Abstract
Despite their close phylogenetic relationship, type A and B influenza viruses exhibit major epidemiological differences in humans, with the latter both less common and less often associated with severe disease. However, it is unclear what processes determine the evolutionary dynamics of influenza B virus, and how influenza viruses A and B interact at the evolutionary scale. To address these questions we inferred the phylogenetic history of human influenza B virus using complete genome sequences for which the date (day) of isolation was available. By comparing the phylogenetic patterns of all eight viral segments we determined the occurrence of segment reassortment over a 30-year sampling period. An analysis of rates of nucleotide substitution and selection pressures revealed sporadic occurrences of adaptive evolution, most notably in the viral hemagglutinin and compatible with the action of antigenic drift, yet lower rates of overall and nonsynonymous nucleotide substitution compared to influenza A virus. Overall, these results led us to propose a model in which evolutionary changes within and between the antigenically distinct ‘Yam88’ and ‘Vic87’ lineages of influenza B virus are the result of changes in herd immunity, with reassortment continuously generating novel genetic variation. Additionally, we suggest that the interaction with influenza A virus may be central in shaping the evolutionary dynamics of influenza B virus, facilitating the shift of dominance between the Vic87 and the Yam88 lineages.
Keywords: Influenza B virus, Phylogeny, Reassortment, Coalescent, Antigenic drift, Epidemiology
Introduction
Influenza B virus is the closest relative of influenza A virus. As such, these viruses possess a similarity in viral structure, genome organization and epidemiology. Both viruses belong to the Orthomyxoviridae family of enveloped, segmented negative-sense RNA viruses, and both have a genome possessing eight segments; PB2, PB1, PA, HA, NP, NA, MP, and NS. However, these viruses also exhibit a number of important differences. First, while influenza A virus infects a variety of vertebrate hosts, including birds, humans, and other mammals, influenza B viruses are predominantly found in humans. Second, although both viruses cause human respiratory infection on a seasonal basis in temperate climate regions, and on a more continual basis in the tropics (Viboud et al. 2006a), influenza B virus normally exists at a lower prevalence and causes less severe disease than influenza A viruses (especially compared to viruses of the A/H3N2 subtype) (Webster et al. 1992). Further, while the hemagglutinin (HA) gene of A/H3N2 viruses continually evolves into new antigenic clusters, and in an apparently episodic manner (Smith et al. 2004), type B viruses are divided into two major and cocirculating antigenically distinct lineages, denoted B/Victoria/2/87-like (‘Vic87’) and B/Yamagata/16/88-like (‘Yam88’) (Kanegae et al. 1990; Rota et al. 1990, 1992; McCullers et al. 2004). These important epidemiological and genetic differences imply that influenza viruses A and B also differ in their underlying evolutionary dynamics. Understanding the different evolutionary behavior of the influenza viruses, and how they might interact, is clearly of importance in predicting their future impact on human populations and in their control.
Previous studies have revealed that influenza B viruses exhibit lower rates of evolutionary change than A/H3N2 viruses (Webster et al. 1992; Lindstrom et al. 1999; Hiromoto et al. 2000). However, the basis of this difference in evolutionary dynamics is not clearly understood. Additionally, isolates of influenza B virus experience relatively frequent segment reassortment (Chi et al. 2003; Matsuzaki et al. 2004; McCullers et al. 2004). As a result, most prior work suggests that reassortment plays a major role in the evolution of influenza B virus, with positive selection for immune escape of less importance, in contrast to the picture seen in influenza A virus (Air et al. 1990; McCullers et al. 1999; Zou et al. 1997). However, while reassortment is evidently of great evolutionary importance for segmented RNA viruses, potentially generating advantageous genomic configurations and removing deleterious ones, its frequency and precise role in the evolution of influenza B viruses have not been fully elucidated. Likewise, the role played by antigenic drift—the ongoing change in the antigenicity of the HA and neuraminidase (NA) surface glycoproteins due to the selectively driven accumulation of amino acid substitutions at antigenic sites—in the evolution of influenza B virus is unclear.
To determine the evolutionary and epidemiological forces that shape the genetic diversity of influenza B virus, we present the first detailed molecular evolutionary analysis of complete genome sequence data. In particular, we used a Bayesian Markov chain Monte Carlo (MCMC) approach to trace the pattern of viral evolution in real time, determined the occurrence of segmental reassortment over a 30-year period, estimated segment-specific rates of nucleotide substitution rate, and measured selection pressures among different segments and branches of the influenza B virus phylogeny. By combing the phylogenetic and epidemiological data we are able to examine, for the first time, how host immune selection and reassortment, as well as the interaction with influenza A viruses, shape the evolutionary dynamics of influenza B virus.
