7*5 polymorphism, p=0.12; rs998210, p=0.29). A logistic regression analysis performed between the MUC7*5 allelic polymorphism and the rs9982010 SNP showed an odd ratio of 66.0 (p<0.0001), CI (5.913 – 736.72), indicating that the African American individuals with the MUC7*5 allelic polymorphism were 66 times more likely to have the T to C conversion in rs9982010, irrespective of asthma.
DISCUSSION
African Americans have an increased incidence of asthma and are three times more likely to be hospitalized with asthma related symptoms. Additionally, African Americans living in inner-cities are two to six times more likely to die from asthma 7;18. An individual’s overall genotype and environment can predispose one to asthma, but the contributions of genetic background to the risk of being diagnosed with asthma are uncertain.
Mucin overproduction is implicated in asthma (Rose and Voynow, 2006). Thus, an association of MUC genes with asthma was carried out by Swallow and co-workers. The data show no association of VNTR numbers in allelic variants in the MUC1, MUC2, MUC4, MUC5AC, MUC5B genes 14. However, this group identified a polymorphism in the MUC7 gene, MUC7*5, with an allelic frequency of 0.10 associated with a decreased risk of being diagnosed with asthma in a Northern European asthmatic cohort 15. This data suggested that the MUC7*5 allele was protective against asthma and a subsequent longitudinal study supported the concept that the MUC7*5 allele had a protective effect on respiratory function.
Since minority pediatric populations in inner-cities have a higher prevalence of asthma, we predicted that our cohort would have a lower prevalence of the MUC7*5 allele if it were protective. Our data showed an allelic frequency of 0.05, which is lower than that observed (0.1) for the MUC7*5 allele in a Northern European cohort 15. Interestingly, this is supported by a small African cohort (n =29) that also shows a reduction in the MUC7*5 allele frequency = 0.052 15. Though each of the two sample sets -- Northern European and inner-city African-American -- are relatively small, the combined data support the hypothesis of a reduction in the frequency of the protective MUC7*5 allelic polymorphism in a population that is of greater risk of being diagnosed with asthma.
In the Northern European asthmatic cohort the rs998210 intronic SNP in the MUC7 gene has been shown to be in 100% LD with the MUC7*5 17. Our data showed that this SNP is significantly associated with the MUC7*5 polymorphism (p=0.0007) in African Americans, but not at 100% LD. Rousseau et al (2006) suggested that this SNP might not be functional as it does not reside in an identified motif region. These data on MUC7 allelic polymorphisms highlight one example where a small genetic difference between ethnically diverse populations could impact the susceptibility of being diagnosed with asthma, especially in a high risk inner-city population.
While MUC7*6 and MUC7*5 appear to be the two most predominant alleles in the human population, unique MUC7 alleles have been identified, e.g. MUC7*8 in a Northern European with atopic asthma 15. Herein, we identified a novel MUC7*4 polymorphism in an African American non-asthmatic individual that resulted in a reduction in the number and rearrangement of the encoded TR domains. Two SNPs within the first TR resulted in a change in the amino acid sequence of the first encoded TR (TR1Δ2). The VNTR domain of MUC7*4 is TR1Δ2, TR2, TR1, TR2, in contrast to TR1-6 of MUC7*6 and TR1,2,3, 5,6 of MUC7*5.
The role of mucins in the mucosal immune system is only beginning to be understood at the molecular level 19. Mucins are overproduced in acute and chronic airway diseases and contribute to the disease morbidity and mortality. MUC7 is a small secreted mucin glycoprotein (180 kDa) expressed predominantly in the submandibular and sublingual glands 20 and salivary secretions 21;22. MUC7 has been shown to bind to bacteria and small recombinant MUC7 peptides exhibit anti-bacterial, anti-fungal properties 23;24,22, as well as anti-viral properties 25. We have recently shown that MUC7 mucin is present in the airway secretions of asthmatic, but not control, pediatric patients 26, suggesting that MUC7 mucin may have a role in the pathophysiology of asthma. MUC7, like all mucins, is highly O-glycosylated and alterations in the sequence and number of encoded VNTR domains could have a significant impact on its biochemical properties and biological functions and thus its role in diseases. Future studies will be needed to determine mechanisms by which polymorphisms in the MUC7 gene alter the host innate immune response of MUC7 mucin and its relevance to asthma.
