Abstract
Although a number of γD-crystallin mutations are associated with cataract formation, there is not a clear understanding of the molecular mechanism(s) that lead to this protein deposition disease. As part of our ongoing studies on crystallins, we investigated the recently discovered Arg76 to Ser (R76S) mutation that is correlated with childhood cataract in an Indian family. We expressed the R76S γD-crystallin protein in E. coli, characterized it by CD, fluorescence, and NMR spectroscopy, and determined its stability with respect to thermal and chemical denaturation. Surprisingly, no significant biochemical or biophysical differences were observed between the wild-type protein and the R76S variant, except a lowered pI (6.8 compared to the wild-type value of 7.4). NMR assessment of the R76S γD-crystallin solution structure, by RDCs, and of its motional properties, by relaxation measurements, also revealed a close resemblance to wild type crystallin. Further, kinetic unfolding/refolding experiments for R76S and wild-type protein showed similar degrees of off-pathway aggregation suppression by αB-crystallin. Overall, our results suggest that neither structural nor stability changes in the protein are responsible for the R76S γD-crystallin variant's association with cataract. However, the change in pI and the associated surface charge or the altered nature of the amino acid could influence interactions with other lens protein species.
Keywords: crystallin mutants, NMR structure, dynamics, cataract
Cataracts, the leading cause of blindness worldwide, are predominantly an affliction of the elderly. Cataracts are associated with a loss of lens transparency, which is related to protein modifications that cause the lens proteins to form insoluble aggregates, resulting in increased light scattering and, hence, loss of vision (1). Protein modifications known to occur with aging include oxidation, predominantly of cysteine and methionine residues (2, 3) but also of tryptophan (4), deamidation of asparagines and glutamines (5–8), glycosylation of lysine residues (9), as well as cleavage of peptide bonds (10–12). While age-related cataracts occur only in adults, inherited cataracts manifest in early childhood (13). At the present time, at least 60 cases of inherited cataracts have been documented; these appear to be caused by a single-gene disorder, and the amino acid substitutions have been mapped (14). Non-syndromic childhood congenital cataracts occur at an estimated frequency of 1–6 per 10000 live births, with at least one third of childhood cataracts having a familial basis (15).
The major pre-requisite for lens transparency and a high refractive index is a quasi-ordered arrangement and distribution of proteins in the lens fiber cells. Crystallins, α, β and γ-types, make up over 90% of eye lens proteins (16) and are thought to be responsible for maintaining the favorable optical properties of the lens (17). β and γ-crystallins are dimeric or monomeric β-sheet proteins, while α-crystallins are multimeric assemblies that function as chaperones (18). Human γD-crystallin (HGD) comprises two structurally homologous domains, with each domain containing two Greek key motifs. Over the last decade, single amino acid changes in crystallins have been extensively studied, with a goal of elucidating mechanisms that lead to cataracts. For example, the R14C mutation causes disulfide cross-linking that results in precipitation of oligomeric species in solution (19, 20). Two other mutants, R58H and R36S, spontaneously crystallized under physiological condition (19, 21). The P23T mutant is extremely insoluble compared to wild-type HGD (WT-HGD) and forms high molecular weight aggregates at physiological concentration (22–24). Interestingly, for all the above mutant proteins, no significant changes have been observed in their secondary and tertiary structures. Smaller structural differences, however, can be detected. For example, in the P23T mutant protein, the imidazole ring of H22 exists predominantly as the Nδ1 tautomer instead of the Nε2 tautomer that is found in the wild-type protein, and this local difference may initiate protein-protein contacts that serve as nuclei for aggregation (25).
Several approaches have been developed to describe and explain the physical basis of lens transparency. One of these involves characterizing the phase behavior of the lens protein solution, interpreted as microscopic and collective protein-protein interactions that may also apply to their behavior in the cataractous lens. For example, several mutations (i.e. R14C, P23T, R36S, R58H) specifically change the homotypic γD-γD interactions and form distinct condensed phases with increased light scattering (26). Based on small angle neutron scattering and molecular dynamics simulation of heterotypic α-γ mixtures, weak attractions between different proteins have been postulated to help maintain lens transparency (27). For example, the E107A mutant exhibited no difference in biophysical properties, compared to WT-HGD, although the interaction between γ and α-crystallin appeared to be altered, as mixtures containing both E107A and α-crystallin consistently exhibited more pronounced light-scattering compared to mixtures of WT-HGD and α-crystallin (28). In addition, in γD and βB1 mixtures, the energy of the γD-βB1 interaction is significantly smaller than that of γD-γD, such that increasing the amount of βB1 in solution lowers the phase separation temperature (29). Indeed, all data available at present support the notion that heterotypic interactions between the different types of crystallins are delicately balanced in the eye lens, such that even slight strengthening or weakening of such interactions may lead to instability in mixtures of these proteins (30), with α-crystallin, by virtue of its chaperone properties, playing a pivotal role in preventing the aggregation and/or precipitation of other lens crystallins.
Here, we present data on another congenital cataract-associated mutation in human γD-crystallin, as part of our ongoing program to elucidate the structural basis of cataract formation involving γD-crystallin. This mutant was identified in an Indian family, with the affected individuals presenting with juvenile autosomal dominant anterior polar and coronary cataracts (18). The mutant is a single A/C change at position 229 in the CRYGD gene, leading to an R76S change in the protein. At present, no information is available as to how the mutation causes cataract.
We carried out biochemical, biophysical, and structural analyses of R76S γD-crystallin (R76S). The overall protein properties were assessed by CD and fluorescence spectroscopy; thermodynamic stability was explored using thermal and chemical denaturation experiments; and details of the three-dimensional structure were investigated using residual dipolar couplings (RDCs). The protein's dynamic properties were probed by 15N NMR relaxation. Kinetic unfolding/refolding experiments were performed and the influence of α-crystallin was investigated. All combined data revealed no substantial differences from WT-HGD in the properties of R76S, except for a change in surface charge. Thus, interactions with other proteins or factors in the lens may be the cause for R76S's association with cataract.
EXPERIMENTAL PROCEDURES
Cloning, Expression and Purification
Wild-type and mutant genes were inserted into pET14b vectors. E. coli BL21 (DE3) cells (EMD Chemicals) were transformed with these vectors. For protein production, cells were grown at 37 °C to an absorbance at 600 nm of 0.6 and induced by the addition of 0.5 mM IPTG at 18 °C for 16 h. For 15N and 13C labeling, cells were grown in modified minimal medium containing 1g/L 15NH4Cl and/or 2 g/L [13C] glucose as the sole nitrogen and/or carbon sources, respectively (25).