Materials and Methods
Sequence Data
Data sets of complete genome sequences, representing all eight gene segments of influenza B virus, were downloaded from GenBank (with all laboratory recombinant isolates excluded). These data sets were then subdivided into three groups to address different evolutionary questions. (1) For estimating rates of nucleotide substitution, only sequences collected from the NIAID Influenza Genome Sequencing Project (http://www.niaid.nih.gov/dmid/genomes/mscs/influenza.htm [Ghedin et al. 2005]), where the exact day of isolation is available, were used. Estimating evolutionary rates using daily sampled data provides more accuracy than the yearly sampled viruses normally used in studies of influenza virus evolution, where it is impossible to distinguish among influenza seasons (which run in the winter across year boundaries in temperate latitudes). This resulted in a data set of 102 complete genomes sampled between February 11, 1989, and April 4, 2006, although in the case of NA a total of 112 dated sequences were available for analysis, while only 98 dated sequences were available for PB2. (2) To better infer the evolutionary history of influenza B virus at the level of individual gene segments (or domains in the case of HA), we constructed a number of larger data sets with yearly isolation dates ranging from 1965 to 2006. Due to the very large size of some of these data sets, highly similar sequences isolated from the same area in the same year were excluded. This resulted in data sets of the following size: PB2, PB1, PA, M1, NP = 143 sequences; HA1 domain (of HA) = 405 sequences (used for the detailed analysis of reassortment patterns); NA = 208 sequences; and NS1 = 182 sequences. (3) Finally, we compiled a data set of 214 (yearly sampled) HA1 domain sequences from which we inferred a maximum a posteriori (MAP) tree on which we could reconstruct the pattern of amino acid change through time (see below). For all three data sets, sequence alignments were created using MUSCLE (Edgar 2004) and adjusted manually using Se-Al (Rambaut 1996). This resulted in alignments of the following size (the major coding region in each segment only): PB2 = 2256 nucleotides (nt), PB1 = 2310 nt, PA = 2178 nt, HA = 1755 nt, HA1 = 1041 nt, NP = 1680 nt, NA = 1401 nt, M1 = 744 nt, and NS1 = 846 nt. All sequence alignments are available from the authors upon request.
Phylogenetic Analysis
For each data set, maximum likelihood (ML) phylogenetic trees were inferred using the PAUP* (version 4.0b10) package (Swofford 2003), based on the best-fit models of nucleotide substitution determined by MODELTEST (Posada and Crandall 1998 [parameter values available from the authors upon request]). For each tree, the reliability of phylogenetic groupings was determined through a bootstrap resampling analysis (1000 replicates of neighbor-joining trees estimated under the ML substitution model). In all cases, tree topologies were compared and reassortment events traced following the pattern of major topological incongruence, in which viruses change phylogenetic position with strong bootstrap support, using FigTree v1.0 (http://www.tree.bio.ed.ac.uk/software/figtree).
Rates of Nucleotide Substitution
Overall rates of evolutionary change (nucleotide substitutions per site, per year) for each segment and the two major lineages (Vic87 and Yam88) within the HA segment of influenza B virus were estimated using the Bayesian MCMC approach available in the BEAST package (http://www.evolve.zoo.ox.ac.uk/Beast/) (Drummond et al. 2002, 2006). The HKY85 + I Γ4 substitution model was used in most cases, as more complex models did not result in statistical convergence (the reverse was true of the HA segment, where the GTR + I Γ4 substitution model was employed). All data sets were analyzed under the Bayesian skyline model, as this allows the fluctuating population dynamics characteristic of influenza virus, and using relaxed (uncorrelated lognormal) molecular clocks. In each case, MCMC chains were run for sufficient time to achieve convergence (assessed using the TRACER program; http://www.evolve.zoo.ox.ac.uk/software.html?id=tracer), with uncertainty in parameter estimates reflected in the 95% highest probability density. Finally, BEAST was also used to infer the MAP tree for the HA1 domain (214 sequences), which depicts phylogenetic relationships against the time (year) of sampling of each isolate. Amino acid substitutions along internal branches of the MAP tree were reconstructed using the parsimony algorithm available in the MacClade package (version 4; Maddison and Maddison 2001). Finally, to document the change in antigenicity through time, the antigenic status of representative isolates from each antigenically distinguishable group of viruses was taken from hemagglutinin inhibition (HI) test results available in regular reports from the Weekly Epidemiological Record (WER).
Selection Pressures
To determine the tempo and mode of natural selection acting on influenza B virus, we estimated the average number of nonsynonymous (dN) and synonymous (dS) substitutions per site (ratio dN/dS) for each gene segment using the ‘one-ratio’ model from the CODEML program available within the PAML package (Yang 2007) and employing the ML tree in each case. Similar results were obtained using the Single Likelihood Ancestor Counting (SLAC) method (under the GTR substitution model) available in the HYPHY package (Pond and Frost 2005) and accessed through the Datamonkey interface (http://www.datamonkey.org; data not shown, available from the authors upon request). Similarly, the numbers and locations of positively and negatively selected sites were estimated using the Internal Fixed Effects Likelihood (IFEL) method available in HYPHY. For an additional examination of selection pressures we employed the ‘two-ratio’ model available in CODEML. This allows dN/dS to be estimated separately for the external and internal branches of each ML segment phylogeny. Because of the presence of overlapping reading frames in the M and NS segments, this analysis was restricted to the nonoverlapping regions of M1 and NS1 in these cases.