Acknowledgments
The authors thank Dr. Anamaris Colberg-Poley and Dr. Joseph Devaney for excellent technical advice and Dr. Eric Hoffman for his insight and careful analysis of this manuscript. This work was supported by the Board of Visitors Grant, Children’s National Medical Center [BLV-08 to AMW], by the National Institutes of Health [HL-33152 to MCR, K23-RR-020069 to RJF], and a pilot grant from the DC-Baltimore Research Center on Child Health Disparities [P20-MD-000198 to RJF]. Alan Watson was a predoctoral student in the Immunology Program of the Institute for Biomedical Sciences at the George Washington University. This work is from a dissertation presented to the above program in partial fulfillment of the requirements for the Ph.D. degree.
Reference List
- 1.Martinez FD. Definition of pediatric asthma and associated risk factors. Pediatr Pulmonol Suppl. 1997;15:9–12. [PubMed] [Google Scholar]
- 2.Adams PF, Hendershot GE, Marano MA. Current estimates from the National Health Survey, 1996. Vital Health Stat. 1999:10. [PubMed] [Google Scholar]
- 3.Wong GW, Chow CM. Childhood asthma epidemiology: insights from comparative studies of rural and urban populations. Pediatr Pulmonol Suppl. 2008;43:107–116. doi: 10.1002/ppul.20755. [DOI] [PubMed] [Google Scholar]
- 4.Eggleston PA. The environment and asthma in US inner cities. Chest. 2007;132:782S–788S. doi: 10.1378/chest.07-1906. [DOI] [PubMed] [Google Scholar]
- 5.Diette GB, Hansel NN, Buckley TJJ, et al. Home indoor pollutant exposures among inner-city children with and without asthma. Environmental Health Perspectives. 2007;115:1665–1669. doi: 10.1289/ehp.10088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eggleston PA. Environmental causes of asthma in inner city children. The National Cooperative Inner City Asthma Study. Clinical Reviews in Allergy & Immunology. 2000;18:311–324. doi: 10.1385/CRIAI:18:3:311. [DOI] [PubMed] [Google Scholar]
- 7.American Lung Association, E. &. S. U. B. P. a. P. S. NIH Web site. 2002. National Center for Health Statistics, National Health Interview Survey, 1999. [Google Scholar]
- 8.Kaliner M, Shelhamer JH, Borson BJ, et al. Human respiratory mucus. Am Rev Respir Dis. 1986;134:612–621. doi: 10.1164/arrd.1986.134.3.612. [DOI] [PubMed] [Google Scholar]
- 9.Blyth DI. The homeostatic role of bronchoconstriction. Respiration. 2001;68:217–223. doi: 10.1159/000050497. [DOI] [PubMed] [Google Scholar]
- 10.Rogers DF. Mucus pathophysiology in COPD: differences to asthma, and pharmacotherapy. Monaldi Arch Chest Dis. 2000;55:324–332. [PubMed] [Google Scholar]
- 11.Holgate ST. The epidemic of allergy and asthma. Nature. 1999;402:B2–B4. doi: 10.1038/35037000. [DOI] [PubMed] [Google Scholar]
- 12.Rose MC, Voynow JA. Respiratory Tract Mucin Genes and Mucin Glycoproteins in Health and Disease. Physiol Rev. 2006;86:245–278. doi: 10.1152/physrev.00010.2005. [DOI] [PubMed] [Google Scholar]
- 13.Vinall LE, Hill AS, Pigny P, et al. Variable number tandem repeat polymorphism of the mucin genes located in the complex on 11p15.5. Hum Genet. 1998;102:357–366. doi: 10.1007/s004390050705. [DOI] [PubMed] [Google Scholar]