Proteins were purified essentially as described previously (25). Cells were lysed by passage through a microfluidizer (Microfluidics, Newton, MA), and cell debris was removed by centrifugation at 100,000g for 1 h at 4 °C. Clarified cell lysate was loaded onto a HiTrap Q XL anion exchange column (GE Healthcare), equilibrated with 50 mM Tris buffer, pH 8.0, 1mM EDTA, 1mM DTT. The flow-though fraction was collected and dialyzed overnight against 10 mM MES buffer, pH 6.2, 1 mM DTT, 2% glycerol. The dialyzed sample was loaded on a HiTrap SP cation exchange column (GE Healthcare) and eluted using a linear NaCl gradient from 0 to 1 M over a 20-column volume. Fractions containing HGD were subjected to gel filtration on a Superdex 75 26/60 column (GE Healthcare) in 10 mM MES buffer, pH 6.2, with 5 mM DTT, for final purification.
Protein concentration was determined spectrophotometrically using an extinction coefficient of 42.86 mM−1 cm−1 at 280 nm. Protein purity and identity was assessed by matrix-assisted laser desorption/ionization mass spectrometry on a Voyager-DE PRO instrument, operated in a linear mode with external calibration. WT-HGD and R76S exhibited experimental molecular masses of 20592.1 Da and 20527.5 Da, respectively (the theoretical molecular masses are 20606.94 Da and 20537.83 Da, respectively).
Circular Dichroism Spectroscopy
CD spectra were recorded at 37 °C using a Jasco J-810 spectropolarimeter. The protein concentration was 100 μg/ml in 10 mM sodium phosphate buffer, pH 7.0. Far-UV CD spectra were collected from 195 to 260 nm in a 1-cm path length quartz cuvette. The buffer baseline signal was subtracted from all spectra.
Fluorescence Emission Spectroscopy
Fluorescence spectra of WT-HGD and R76S were recorded at 37 °C using a Cary Eclipse fluorescence spectrometer. Protein concentrations were 0.5 μM (10 μg/ml) in 100 mM sodium phosphate buffer, pH 7.0, 5mM DTT, 1mM EDTA. An excitation wavelength of 295 nm was used and emission spectra were recorded over a range of wavelengths from 305 to 400 nm with slit widths of 5nm for excitation and 10 nm for emission. All spectra were corrected for the buffer background.
Thermal and Chemical Denaturation
Samples for denaturation contained 40 μg/mL protein in 10mM sodium phosphate buffer, pH 7.0 and were placed in an air-tight, 1 cm path length, quartz cuvette. Thermal denaturation was followed by CD spectroscopy, monitoring the molar ellipticity at 218 nm in 1°C intervals over the temperature range of 25 to 95°C. Samples were equilibrated at each temperature for 1 min prior to the measurements and each spectrum was an average of three consecutive scans.
Chemical unfolding by guanidinium hydrochloride (GdnHCl) was monitored by tryptophan fluorescence. Samples contained 10 μg/mL protein in 100 mM sodium phosphate buffer, pH 7.0, 5 mM DTT, 1 mM EDTA and 0–5.5 M GdnHCl. Each sample was equilibrated at 37 °C for 24 h prior to the measurements. Spectra were corrected for background fluorescence from GdnHCl and unfolding was monitored by using the ratio of the fluorescence intensities at 360 nm and 320 nm.
Determination of Isoelectric Points
WT-HGD and R76S proteins were subjected to electrophoresis on PhastGelTM IEF gels (pH range, 3–9; GE Healthcare) using the AmershamTM Isoelectric Focusing Calibration Kit (High Range pI, pH 5–10.5) for pI estimation. Protein bands were detected with Coomassie Brilliant Blue R-250 staining.
Kinetics of Unfolding/Refolding
The time course of unfolding in 5.5 M GdnHCl, 100 mM sodium phosphate buffer, pH 7.0, 5 mM DTT, 1 mM EDTA was followed by fluorescence emission at 350 nm using an excitation wavelength of 295 nm on a Hitachi F-4500 fluorometer. Protein at 100 μg/mL was injected into the cuvette yielding a final protein concentration of 10 μg/mL at 37 °C, with constant stirring. Fluorescence was monitored until no further changes were observed. Each unfolding experiment was carried out in triplicate. Refolding experiments were performed in an analogous fashion. 100 μg/mL of protein that had been incubated in 5.5 M GdnHCl, 100 mM sodium phosphate buffer, pH 7.0, 5 mM DTT, 1 mM EDTA at 37 °C for 5 h, was injected into 0.5 M GdnHCl in the same buffer. The final GdnHCl concentration was 1.0 M.
Turbidity Measurements
Protein samples at a concentration of 0.5 mg/mL were unfolded in 5 M GdnHCl, 100 mM sodium phosphate buffer, pH 7.0, 5 mM DTT, 1 mM EDTA overnight at 37 °C. Aggregation was initiated by diluting the GdnHCl to 0.5 M with refolding buffer, yielding a final protein concentration of 50 μg/mL. Immediately after the dilution step the samples were thoroughly mixed and turbidity was measured at 37 °C by absorbance at 350nm using a temperature-controlled Cary 50 UV/Vis spectrophotometer (Varian Inc.).
For the aggregation suppression assays, the refolding buffer also contained αB-crystallin at 250 μg/mL. The final concentration of WT-HGD and R76S were 50 μg/mL. Turbidity of the solutions was measured as described above.
NMR Spectroscopy
All NMR spectra for resonance assignments and backbone 15N relaxation data collection were acquired at 25 °C, using either 15N-labeled or 13C,15N-labeled samples on Bruker AVANCE 800, AVANCE 700, or AVANCE 600 spectrometers, equipped with 5 mm triple-resonance, three-axis gradient probes or a z-axis gradient cryoprobe. Temperature calibration was performed using the chemical shift differences between methyl and hydroxyl protons of 100% methanol. All samples contained protein at a concentration of ~1 mM in NMR buffer (10mM MES buffer, pH 6.2, 0.02% NaN3, 5mM DTT, 5% (v/v) D2O). HNCA, HNCACB and CBCA(CO)NH experiments were recorded for backbone, Cβ, and Hβ atom resonance assignments (31, 32). Residual 1J 1H-15N dipolar couplings were measured on protein samples in C12E5/hexanol mixtures (5% w/v, molar ratio 0.96) (33) as the alignment medium, using in-phase/anti-phase 2D 1H,15N-HSQC spectroscopy (34). Samples were kept in the magnet prior to RDC measurements for at least two hours to ensure complete alignment. Measured RDC values were compared with predicted values based on the crystal structure of the WT-HGD (PDB: 1hk0). The alignment tensor was determined using single value decomposition for calculating the Saupe order matrix with the program PALES (35). Backbone 15N T1, T2 relaxation data were recorded at 25°C on 15N-labeled 1mM protein samples in NMR buffer at 600 MHz. Relaxation delays in the T1 experiments were 20, 50, 100, 300, 700, 900, 1200, 1500, 2000 and 2500 ms. The experiments with 20 ms and 300 ms delays were repeated twice for error estimation. CPMG mixing times in the T2 experiments were 15.84, 31.68, 47.52, 63.36, 79.19, 95.04, 110.88, 126.72, 142.56 and 158.39 ms using 1024*128 complex points. The experiments with 31.68 ms and 47.52 ms mixing times were recorded twice for error estimation. Repetition delays in both experiments were 2 s. Peak intensities were fitted using the CURVEFIT program (A.G. Palmer III, Columbia University). All spectra were processed with NMRPipe (36) and analyzed using CARA (37) or SPARKY3 (version 3.113) (38). Amide 1H,15N- combined chemical shift differences were calculated using Δδ=√((0.15*ΔδN)2+ΔδH2).