Results
Evolutionary Rates and Selection Pressures in Influenza B Virus
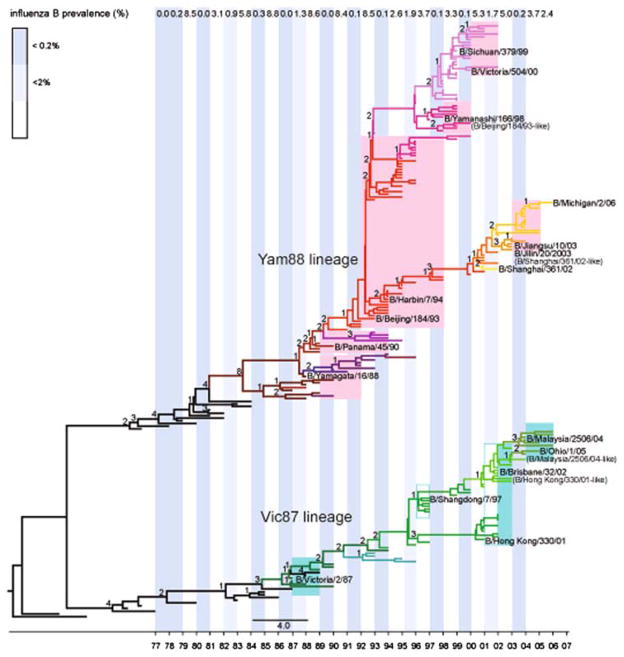
Estimates of rates of nucleotide substitution in influenza B virus ranged from 0.14 ×10−3 to 3.32 ×10−3 substitutions/site/year (Table 1). Although these rates are within the normal range observed in RNA viruses (Hanada et al. 2004; Jenkins et al. 2002), they are generally lower than those of H3N2 human influenza A virus (estimated at 2.68 ×10−3 to 12.50 ×10−3 subs/site/year [Nelson et al. 2006]) determined using similar methodology. Also of note was that the viral HA experienced an elevated rate of nucleotide substitution relative to those seen in other segments, especially on the Vic87 lineage, suggesting that it is subject to at least some degree of positive selection, as is well documented in influenza A virus (Fitch et al. 1991, 1997). The phylogenetic tree of the HA1 domain also presents an evolutionary pattern compatible with the continual action of antigenic drift; that is, the accumulation of amino acid changes along the main trunk in a progressive temporal manner (all amino acid changes have been mapped to internal branches of the HA1 tree presented in Fig. 1). Further, some of these sites were identified as being subject to positive selection (see below). This is consistent with the continuous change of antigenicity as depicted by HI tests from Weekly Epidemiology Records.
Table 1.
Rates of nucleotide substitution and dN/dS values for gene segments of influenza B virus
| Data set | Substitution rate (×10−3 sub/site/year)
|
dN/dS |
||||||
|---|---|---|---|---|---|---|---|---|
| Mean | 95% HPD
|
Overall mean | Internal | External | Internal/external | A/H3N2 overalla | ||
| Lower | Higher | |||||||
| PB2 | 0.14 | 0.01 | 2.99 | 0.04 | 0.03 | 0.05 | 0.60 | 0.10 |
| PB1 | 0.27 | 0.12 | 0.41 | 0.04 | 0.02 | 0.05 | 0.40 | 0.09 |
| PA | 1.05 | 0.70 | 1.41 | 0.07 | 0.06 | 0.08 | 0.75 | 0.13 |
| HA | 2.15 | 1.85 | 2.46 | 0.22 | 0.22 | 0.22 | 1.00 | 0.37 |
| HA–Vic87 | 3.32 | 2.31 | 6.48 | 0.25 | 0.17 | 0.47 | 0.36 | N/A |
| HA–Yam88 | 1.32 | 0.76 | 1.79 | 0.22 | 0.25 | 0.21 | 1.19 | N/A |
| NP | 1.06 | 0.75 | 1.40 | 0.07 | 0.05 | 0.09 | 0.56 | 0.10 |
| NA | 1.29 | 0.09 | 1.72 | 0.20 | 0.19 | 0.20 | 0.95 | 0.27 |
| Mb | 1.11 | 0.71 | 1.58 | 0.03 | 0.01 | 0.05 | 0.20 | 0.07 |
| NSb | 1.39 | 0.95 | 1.88 | 0.29 | 0.28 | 0.30 | 0.93 | 0.24 |
HPD, highest probability density; N/A, not applicable
dN/dS values of human influenza A/H3N2 virus were estimated based on the data set used by Holmes et al. (2005)
Overlapping reading frames in the M and NS segments mean that this analysis was restricted to the nonoverlapping regions of M1 and NS1
Fig. 1.