- 14.Vinall LE, Fowler JC, Jones AL, Kirkbride HJ, et al. Polymorphism of human mucin genes in chest disease: possible significance of MUC2. Am J Respir Cell Mol Biol. 2000;23:678–686. doi: 10.1165/ajrcmb.23.5.4176. [DOI] [PubMed] [Google Scholar]
- 15.Kirkbride HJ, Bolscher JG, Nazmi K, et al. Genetic polymorphism of MUC7: Allele frequencies and association with asthma. Eur J Hum Gen. 2001;9:347–354. doi: 10.1038/sj.ejhg.5200642. [DOI] [PubMed] [Google Scholar]
- 16.Biesbrock AR, Bobek LA, Levine MJ. MUC7 gene expression and genetic polymorphism. Glycoconjugate J. 1997;14:415–422. doi: 10.1023/a:1018587031814. [DOI] [PubMed] [Google Scholar]
- 17.Rousseau K, Vinall LE, Butterworth SL, et al. MUC7 Haplotype Analysis: Results from a Longitudinal Birth Cohort Support Protective Effect of the MUC7*5 Allele on Respiratory Function. Ann Hum Genet. 2006;70:417–427. doi: 10.1111/j.1469-1809.2006.00250.x. [DOI] [PubMed] [Google Scholar]
- 18.Beckett WS, Belanger K, Gent JF, et al. Asthma among Puerto Rican Hispanics: a multi-ethnic comparison study of risk factors. Am J Respir Crit Care Med. 1996;154:894–899. doi: 10.1164/ajrccm.154.4.8887582. [DOI] [PubMed] [Google Scholar]
- 19.Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008;1:183–197. doi: 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piludu M, Rayment SA, Liu B, et al. Electron microscopic immunogold localization of salivary mucins MG1 and MG2 in human submandibular and sublingual glands. J Histochem Cytochem. 2003;51:69–79. doi: 10.1177/002215540305100109. [DOI] [PubMed] [Google Scholar]
- 21.Prakobphol A, Levin MJ, Tabak LA, et al. Purification of a low-molecular-weight, mucin-type glycoprotein from human submandibular-sublingual saliva. Carbohydr Res. 1982;108:111–122. doi: 10.1016/s0008-6215(00)81896-0. [DOI] [PubMed] [Google Scholar]
- 22.Liu B, Rayment S, Oppenheim FG, et al. Isolation of human salivary mucin MG2 by a novel method and characterization of its interactions with oral bacteria. Archives of Biochem Biophys. 1999;364:286–293. doi: 10.1006/abbi.1999.1141. [DOI] [PubMed] [Google Scholar]
- 23.Situ H, Wei G, Smith CJ, et al. Human salivary MUC7 mucin peptides: effect of size, charge and cysteine residues on antifungal activity. Biochem J. 2003;375:175–182. doi: 10.1042/BJ20030779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei GX, Bobek LA. Human salivary mucin MUC7 12-mer-L and 12-mer-D peptides: antifungal activity in saliva, enhancement of activity with protease inhibitor cocktail or EDTA, and cytotoxicity to human cells. Antimicrobial Agents & Chemotherapy. 2005;49:2336–2342. doi: 10.1128/AAC.49.6.2336-2342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Habte HH, Mall AS, de Beer C, et al. The role of crude human saliva and purified salivary MUC5B and MUC7 mucins in the inhibition of Human Immunodeficiency Virus type 1 in an inhibition assay. Virol J. 2006;3:99. doi: 10.1186/1743-422X-3-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watson A, Troxler RF, Pena, et al. MUC7 mucin glycoprotein is present in airway secretions of asthmatic, but not control, patients. Am J Respir Crit Care Med. 2003;167(A465) [Google Scholar]