RESULTS
Protein Solubility
Since previous work established that some cataract-associated mutations result in dramatically lower solubility of the mutant proteins (23, 25, 39), we evaluated the solubility of the R76S variant and compared it to that of WT-HGD. The initial sample was concentrated at 4°C and subsequently kept at 25 °C. Even for the highest concentration tested (320 mg/mL), no differences were noted between the mutant and wild-type protein, suggesting that the R76S mutant is extremely soluble and that reduced solubility cannot play a role in cataract formation.
Conformational properties
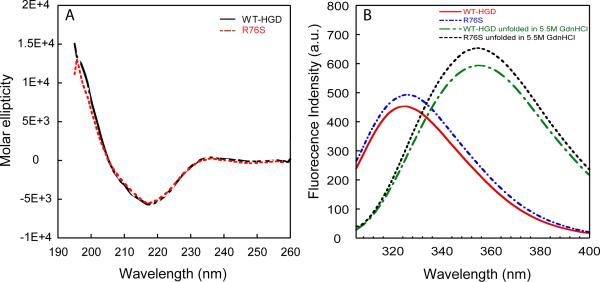
The overall structure of the R76S protein was assessed by CD and fluorescence spectroscopy. As evidenced by the data displayed in Figure 1, R76S and WT-HGD exhibit identical far-UV CD and fluorescence spectra. The CD spectra show distinct minima at 218 nm, indicative of the proteins' high β-sheet content, and a small shoulder at 208 nm (40). The fluorescence spectrum of native WT-HGD exhibits an emission maximum at 326 nm that shifts to 353 nm after denaturation in 5.5 M GdnHCl. The same behavior is seen for the mutant protein. Since there are no changes in tryptophan or tyrosine content (4 Trp, 14 Tyr), it is clear from this data that no changes in the secondary structure and overall conformation result from the R76S mutation.
Figure 1.
Far-UV CD and fluorescence spectra of R76S and WT-HGD. (A) Far-UV CD spectra of R76S (dashed red line) and WT-HGD (solid black line). All samples contained 100 μg/mL protein in 10mM sodium phosphate buffer, pH 7.0 at 37 °C. (B) Fluorescence spectra of native WT-HGD (red solid line) and R76S mutant (blue dashed line) at pH7.0 and denatured WT-HGD (green dashed line) and R76S mutant (black short dotted line) in 5.5 M GdnHCl. Samples contained 10 μg/mL protein in 100 mM sodium phosphate buffer, pH 7.0, 5 mM DTT, 1mM EDTA at 37 °C.
Thermodynamic Stability
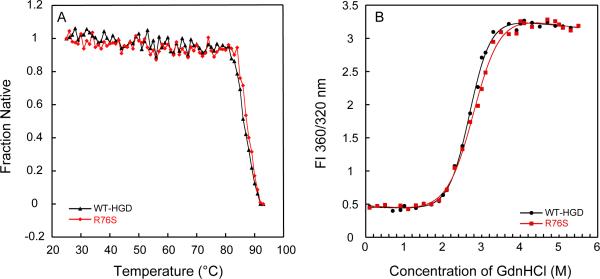
The thermal stability of R76S was investigated by monitoring the ellipticity at 218 nm over the temperature range from 25 to 95°C. The data in Figure 2A suggest that the transitions are two state, without any major intermediates. The melting temperatures (Tm) for both proteins (midpoint of the transition) were ~85.5 °C and ~86 °C, indistinguishable within the error of the measurement. Figure 2B displays GdnHCl unfolding curves for WT-HGD and R76S, monitored by tryptophan fluorescence at 37 °C. Again, no differences between WT-HGD and R76S are present. The data were fit using a two-state model and, for R76S, yielded an unfolding midpoint of 2.9 ± 0.1 M GdnHCl and an apparent of 5.0 ± 0.4 kcal/mol. For WT-HGD, the unfolding midpoint was 2.8 ± 0.1 M GdnHCl, and the apparent was 6.0 ± 0.3 kcal/mol. These values are very similar to those previously calculated for WT-HGD (41). These data clearly indicate that the thermodynamic stability of the mutant protein is unchanged from wild-type crystallin.
Figure 2.
Thermal and chemical unfolding of R76S and WT-HGD. (A) Thermal denaturation of R76S (red) and WT-HGD (black) monitored by CD spectroscopy. All samples contained 40 μg/mL purified protein in 10 mM sodium phosphate buffer, pH 7.0. (B) Chemical unfolding of R76S (red) and WT-HGD (black). All samples contained 10 μg/mL protein in 100mM sodium phosphate buffer, pH 7.0, 5 mM DTT, 1mM EDTA and GdnHCl from 0 to 5.5 M at 37 °C.
Net Surface Charge
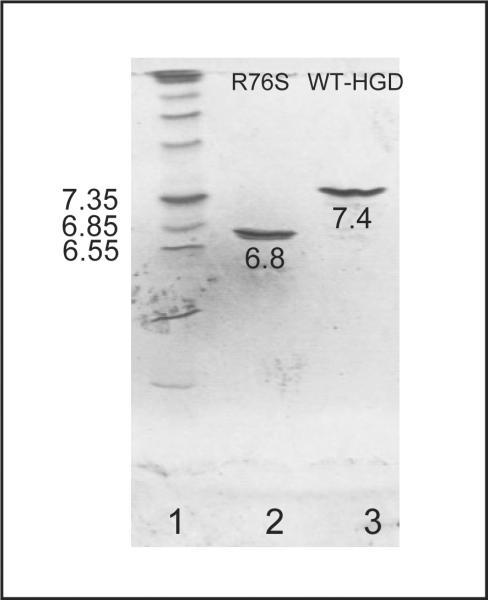
Since the change from a positive arginine side chain to a polar, but neutral serine is expected to alter the pI of the mutant, we determined experimental pI values by isoelectric-focusing (Figure 3). For WT-HGD and R76S, pI values of 7.4 ± 0.1 and 6.8 ± 0.1, respectively, were measured. The former is in excellent agreement with a previous report (28) and the decrease by ~0.6 units for R76S is in accord with the loss of one positive charge on the surface of the protein.