Maximum a posteriori (MAP) tree of the influenza B virus HA1 domain based on 214 sequences sampled annually between 1970 and 2006. Dominant antigenic clusters (names in roman font) within the Vic87 lineage are shaded green, while those in the Yam88 lineage are shaded pink. Open blocks denote viral clusters that did not dominate in the United States. The appearances of clades related to reassortment are represented by different colored branches on the tree, as are the two antigenically distinct clades (B/Beijing/184/93-like and B/Malaysia/2506/04-like viruses) whose appearance is not related to reassortment. Representative vaccine strains are also shown. The numbers of amino acid substitutions are given next to the internal branches of both the Vic87 and the Yam88 lineages. The prevalence of influenza B virus in the United States is also shown; data taken from Viboud et al. (2006b) and updated with data from the CDC Web site (http://www.cdc.gov/flu/weekly/fluactivity.htm) (see Supplementary Material)
More direct evidence for the occurrence of adaptive evolution is provided by an analysis of selection pressures through the relative rates of nonsynonymous and synonymous substitution per site (dN/dS ratio). First, the HA is characterized by a high overall mean dN/dS value—0.22—compared to most of the other segments. Notably, the mean dN/dS ratio is also elevated in NA (0.20), the second viral surface antigen, as well as NS1 (0.29), which may reflect its role in evading innate immune responses (Baigent and McCauley 2003). Second, HA, NA, and NS1 also have an elevated dN/dS ratio relative to other genes on internal compared to external branches (internal/internal dN/dS ratios of 1.00, 0.95, and 0.93, respectively), most likely reflecting a high rate of amino acid fixation as expected under regular positive selection (Table 1) (Pybus et al. 2007). This elevation is especially notable in the HA of viruses from the Yam88 lineage (internal/external dN/dS ratio of 1.19), although a reduced value is seen on the HA from the Vic87 lineage, suggesting that many of the nonsynonymous mutations observed in this lineage represent transient deleterious mutations. Finally, a small number of positively selected sites were detected in each segment data set (p < 0.1) (Table 2). Again, this selection was strongest in HA, which contained nine positively selected sites across all those data sets analyzed. Similarly, three and two positively selected sites were found in the NA and the NS1 data sets, respectively. Further, some of these selected sites —164 in HA and 345, 373, and 396 in NA—have previously been documented as the location of escape mutants from neutralizing antibodies (Air et al. 1990; Nakagawa et al. 2005). However, despite this evidence for positive selection, the dN/dS values observed in influenza B virus are generally lower than those documented in human influenza A H3N2 viruses (Table 1) (Holmes et al. 2005), revealing an overall difference in selection pressure between these two viruses.
Table 2.
Selection pressures acting on influenza B virus
| Data set | No. unique sequences | No. codons | No. negatively selected | No. positively selected | Positively selected codon positions |
|---|---|---|---|---|---|
| HA | 94 | 585 | 58 | 5 | 90, 131, 179, 180, 277 |
| HA–Vic87 | 40 | 585 | 29 | 0 | |
| HA1–Vic87 (MAP)a,b | 63 | 347 | 15 | 3 | 131, 179, 214 |
| HA–Yam88 | 54 | 585 | 10 | 2 | 71, 90 |
| HA1–Yam88 (MAP)b,c | 146 | 347 | 53 | 3 | 164, 212, 214 |
| NA | 98 | 467 | 43 | 3 | 345, 373, 396 |
| PB2 | 90 | 752 | 72 | 1 | 391 |
| PB1 | 97 | 770 | 81 | 0 | |
| PA | 91 | 726 | 21 | 1 | 258 |
| NP | 90 | 560 | 52 | 2 | 513, 520 |
| M1 | 66 | 248 | 24 | 0 | |
| NS1 | 71 | 218 | 10 | 2 | 115, 195 |
Codon positions in HA1 were corrected to match their positions in the HA0 precursor gene
The maximum a posteriori (MAP) tree data sets were used in these cases. Maximum likelihood tree sets were used in all other cases
The SLAC method was used due to the large sample size. In all other cases the IFEL method was used
Reassortment Among Lineages of Influenza B Virus
According to our expansive HA1 domain phylogeny (Supplementary Fig. 1), two major clades of influenza B virus have been present since the mid-1970s, and since 1987–1988, HI tests reveal that these represent the antigenically distinct ‘Vic87’ and ‘Yam88’ lineages (Kanegae et al. 1990; Rota et al. 1990, 1992; WER February 1998). As this major phylogenetic division is present in all segments, it evidently reflects a large-scale genomic event, although frequent reassortment complicates lineage definitions in segments other than HA (such that we classify the two major lineages as ‘I’ and ‘II’ in all segments other than HA).
Our phylogenetic analysis further reveals that the Vic87 and Yam88 lineages have developed into a series of component clades, which are also antigenically distinct as determined by HI tests (WER 1988–2007; http://www.who.int/wer/archives/en/) and denoted by different colored background blocks in Fig. 1 (with the Vic87 clades in green and the Yam88 clades in pink). Each clade dominates, or codominates, the viral population for a particular period, the timescale of which is also depicted in the MAP tree (also see the Discussion). The Yam88 lineage has diverged into the B/Yamagata/16/88-like viruses (the original strain), the B/Panama/45/90-like viruses, the B/Beijing/184/93-like viruses, the B/Sichuan/379/99-like viruses, and the B/Shanghai/361/02-like viruses, while the Vic87 lineage has diversified into the B/Victoria/2/87-like viruses (the original strain), the B/Shangdong/7/97-like viruses, the B/Hong Kong/330/2001-like viruses, and the B/Malaysia/2506/2004-like viruses (Fig. 1).
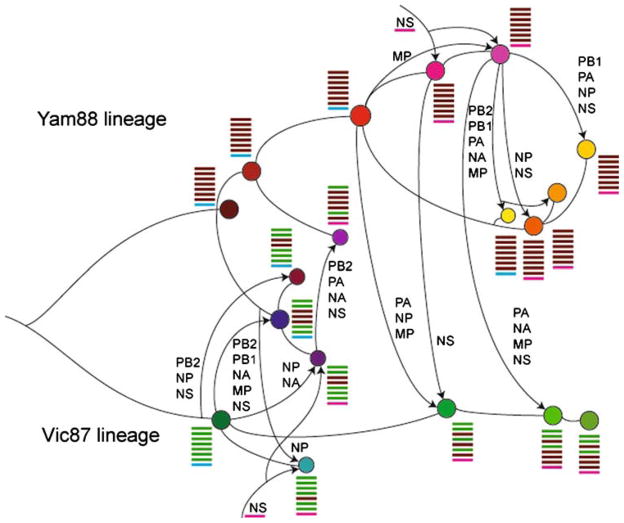
A more detailed analysis of the trees for each segment (presented in Supplementary Figs. 1–8) reveals that there are frequent, and sometimes complex, reassortment events involving multiple viral lineages of influenza B virus and manifest as widespread phylogenetic incongruence. To highlight the importance of this process, clusters of sequences whose appearance is related to reassortment are depicted by different branch colors in each segment tree (Fig. 1 and Supplementary Figs. 1–8) and the direction of these reassortment events, reflected in the pattern of incongruence among segment trees mapped onto the HA1 phylogeny, are illustrated in Fig. 2. Interestingly, the genesis of most new HA clades is associated with reassortment, which can involve parental viruses from either different antigenic lineages (Vic87 and Yam88) or a single lineage. Further, some of the reassortment events involve a new combination of HA and NA segments, with potentially important effects on viral fitness. For example, the B/Brisbane/32/02 group of viruses (Vic87 lineage) is a reassortant composed of the PB2, PB1, HA, and NP segments derived from this lineage (from B/Shangdong/7/97-like viruses) and the PA, NA, M, and NS segments derived from B/Sichuan/379/99-like viruses of the Yam88 lineage. Similarly, although the group of viruses represented by B/Yamanashi/166/98 is commonly referred to as the ‘B/Beijing/184/93-like viruses,’ this isolate is actually a recombinant involving a new NS segment (Fig. 2). However, not all changes in antigenic group can be associated with reassortment. For example, the direct phylogenetic link between the antigenically distinct B/Beijing/184/93-like and B/Panama/45/90-like viruses indicates that they evolved in a progressive manner without the assistance of reassortment, while the Malaysia/2506/04-like viruses originated directly from a group of B/Hong Kong/330/01-like viruses (although the latter virus is itself is a reassortant).
Fig. 2.
Reassortment among the major clades of influenza B viruses. The phylogenetic background for this representation of evolutionary history is those viral clades identified in the HA1 tree shown in Fig. 1. The curved lines without arrows indicate a direct ancestor-descendent relationship (i.e., without reassortment) between viruses in the HA1 tree, while the lines with arrows depict reassortment events, with the names of new segments also listed. Due to uncertainty in some of the segment phylogenies, it is not possible to fully determine the ancestry of all reassortment events. Genome compositions (that is, the origin of the eight segments) are illustrated besides each clade by colored bars, with the order (from top to bottom) corresponding to the segment number (largest to smallest: PB2, PB1, PA, HA, NP, NA, M, NS). As the B/Victoria/02/87 (Vic87) and B/Yamagata/166/88 (Yam88) viruses fall in the same branch of the NS tree (contrary to their distinction in other gene segments), the major lineages within NS are colored differently from those in other segments. The detailed trees from which this representation was inferred are shown in Supplementary Figs. 1–8
Some of the reassortment events in influenza B virus are also complex. This is best documented in the case of the B/Shanghai/361/02-like viruses that are composed of at least four groups of reassortants from Yam88 viruses (Figs. 1 and 2). Specifically, group 1 viruses (yellow) acquired the PB2, PB1, PA, NA, and M segments, and group 2 viruses (dark-orange) acquired the NP and NS segments, from B/Sichuan/379/99-like viruses. Group 3 viruses (light-orange) appear to be a reassortant between the first two groups, with the NP and NS segments acquired from group 2 viruses. Therefore, with the exception of HA, all segments in group 3 viruses were acquired indirectly from B/Sichuan/379/99-like viruses. Finally, group 4 viruses (gold) were generated by a reassortment event involving group 2 and B/Sichuan/379/99-like viruses, with the PB1, PA, NP, and NS segments obtained from the latter.