Figure 3.
Isoelectric-focusing of R76S and WT-HGD. Marker proteins with pI values ranging from 5 to 10.5 are shown in lane 1, lanes 2 and 3 contain R76S and WT-HGD, respectively.
Kinetics of Unfolding/Refolding
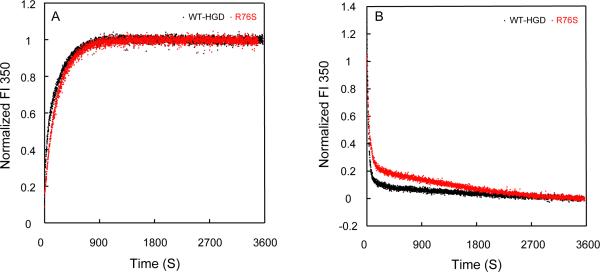
Having found no differences in equilibrium parameters when comparing WT-HGD and R76S, we next examined whether any differences may be present in the kinetics of un/folding. As can be appreciated from the kinetic traces displayed in Figure 4, no substantive differences are apparent. This makes it unlikely that the amino acid substitution is acting through changing the properties and/or presence of distinct intermediates in the un/folding pathway.
Figure 4.
Kinetic unfolding/refolding experiments of R76S and WT-HGD. (A) Kinetic unfolding for R76S (red) and WT-HGD (black) at 37 °C. All samples contained 10 μg/mL protein in 100 mM sodium phosphate buffer, pH 7.0, 5 mM DTT, 1 mM EDTA. Unfolding was initiated by injecting folded native protein into a solution containing 5.5 M GdnHCl. (B) Kinetic refolding for R76S (red) and WT-HGD (black). All samples contained 10 μg/mL protein in 100 mM sodium phosphate buffer, pH 7.0, 5 mM DTT, 1mM EDTA at 37 °C. Refolding was initiated by injecting unfolded protein in 5.5 M GdnHCl into a solution containing 1.0 M GdnHCl. Fluorescence was monitored at 350 nm and the intensity was normalized with respect to native and unfolded protein controls.
Suppression of Aggregate formation by the Chaperone αB-crystallin
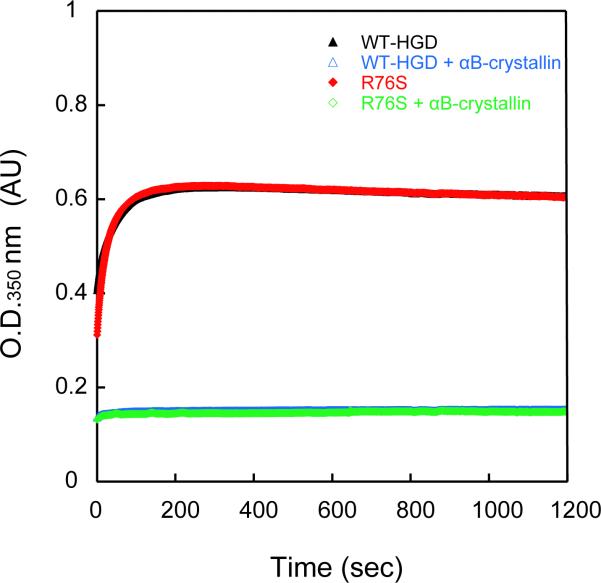
It is generally accepted that α-crystallins act as chaperones to keep γ– and β–crystallins soluble in the highly concentrated protein environment of the eye lens. We, therefore, investigated whether any differences in heterotypic interactions between HGD and the αB-crystallin chaperone could be detected for R76S compared to WT-HGD. The influence of αB-crystallin on un/folding was tested in an aggregation assay (42). Aggregation during refolding of denatured WT-HGD and R76S at 37 °C was used to monitor αB-crystallin chaperone activity. HGD aggregation reached a plateau after 3 minutes in the absence of αB-crystallin (Figure 5). Addition of the chaperone reduced protein aggregation, and, for a molar ratio of 5:1 of αB-crystallin to HGD, the light-scattering intensity after 2 minutes was decreased to 25%, for both WT-HGD and the R76S. The almost perfect superposition of the kinetic traces displayed in Figure 5 clearly demonstrates that the αB-crystallin chaperone has essentially identical effects on WT-HGD and R76S refolding, suppressing aggregation completely.
Figure 5.
Refolding of R76S and WT-HGD in the presence of the αB-crystallin chaperone. R76S and WT-HGD were unfolded in 5.0 M GdnHCl for 24 h at 37 °C. Refolding was initiated by dilution to 0.5M GdnHCl at 37 °C. The final HGD protein concentration was 50 μg/mL in all samples and the final αB-crystallin protein concentration was 250 μg/mL.