Evolutionary Interactions Among Influenza A and B Viruses
To determine whether larger-scale epidemiological processes might also shape the evolution of influenza B virus, we examined, in combination with our phylogenetic analyses, the changing patterns of prevalence for influenza viruses A and B in the United States over a period of almost 30 years (Fig. 1). As expected, the prevalence of influenza B virus fluctuates on an annual basis. In particular, some years represent strong population bottlenecks (<0.2% prevalence) for influenza B virus, including the 1993–1994, 1997–1998, 1999–2000, and 2003–2004 epidemic seasons. In turn, these population bottlenecks in influenza B virus are always associated with a high prevalence of influenza A virus (Supplementary Table 1). As these two viruses are clearly competing to fill the same niche—the susceptible human population—it is likely that infection by one type of virus will inhibit either the infection or the replication of the other (Anestad 1987; Mikheeva and Ghendon 1982). Hence, when there is a broad and long-lasting epidemic of influenza A virus, type B viruses will necessarily suffer a major population bottleneck. More striking was the observation that changes in the dominant antigenic cluster of influenza B virus always occur during, or immediately following, a population bottleneck, or in years with greatly reduced prevalence (for example, during the 1995–1996 and 2001–2002 seasons, when the prevalence was 1.9% and 1.7%, respectively; Fig. 1). Hence, there appears to be a relationship between population prevalence and the pattern of viral evolution.
Discussion
Our phylogenetic analysis reveals that the evolution of influenza B virus is characterized by a continuous turnover of antigenically distinct lineages. Each dominant antigenic cluster is usually only transitory, existing for a short period of time before being replaced by a new group of viruses. This process, equivalent to antigenic drift, is clearly a selectively driven process, as shown by the increased rate of evolutionary change in HA, including that at nonsynonymous sites, and the direct identification of a number of amino acid sites experiencing positive selection (and which are likely to be underestimates given the difficulties in detecting site-specific positive selection). Hence, immune selection on the HA, and perhaps also the NA and NS proteins, plays a major role in determining patterns of evolutionary change in influenza B virus.
The importance of immune selection is also reflected in the alternation in dominance between the Vic87 and the Yam88 lineages. The Vic87 lineage was the major lineage isolated during the period 1987 to 1989, while the Yam88 lineage was dominant for the majority of the 1990s. Although it caused an epidemic in Asia in the 1996/1997 season, Vic87 viruses did not begin to regain dominance globally until the 2001/2002 season. Apart from a short break in the 2003/2004 season, the Vic87 lineage was dominant until the 2006/2007 season, when an increase in the number of Yam88 viruses was observed (WER 1987–2007). This pattern of alternating dominance is compatible with a model in which one of these two lineages dominates the viral population until sufficient herd immunity is generated against it, either through recovery from infection or vaccination, at which point the other lineage is able to increase in frequency. Notably, the highest dN/dS ratio on internal branches is observed in the HA1 domain from Yam88 viruses, suggesting that the long period of Yam88 circulation during the 1990s may have been associated with stronger antigenic drift on this lineage.
Further support for the overarching action of immune selection is provided by the association between the temporal progression of dominant strains and changing patterns of antigenicity. Specifically, when dominant clusters from successive seasons belong to the same viral lineage (Vic87 or Yam88), significant antigenic differences are always observed between viruses because, in this case, there must be direct selection to evade herd immunity. In contrast, where a new dominant cluster is derived from a different viral lineage to the previous dominant strain, we observe no progressive change in antigenicity. We suggest that this is because there is no selective requirement to be antigenically distinct in the latter case, since the Vic87 and Yam88 lineages are already differ antigenically. As an example, the antigenic difference (as revealed by HI data) between the B/Brisbane/32/02 (HongKong/330/2001-like) and the Shang-dong/7/97-like viruses is smaller than that between the B/Yamanashi/166/98-like (B/Beijing/184/93-like) and B/Sichuan/379-like viruses; although both belong to the Vic87 lineage, the former viruses are not directly and temporally related to one another, whereas the latter viruses represent successive clusters from the same (Yam88) lineage (Fig. 1).
Our genome-wide analysis also reveals that the process of lineage turnover is often, although not exclusively, associated with segmental reassortment. Although this indicates that reassortment plays an important role on the evolution of influenza B virus, it is unclear whether the large-scale patterns of reassortment observed (that is, whether specific segments tend to reassort together) reflect major functional constraints, such as compatibility in packaging, or are simply random events. Further, that reassortant clades often go on to dominate the viral population suggests that reassortment may sometimes enhance viral fitness, perhaps by placing segments, particular the HA and NA, in new, advantageous genetic backgrounds.