Structural Differences between WT-HGD and R76S
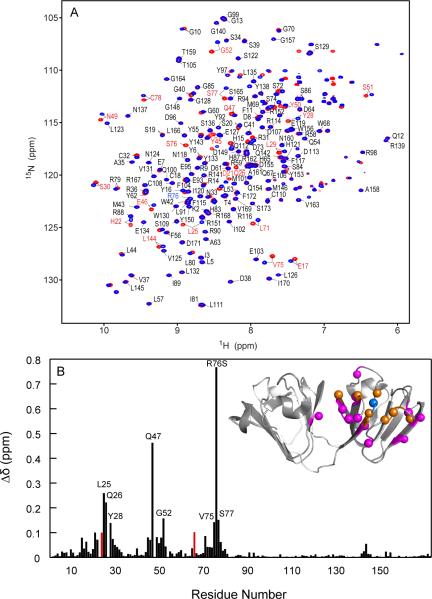
To evaluate whether a protein undergoes small or significant structural changes upon mutation, 1H-15N HSQC spectroscopy is a useful tool and can be used as a “fingerprint” of the three-dimensional structure (43). As is evident from the superposition of the two 1H-15N HSQC spectra provided in Figure 6, most resonances, especially for residues in the C-terminal domain, superimpose extremely well, suggesting a similar structure for the mutant. The overall appearance of the spectrum, well dispersed and sharp cross-peaks, also confirms that no significant unfolding or destabilization of the protein conformation is induced by the mutation, in agreement with the data discussed above. In the R76S mutant spectrum, 162 amide resonances were assigned (of 168 possible). Assignments for N24 and Q66 could not be obtained due to extreme broadening of the resonances in agreement with the R2 data (see below). Most chemical shift differences pertain to residues in the N-terminal domain, with the largest differences observed for resonances of amino acids in the vicinity of position 76, in particular L71–S77. In addition, Q47 and L25 also exhibit substantial differences in shifts, with Δδ values of 0.46 and 0.26 ppm, respectively. Overall, 18 resonances of residues located in the N-terminal domain exhibited chemical shift differences larger than 0.05 ppm. In contrast, resonances associated with residues in the C-terminal domain exhibit average chemical shift differences of ±0.006 ppm, with differences <0.02 ppm, except L144 and Y143 for which Δδ values of 0.054 and 0.024 ppm, respectively, were observed. Two of the affected areas in the N-terminal domain are close to the site of mutation, centered around L25 and Q47, respectively. The Q47 amide proton exhibited an exceptionally large perturbation. In the WT-HGD crystal structure, the backbone carbonyl atom of E46 is located only 2.8 Å away from the R76 Nε atom, suggesting that replacement of the arginine side chain with that of serine removes the interaction between the R76 guanidinium group and the E46 carboxyl group. R76 is also located at the start of the last β-strand of the N-terminal domain and participates in a hydrogen bonding network of the Greek-key motif. Therefore, the effect of the R76S mutation could propagate to other parts of the polypeptide chain via perturbations in this hydrogen bond network.
Figure 6.
Superposition of the 1H-15N HSQC spectra and chemical shift differences between R76S and WT-HGD. (A) 1H-15N HSQC spectra of ~1 mM R76S (red contours) and WT-HGD (blue contours) at 25 °C. 162/168 amide resonances were assigned and are labeled by amino acid name and number. Residues with 1H,15N chemical shift differences > 0.05 ppm compared to WT-HGD are labeled in red. The amide resonance of R76 in WT-HGD is labeled in blue. (B) Combined amide 1H,15N chemical shift differences versus residue number. Unassigned resonances are shown with an arbitrary value of 0.1 ppm. The inset depicts the backbone structure of WT-HGD (1HK0) onto which the chemical differences are mapped. The location of R76 is marked by a blue sphere. Positions of residues whose amide resonances exhibit Δδ > 0.1 ppm and 0.1 ppm > Δδ > 0.05 ppm are shown with orange and magenta spheres, respectively.
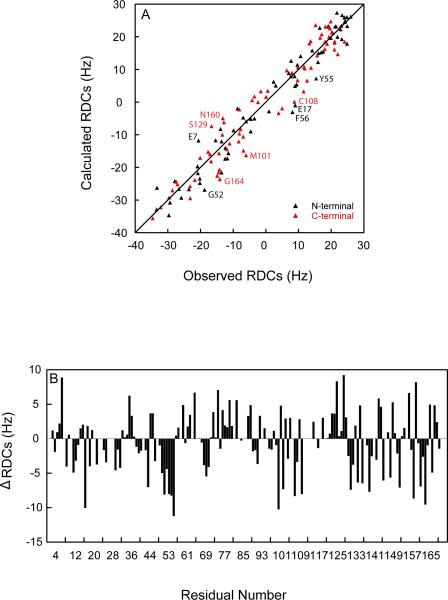
Although chemical shifts can provide a qualitative assessment of where and why potential conformational changes are found, residual dipolar couplings (RDCs) allow for a quantitative evaluation of differences. We measured 137 1H-15N RDCs and compared the experimental values with predicted values based on the structure of WT-HGD. The correlation between observed and calculated RDC values is provided in Figure 7A. No significant outliers are noted, and excellent agreement between measured and predicted RDC values exists, with average r.m.s. deviations of 4.57 Hz, a Q-factor of 0.19 and a correlation coefficient of 0.97. Residues that exhibited differences >8 Hz are labeled and most of them are located on the surface of protein and engaged in crystal contacts. Thus, they reflect differences between the solution and crystal states and not necessarily differences in structure between WT-HGD and the R76S mutant. For some residues, the RDC data suggest that moderate structural changes are present in regions around Q47 and R76, residues that also exhibited chemical shift perturbation, since some clustered and systematic difference between observed and calculated RDC value were seen (Figure 7B).
Figure 7.
Comparison between observed and calculated 1H,15N RDCs. (A) Calculated RDCs were predicted based on the crystal structure of HGD (PDB: 1hk0) and experimental values were measured in 5% C12E5/hexanol (r=0.96) at 25°C. Red and black data points relate to resonances of residues in the N-terminal (3–81) and C-terminal (89–171) domains, respectively. (B) Difference between observed and calculated RDC values are plotted versus residue number.
To assess whether the mutation affected the domain-domain orientation in the structure, RDCs of each domain were back-calculated from the Saupe matrix obtained from the other domain. Q-factors of 0.20 and 0.23 were obtained for the N-terminal and C-terminal domains, respectively. No significant differences between two Saupe frames were seen. Therefore, the mutation does not cause any significant change in the relative domain orientation.
Dynamics of WT-HGD and R76S
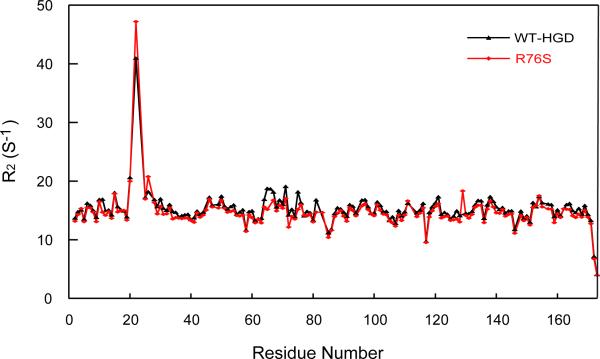
Even if no significant structural differences exist between WT-HGD and R76S, the motional properties of the protein may be affected by the mutation. We measured R1, R2, and steady-state heteronuclear {1H}-15N NOE data to evaluate whether the observed chemical shift changes are associated with local unfolding. Figure 6 summarizes the experimental values versus the residue number. For WT-HGD and R76S, average R2 values of 14.62 ± 0.29 s−1 and 15.23 ± 0.28 s−1 were obtained. Only for H22 in the R76S mutant a slightly larger R value of 47.19 s−1 compared to 40.98 s−1 for WT-HGD was measured, indicating a slight local increase in motion for the amide group. Overall, however, very similar dynamics throughout both proteins prevail.