At a larger, epidemiological scale, our analysis indicates that the changing prevalence of influenza B virus, particularly its interaction with influenza A virus, also plays a key role in shaping evolutionary dynamics. Specifically, we propose that during population bottlenecks of influenza B virus, which correspond to a high prevalence of influenza A virus, an important stochastic element into influenza dynamics. The evolutionary fate of new genetic variants is therefore determined by their initial frequency of in the population, their antigenic difference from existing viruses, and the randomizing effect of large-scale population bottlenecks. This mix of stochastic and deterministic factors may explain why new dominant clusters can arise from either the same or different lineages. In sum, the evolutionary dynamics of influenza B virus are shaped by a complex set of evolutionary interactions, both among the various antigenically distinct clades of this virus and with the largely dominant influenza A viruses that cocirculate within the same human population.
The difference in epidemiological dynamics between influenza viruses types A and B is also reflected in their rates of evolutionary change and selection pressures. Although there is evidence of a lower mutation rate in type B compared to type A viruses (Nobusawa and Sato 2006), it is likely that differences in the strength of immune selection, itself a function of overall prevalence, the level of herd immunity (in part due to vaccination), also play a key role in shaping substitution rates in the long term. Such differences are reflected in the lower rates of nonsynonymous substitution in influenza B virus compared to A/H3N2. At the broadest level, our study shows that patterns and processes of evolutionary change in influenza viruses not only are determined by the nature of their interaction with the human immune system, but also reflect how these viruses interact with each other.
Supplementary Material
Acknowledgments
We thank Dr. J. K. Taubenberger for constructive comments on an earlier version of the manuscript and Dr. C. Viboud for assistance with the epidemiological data. We thank all those involved in the Influenza Genome Sequencing Project for contributing their viruses or obtaining genome sequence data. This work was supported in part by NIH Grant GM080533-01.
Footnotes
Electronic supplementary material: The online version of this article (doi:10.1007/s00239-008-9119-z) contains supplementary material, which is available to authorized users.
Contributor Information
Rubing Chen, Center for Infectious Disease Dynamics, Department of Biology, The Pennsylvania State University, Mueller Laboratory, University Park, PA 16802, USA.
Edward C. Holmes, Email: ech15@psu.edu, Center for Infectious Disease Dynamics, Department of Biology, The Pennsylvania State University, Mueller Laboratory, University Park, PA 16802, USA, Fogarty International Center, National Institutes of Health, Bethesda, MD 20892, USA
References
- Air GM, Gibbs AJ, Laver WG, Webster RG. Evolutionary changes in influenza B are not primarily governed by antibody selection. Proc Natl Acad Sci USA. 1990;87:3884–3888. doi: 10.1073/pnas.87.10.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anestad G. Surveillance of respiratory viral infections by rapid immunofluorescence diagnosis, with emphasis on virus interference. Epidemiol Infect. 1987;99:523–531. doi: 10.1017/s0950268800068023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baigent SJ, McCauley JW. Influenza type A in humans, mammals and birds: determinants of virus virulence, host-range and interspecies transmission. BioEssays. 2003;25:657–671. doi: 10.1002/bies.10303. [DOI] [PubMed] [Google Scholar]
- Chi XS, Bolar TV, Zhao P, Rappaport R, Cheng SM. Cocirculation and evolution of two lineages of influenza B viruses in Europe and Israel in the 2001–2002 season. J Clin Microbiol. 2003;41:5770–5773. doi: 10.1128/JCM.41.12.5770-5773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM, Leiter JME, Li X, Palese P. Positive Darwinian evolution in human influenza A viruses. Proc Natl Acad Sci USA. 1991;88:4270–4274. doi: 10.1073/pnas.88.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM, Bush RM, Bender CA, Cox NJ. Long term trends in the evolution of H(3) HA1 human influenza type A. Proc Natl Acad Sci USA. 1997;94:7712–7718. doi: 10.1073/pnas.94.15.7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E, Sengamalay N, Shumway M, Zaborsky J, Feldblyum T, Subbu V, Spiro D, Sitz J, Koo H, Bolotov P, Dernovoy D, Tatusova T, Bao Y, St George K, Taylor J, Lipman D, Fraser C, Taubenberger J, Salzberg S. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature. 2005;437:1162–1166. doi: 10.1038/nature04239. [DOI] [PubMed] [Google Scholar]
- Hanada K, Suzuki Y, Gojobori T. A large variation in the rates of synonymous substitution for RNA viruses and its relationship to a diversity of viral infection and transmission modes. Mol Biol Evol. 2004;21:1074–1080. doi: 10.1093/molbev/msh109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromoto Y, Saito T, Lindstrom SE, Li Y, Nerome R, Sugita S, Shinjoh M, Nerome K. Phylogenetic analysis of the three polymerase genes (PB1, PB2 and PA) of influenza B virus. J Gen Virol. 2000;81:929–937. doi: 10.1099/0022-1317-81-4-929. [DOI] [PubMed] [Google Scholar]