We also investigated whether any possible protein-protein association of the R76S mutant protein was manifest in the T1 and T2 15N relaxation data. The rotational correlation time τc for each residue was calculated, and, after discarding the top and bottom 10%, an average τc of 11.61 ± 1.61 ns was obtained. Based on the Stokes-Einstein relationship, this τc value translates to a molecular mass of 19.35 kDa, indicating that the R76S mutant protein is monomeric, with no indication of multimerization.
DISCUSSION
Characterization of proteins isolated from cataractous lenses suggested that increased light scattering is associated with accumulation of high molecular mass aggregates lens crystallins. In the lens fiber cell where protein catabolism and novel synthesis are limited, protein depositions can cause serious problems. Given that the R76S mutation was found to be associated with juvenile cataracts, it is important to consider how any mutation could cause the cataract. First, the mutation could destroy the protein's native, 3D fold, resulting in aggregation of the unfolded polypeptide. This scenario appears to be the case for W158X mutations of the CRYGD gene. Second, a mutation can partially and locally destabilize the protein, thereby increasing the population of a less soluble, partially unfolded species, which deposits gradually. This may be the case for the V75D mutant (44). Third, a mutation may have no effect on protein folding or stability, but simply decreases the folded protein's solubility. R14C, P23T, R36S, and R58H mutants appear to belong to this class.
We thoroughly investigated the biochemical and biophysical properties of the cataract associated R76S variant in parallel with WT-HGD protein in order to derive a classification for this mutant. Intriguingly, none of the above scenarios appears to apply to the R76S. The R76S protein is as soluble as WT-HGD (>300 mg/ml). In addition, no differences in thermodynamic stability were noted, with no change in melting temperature or other thermodynamic parameters (unfolding transition midpoint of ~2.8 M GdnHCl). Likewise, no significant structural differences between WT-HGD and the mutant could be detected, using a variety of spectroscopic methods, including NMR.
Even the heterotypic interaction with αB-crystallin did not reveal any differences, and mixtures of R76S with WT-HGD at high concentration, also did not result in aggregation (data not shown).
The only difference found between HGD and the R76S mutant was their isoelectric points. However, the isoelectric point reflects a macroscopic property and does not inform about changes at the atomic level. In order to evaluate whether any significant changes in surface charge properties may have been caused by the mutation, we calculated the electrostatic potential for both WT-HGD and the R76S protein, using APBS (Adaptive Poisson-Boltzmann Solver) (45). Only a very localized effect was noticed and this change did not result in an overall significant difference, such as creating a larger area or patch of one kind of charge, positive or negative, and therefore cannot explain the difference associating with cataract formation.
These findings indicate that the proposed relationship between the R76S mutation and the occurrence of cataract, is not simply a property of the mutant protein itself, but most likely depends on interactions with other lens molecules. Since the replacement of arginine by serine changes the pI of the protein, but the net γD–γD interactions are essentially unaltered in R76S relative to HGD, interactions with other proteins in the lens could play a role. Alternatively, the different nature of the amino acid side chain could cause a loss of contact with other lens components. The direct measurement of aggregation suppression by the chaperone αB-crystallin argues against αB-crystallin being involved in this interaction.
In addition to the chaperones, the lens contains high concentrations of structurally homologous crystallins, including γC- and γS-crystallins that belong to the β-subfamily. The diversity of these closely related β-sheet proteins may provide a means to suppress crystallization or aggregation of the very concentrated crystallins. Indeed, Pande et al. (2001) showed that the replacement of arginine 36 by serine removed a kinetic barrier to nucleation of crystallization (46).
We, therefore, hypothesize that R76S may cause cataract through interactions with other structural crystallins. In human lens cells, all crystallins pack into a liquid crystal-like material, exhibiting strong short-range order, thereby suppressing fluctuations in refractive index and reducing light scattering (47). This ordering seems maintained by heterotypic interactions of α-, β-, γ- crystallins, and it could be affected by even small changes on any of the proteins' surfaces. Juvenile onset cataracts have also been associated with mutations in genes encoding other lens proteins, such as the connexins (48, 49), and very little is known about the interplay between the soluble crystallins of the fiber cell cytoplasm and these cell envelope proteins.
Since most models of cataract focus on mature onset forms in which the crystallin proteins have aged and been damaged by oxidation or other chemical modifications, these models may not be appropriate for juvenile onset cataracts, for which a different mechanism could apply. In that regard, it is interesting to speculate that the defect could reside in the initial folding of the polypeptide chain after synthesis on the ribosome. Recent evidence indicates that a group II chaperonin recognizes and aids HGD folding. In particular, Mn-Cpn, a homologue of the group II chaperonin from methanogenic archaea M. maripalidus can suppress aggregation during refolding and can refold HGD actively in vitro (50). Thus, the R76S substitutions could cause a defect in productive folding of the newly synthesized chain that is not corrected by interaction with the human CCT chaperonin in the lens fiber cells.
Another possibility could be a mutant-only protein modification. A recent report indicated that crystallins can be phosphorylated, with S74 of HGD as one of the dominant phosphorylation sites. Given the close proximity of residue R76 to this phosphorylation site, S76, in the mutant, could also become phosphorylated, causing unexpected changes in the protein. Further studies are necessary to address these possibilities and aid in deciphering the structural basis of the R76S mutant crystallin pathogenesis.
Figure 8.
Backbone 15N R2 transverse relaxation rates for R76S (red) and WT-HGD (black). All NMR samples contained 1 mM protein in 10 mM MES buffer, pH 6.2, 5 mM DTT. All data were collected at 600 MHz and 25 °C.
ACKNOWLEDGEMENTS
F.J. would like to thank her advisor Prof. Yongming Bao at Dalian University of Technology for training and support. We are indebted to Prof. Jonathan A. King for invaluable discussions and guidance on our crystallin work and for hosting F.J. in his laboratory for conducting the kinetics folding/unfolding experiments. We thank Fanrong Kong for help in performing the kinetics experiments, and Mike Delk for NMR technical support. This work was supported by the scholarship from China Scholarship Council (F.J) and a National Institutes of Health Grant EY021193 (A.M.G).
ABBREVIATIONS
- HGD
human γD-crystallin
- WT-HGD
wild-type human γD-crystallin
- CD
circular dichroism
- DTT
dithiothreitol [(2S, 3S)-1,4-bissulfanylbutane-2, 3-diol]
- EDTA
2-_2-[bis(carboxymethyl)-amino]ethyl(carboxymethyl)amino}acetic acid
- IPTG
isopropyl τ-D-1-thiogalactopyranoside
- MES
2-(N-morpholino)ethanesulfonic acid
- GdnHCl
guanidinium hydrochloride
- HSQC
heteronuclear single quantum correlation
- RDCs
residual dipolar couplings
- IPAP
in-phase anti-phase
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- CCT
cytosolic chaperonin-containing t-complex polypeptide 1
- C12E5
2-[2-[2-[2-(2-dodecoxyethoxy)ethoxy]ethoxy]ethoxy]ethanol
REFERENCES
- 1.Sharma KK, Santhoshkumar P. Lens aging: effects of crystallins. Biochim Biophys Acta. 2009;1790:1095–1108. doi: 10.1016/j.bbagen.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Truscott RJ, Augusteyn RC. Oxidative changes in human lens proteins during senile nuclear cataract formation. Biochim Biophys Acta. 1977;492:43–52. doi: 10.1016/0005-2795(77)90212-4. [DOI] [PubMed] [Google Scholar]
- 3.Truscott RJ, Augusteyn RC. The state of sulphydryl groups in normal and cataractous human lenses. Exp Eye Res. 1977;25:139–148. doi: 10.1016/0014-4835(77)90126-9. [DOI] [PubMed] [Google Scholar]
- 4.Hains PG, Truscott RJ. Post-translational modifications in the nuclear region of young, aged, and cataract human lenses. J Proteome Res. 2007;6:3935–3943. doi: 10.1021/pr070138h. [DOI] [PubMed] [Google Scholar]
- 5.Ma Z, Hanson SR, Lampi KJ, David LL, Smith DL, Smith JB. Age-related changes in human lens crystallins identified by HPLC and mass spectrometry. Exp Eye Res. 1998;67:21–30. doi: 10.1006/exer.1998.0482. [DOI] [PubMed] [Google Scholar]
- 6.Lampi KJ, Ma Z, Hanson SR, Azuma M, Shih M, Shearer TR, Smith DL, Smith JB, David LL. Age-related changes in human lens crystallins identified by two-dimensional electrophoresis and mass spectrometry. Exp Eye Res. 1998;67:31–43. doi: 10.1006/exer.1998.0481. [DOI] [PubMed] [Google Scholar]
- 7.Hanson SR, Smith DL, Smith JB. Deamidation and disulfide bonding in human lens gamma-crystallins. Exp Eye Res. 1998;67:301–312. doi: 10.1006/exer.1998.0530. [DOI] [PubMed] [Google Scholar]
- 8.Hanson SR, Hasan A, Smith DL, Smith JB. The major in vivo modifications of the human water-insoluble lens crystallins are disulfide bonds, deamidation, methionine oxidation and backbone cleavage. Exp Eye Res. 2000;71:195–207. doi: 10.1006/exer.2000.0868. [DOI] [PubMed] [Google Scholar]
- 9.Chiou SH, Chylack LT, Jr., Tung WH, Bunn HF. Nonenzymatic glycosylation of bovine lens crystallins. Effect of aging. J Biol Chem. 1981;256:5176–5180. [PubMed] [Google Scholar]
- 10.Takemoto L, Emmons T. Truncation of alpha A-crystallin from the human lens. Exp Eye Res. 1991;53:811–813. doi: 10.1016/0014-4835(91)90119-y. [DOI] [PubMed] [Google Scholar]
- 11.Ajaz MS, Ma Z, Smith DL, Smith JB. Size of human lens beta-crystallin aggregates are distinguished by N-terminal truncation of betaB1. J Biol Chem. 1997;272:11250–11255. doi: 10.1074/jbc.272.17.11250. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, Smith DL, Smith JB. Human beta-crystallins modified by backbone cleavage, deamidation and oxidation are prone to associate. Exp Eye Res. 2003;77:259–272. doi: 10.1016/s0014-4835(03)00159-3. [DOI] [PubMed] [Google Scholar]
- 13.Reddy MA, Francis PJ, Berry V, Bhattacharya SS, Moore AT. Molecular genetic basis of inherited cataract and associated phenotypes. Surv Ophthalmol. 2004;49:300–315. doi: 10.1016/j.survophthal.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Shiels A, Bennett TM, Hejtmancik JF. Cat-Map: putting cataract on the map. Mol Vis. 2010;16:2007–2015. [PMC free article] [PubMed] [Google Scholar]
- 15.Francis PJ, Berry V, Bhattacharya SS, Moore AT. The genetics of childhood cataract. J Med Genet. 2000;37:481–488. doi: 10.1136/jmg.37.7.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bron AJ, Vrensen GF, Koretz J, Maraini G, Harding JJ. The ageing lens. Ophthalmologica. 2000;214:86–104. doi: 10.1159/000027475. [DOI] [PubMed] [Google Scholar]
- 17.Andley UP. Crystallins and hereditary cataracts: molecular mechanisms and potential for therapy. Expert Rev Mol Med. 2006;8:1–19. doi: 10.1017/S1462399406000111. [DOI] [PubMed] [Google Scholar]
- 18.Roshan M, Vijaya PH, Lavanya GR, Shama PK, Santhiya ST, Graw J, Gopinath PM, Satyamoorthy K. A novel human CRYGD mutation in a juvenile autosomal dominant cataract. Mol Vis. 2010;16:887–896. [PMC free article] [PubMed] [Google Scholar]
- 19.Pande A, Pande J, Asherie N, Lomakin A, Ogun O, King JA, Lubsen NH, Walton D, Benedek GB. Molecular basis of a progressive juvenile-onset hereditary cataract. Proc Natl Acad Sci U S A. 2000;97:1993–1998. doi: 10.1073/pnas.040554397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pande J, Pande A, Gillot D. The Cataract-Associated R14C Mutant of Human gamma D-Crystallin Shows a Variety of Intermolecular Disulfide Cross-Links: A Raman Spectroscopic Study. Biochemistry. 2009;48:4937–4945. doi: 10.1021/bi9004182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kmoch S, Brynda J, Asfaw B, Bezouska K, Novak P, Rezacova P, Ondrova L, Filipec M, Sedlacek J, Elleder M. Link between a novel human gammaD-crystallin allele and a unique cataract phenotype explained by protein crystallography. Hum Mol Genet. 2000;9:1779–1786. doi: 10.1093/hmg/9.12.1779. [DOI] [PubMed] [Google Scholar]
- 22.Pande A, Annunziata O, Asherie N, Ogun O, Benedek GB, Pande J. Decrease in protein solubility and cataract formation caused by the Pro23 to Thr mutation in human gamma D-crystallin. Biochemistry. 2005;44:2491–2500. doi: 10.1021/bi0479611. [DOI] [PubMed] [Google Scholar]