- Holmes EC, Ghedin E, Miller N, Taylor J, Bao Y, St George K, Grenfell BT, Salzberg SL, Fraser CM, Lipman DJ, Taubenberger JK. Whole-genome analysis of human influenza A virus reveals multiple persistent lineage and reassortment among recent H3N2 viruses. PLoS Biol. 2005;3:e300. doi: 10.1371/journal.pbio.0030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GM, Rambaut A, Pybus OG, Holmes EC. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J Mol Evol. 2002;54:156–165. doi: 10.1007/s00239-001-0064-3. [DOI] [PubMed] [Google Scholar]
- Kanegae Y, Sugita S, Endo A, Ishida M, Senya S, Osako K, Nerome K, Oya A. Evolutionary pattern of the hemagglutinin gene of influenza B viruses isolated in Japan: cocirculating lineages in the same epidemic season. J Virol. 1990;64:2860–2865. doi: 10.1128/jvi.64.6.2860-2865.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom SE, Hiromoto Y, Nishimura H, Saito T, Nerome R, Nerome K. Comparative analysis of evolutionary mechanisms of the hemagglutinin and three internal protein genes of influenza B virus: multiple cocirculating lineages and frequent reassortment of the NP, M, and NS genes. J Virol. 1999;73:4413–4426. doi: 10.1128/jvi.73.5.4413-4426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP. MacClade 4: analysis of phylogeny and character evolution. Version 4.03. Sinauer Associates; Sunderland, MA: 2001. [Google Scholar]
- Matsuzaki Y, Sugawara K, Takashita E, Muraki Y, Hongo S, Katsushima N, Mizuta K, Nishimura H. Genetic diversity of influenza B virus: the frequent reassortment and cocirculation of the genetically distinct reassortant viruses in a community. J Med Virol. 2004;74:132–140. doi: 10.1002/jmv.20156. [DOI] [PubMed] [Google Scholar]
- McCullers JA, Wang GC, He S, Webster RG. Reassortment and insertion-deletion are strategies for the evolution of influenza B viruses in nature. J Virol. 1999;73:7374–7348. doi: 10.1128/jvi.73.9.7343-7348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers JA, Saito T, Iverson AR. Multiple genotypes of influenza B virus circulated between 1979 and 2003. J Virol. 2004;78:12817–12828. doi: 10.1128/JVI.78.23.12817-12828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheeva A, Ghendon YZ. Intrinsic interference between influenza A and B viruses. Arch Virol. 1982;73:287–294. doi: 10.1007/BF01318082. [DOI] [PubMed] [Google Scholar]
- Nakagawa N, Kubota R, Okuno Y. Variation of the conserved neutralizing epitope in influenza B virus Victoria group isolates in Japan. J Clin Microbiol. 2005;43:4212–4214. doi: 10.1128/JCM.43.8.4212-4214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Simonsen L, Viboud C, Miller MA, Taylor J, St George K, Griesemer EG, Ghedin E, Sengamalay NA, et al. Stochastic processes are key determinants of short-term evolution in Influenza A virus. PLoS Pathog. 2006;2:e125. doi: 10.1371/journal.ppat.0020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobusawa E, Sato K. Comparison of the mutation rates of human influenza A and B viruses. J Virol. 2006;80:3675–3678. doi: 10.1128/JVI.80.7.3675-3678.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SL, Frost SDW. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Pybus OG, Rambaut A, Freckleton RP, Belshaw R, Drummond AJ, Holmes EC. Phylogenetic evidence for deleterious mutation load in RNA viruses and its contribution to viral evolution. Mol Biol Evol. 2007;24:845–852. doi: 10.1093/molbev/msm001. [DOI] [PubMed] [Google Scholar]
- Rambaut A. Se–Al: Sequence Alignment Editor. 1996 Available at: http://www.evolve.zoo.ox.ac.uk/
- Rota PA, Wallis TR, Harmon MW, Rota JS, Kendal AP, Nerome K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. 1990;175:59–68. doi: 10.1016/0042-6822(90)90186-u. [DOI] [PubMed] [Google Scholar]
- Rota PA, Hemhill ML, Whistler T, Regnery HL, Kendal AP. Antigenic and genetic characterization of the haemagglutinins of recent cocirculating strains of influenza B virus. J Gen Virol. 1992;73:2737–2742. doi: 10.1099/0022-1317-73-10-2737. [DOI] [PubMed] [Google Scholar]
- Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- Swofford DL. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates; Sunderland, MA: 2003. PAUP*. [Google Scholar]
- Viboud C, Alonso WJ, Simonsen L. Influenza in tropical regions. PLOS Med. 2006a;3:461–471. doi: 10.1371/journal.pmed.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viboud C, Bjørnstad ON, Smith DL, Simonsen L, Miller MA, Grenfell BT. Synchrony, waves, and spatial hierarchies in the spread of influenza. Science. 2006b;312:447–451. doi: 10.1126/science.1125237. [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zou S, Prud’Homme I, Weber JM. Evolution of the hemagglutinin gene of influenza B virus was driven by both positive and negative selection pressures. Virus Genes. 1997;14:181–185. doi: 10.1023/a:1007927725332. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.