- 23.Pande A, Zhang J, Banerjee PR, Puttamadappa SS, Shekhtman A, Pande J. NMR study of the cataract-linked P23T mutant of human gammaD-crystallin shows minor changes in hydrophobic patches that reflect its retrograde solubility. Biochem Biophys Res Commun. 2009;382:196–199. doi: 10.1016/j.bbrc.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pande A, Ghosh KS, Banerjee PR, Pande J. Increase in surface hydrophobicity of the cataract-associated P23T mutant of human gammaD-crystallin is responsible for its dramatically lower, retrograde solubility. Biochemistry. 2010;49:6122–6129. doi: 10.1021/bi100664s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung J, Byeon IJ, Wang Y, King J, Gronenborn AM. The structure of the cataract-causing P23T mutant of human gammaD-crystallin exhibits distinctive local conformational and dynamic changes. Biochemistry. 2009;48:2597–2609. doi: 10.1021/bi802292q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benedek GB. Cataract as a protein condensation disease: the Proctor Lecture. Invest Ophth Vis Sci. 1997;38:1911–1921. [PubMed] [Google Scholar]
- 27.Stradner A, Foffi G, Dorsaz N, Thurston G, Schurtenberger P. New insight into cataract formation: enhanced stability through mutual attraction. Phys Rev Lett. 2007;99:198103. doi: 10.1103/PhysRevLett.99.198103. [DOI] [PubMed] [Google Scholar]
- 28.Banerjee PR, Pande A, Patrosz J, Thurston GM, Pande J. Cataract-associated mutant E107A of human gammaD-crystallin shows increased attraction to alpha-crystallin and enhanced light scattering. Proc Natl Acad Sci U S A. 2011;108:574–579. doi: 10.1073/pnas.1014653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Lomakin A, McManus JJ, Ogun O, Benedek GB. Phase behavior of mixtures of human lens proteins Gamma D and Beta B1. Proc Natl Acad Sci U S A. 2010;107:13282–13287. doi: 10.1073/pnas.1008353107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorsaz N, Thurston GM, Stradner A, Schurtenberger P, Foffi G. Colloidal characterization and thermodynamic stability of binary eye lens protein mixtures. J Phys Chem B. 2009;113:1693–1709. doi: 10.1021/jp807103f. [DOI] [PubMed] [Google Scholar]
- 31.Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 32.Yam AY, Xia Y, Lin HT, Burlingame A, Gerstein M, Frydman J. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol. 2008;15:1255–1262. doi: 10.1038/nsmb.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruckert M, Otting G. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J Am Chem Soc. 2000;122:7793–7797. [Google Scholar]
- 34.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J Magn Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 35.Zweckstetter M. NMR: prediction of molecular alignment from structure using the PALES software. Nat Protoc. 2008;3:679–690. doi: 10.1038/nprot.2008.36. [DOI] [PubMed] [Google Scholar]
- 36.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 37.Keller B, Christen M, Oostenbrink C, van Gunsteren WF. On using oscillating time-dependent restraints in MD simulation. J Biomol NMR. 2007;37:1–14. doi: 10.1007/s10858-006-9081-2. [DOI] [PubMed] [Google Scholar]
- 38.Goddard TD, Kneller DG. SPARKY 3. 3.110 ed. University of California; San Francisco: 2004. [Google Scholar]
- 39.McManus JJ, Lomakin A, Ogun O, Pande A, Basan M, Pande J, Benedek GB. Altered phase diagram due to a single point mutation in human gammaD-crystallin. Proc Natl Acad Sci U S A. 2007;104:16856–16861. doi: 10.1073/pnas.0707412104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flaugh SL, Mills IA, King J. Glutamine deamidation destabilizes human gammaD-crystallin and lowers the kinetic barrier to unfolding. J Biol Chem. 2006;281:30782–30793. doi: 10.1074/jbc.M603882200. [DOI] [PubMed] [Google Scholar]
- 41.Flaugh SL, Kosinski-Collins MS, King J. Contributions of hydrophobic domain interface interactions to the folding and stability of human gammaD-crystallin. Protein Sci. 2005;14:569–581. doi: 10.1110/ps.041111405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Acosta-Sampson L, King J. Partially folded aggregation intermediates of human gammaD-, gammaC-, and gammaS-crystallin are recognized and bound by human alphaB-crystallin chaperone. J Mol Biol. 2010;401:134–152. doi: 10.1016/j.jmb.2010.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forman-Kay JD, Clore GM, Stahl SJ, Gronenborn AM. 1H and 15N resonance assignments and secondary structure of the human thioredoxin C62A, C69A, C73A mutant. J Biomol NMR. 1992;2:431–445. doi: 10.1007/BF02192807. [DOI] [PubMed] [Google Scholar]
- 44.Moreau KL, King J. Hydrophobic core mutations associated with cataract development in mice destabilize human gammaD-crystallin. J Biol Chem. 2009;284:33285–33295. doi: 10.1074/jbc.M109.031344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pande A, Pande J, Asherie N, Lomakin A, Ogun O, King J, Benedek GB. Crystal cataracts: human genetic cataract caused by protein crystallization. Proc Natl Acad Sci U S A. 2001;98:6116–6120. doi: 10.1073/pnas.101124798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delaye M, Gromiec A. Mutual diffusion of crystallin proteins at finite concentrations: a light-scattering study. Biopolymers. 1983;22:1203–1221. doi: 10.1002/bip.360220413. [DOI] [PubMed] [Google Scholar]
- 48.Shiels A, Mackay D, Ionides A, Berry V, Moore A, Bhattacharya S. A missense mutation in the human connexin50 gene (GJA8) underlies autosomal dominant “zonular pulverulent” cataract, on chromosome 1q. Am J Hum Genet. 1998;62:526–532. doi: 10.1086/301762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mackay D, Ionides A, Kibar Z, Rouleau G, Berry V, Moore A, Shiels A, Bhattacharya S. Connexin46 mutations in autosomal dominant congenital cataract. Am J Hum Genet. 1999;64:1357–1364. doi: 10.1086/302383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knee KM, Goulet DR, Zhang J, Chen B, Chiu W, King JA. The group II chaperonin Mm-Cpn binds and refolds human gammaD crystallin. Protein Sci. 2011;20:30–41. doi: 10.1002/pro.531. [DOI] [PMC free article] [PubMed] [Google Scholar]