Abstract
Once stimulated, the epidermal growth factor receptor (EGFR) undergoes self-phosphorylation, which, on the one hand, instigates signaling cascades, and on the other hand, recruits CBL ubiquitin ligases, which mark EGFRs for degradation. Using RNA interference screens, we identified a deubiquitinating enzyme, Cezanne-1, that opposes receptor degradation and enhances EGFR signaling. These functions require the catalytic and ubiquitin-binding domains of Cezanne-1, and they involve physical interactions and trans-phosphorylaton of Cezanne-1 by EGFR. In line with the ability of Cezanne-1 to augment EGF-induced growth and migration signals, the enzyme is overexpressed in breast cancer. Congruently, the corresponding gene is amplified in approximately one third of mammary tumors, and high transcript levels predict an aggressive disease course. In conclusion, deubiquitination by Cezanne-1 curtails degradation of growth factor receptors, thereby promotes oncogenic growth signals.
Keywords: gene amplification, deubiquitination, endocytosis, growth factor
INTRODUCTION
Binding of EGF to the cognate transmembrane receptor, EGFR, stimulates the intrinsic tyrosine kinase activity and culminates in cell fate decisions. Importantly, these positively acting processes are coupled to a web of negatively acting feedback loops (Avraham and Yarden, 2010). One robust and early feedback loop entails targeting activated EGFRs to degradation in lysosomes, or to an escape recycling route (Sorkin and von Zastrow, 2009). EGFR transfer from endosomes back to the plasma membrane is thought to enhance intracellular signaling (Fehrenbacher et al., 2009; Goh et al.; Vieira et al., 1996). The regulation of receptor trafficking and the underlying intracellular signals appear complex and redundant. Sorting at the plasma membrane is executed by adaptors of clathrin, the phosphotyrosine-binding protein GRB2 (Goh et al., 2010) and the kinase inhibitor RALT/MIG6 (Frosi et al., 2010). By contrast, sorting at the multi-vesicular body (MVB), a pre-lysosomal compartment, depends on an E3 ligase molecule called CBL (Huang et al., 2006; Levkowitz et al., 1999). Upon tyrosine phosphorylation, CBL modifies multiple lysine residues, located within the kinase domain of EGFR, with monomeric (Haglund et al., 2003; Mosesson et al., 2003), as well as with polymeric ubiquitin molecules (Huang et al., 2006). Ubiquitin moieties attached to EGFR are then recognized by a set of ubiquitin-binding adaptors of the clathrin coat (e.g., Epsin). These adaptors harbor ubiquitin-binding domains (UBDs), and thereby assemble active receptors at clathrin-coated regions of the plasma membrane, endosomes and the MVB (Katzmann et al., 2002). Although they are topologically different, the sorting assemblies (called: endosomal sorting complexes required for transport; ESCRTs) share a mechanism of cargo dissociation and reloading: at the endosome, E3 ligases of the AIP4/Nedd4 family ubiquitinate the ubiquitin binders, for example HRS, to promote their intramolecular folding in a way that dissociates the cargo and enables loading of a new receptor upon their de-ubiquitination (Katz et al., 2002; Polo et al., 2002).
Enzymatic deubiquitination of the endocytic machinery, as well as deubiquitination of receptor molecules, play essential regulatory roles (Sacco et al., 2010). Approximately 100 deubiquitinating enzymes (DUBs) are known that fall into five classes: ubiquitin C-terminal hydrolases (UCHs), the ubiquitin-specific proteases (UBPs), Josephins, the JAB1/MPN/MOV3 metalloenzymes (JAMM), and the ovarian tumor proteases (OTUDs). At least three DUBs are involved, either directly or indirectly, in EGFR endocytosis. The best characterized is AMSH, a JAMM family member whose knockdown leads to enhanced degradation of EGFR (Bowers et al., 2006; McCullough et al., 2004). Targeting of AMSH to endosomes may occur via clathrin (Nakamura et al., 2006) or by ESCRT-III (Ma et al., 2007). The roles for another endosomal DUB, UBPY (USP8), deviate from a simple model of receptor deubiquitination, as this enzyme is involved in stabilization of two endosomal sorting components, HRS and STAM, and it destabilizes EGFR (Row et al., 2006). A third DUB regulating EGFR, namely USP18, acts at the level of protein translation (Duex and Sorkin, 2009). The present study assumed that yet uncovered DUBs regulate EGFR endocytosis, hence applied screens of siRNA libraries to EGF-stimulated cells. In addition to AMSH, our screen identified an OTUD family member, Cezanne-1, as an enzyme able to stabilize ligand-activated EGFRs. In agreement with the ability of Cezanne-1 to enhance receptor signaling, we found that the expression of the corresponding gene, from a locus frequently amplified in tumors, predicts short survival of breast cancer patients.
RESULTS
siRNA screens identify Cezanne proteins as EGFR-specific DUBs
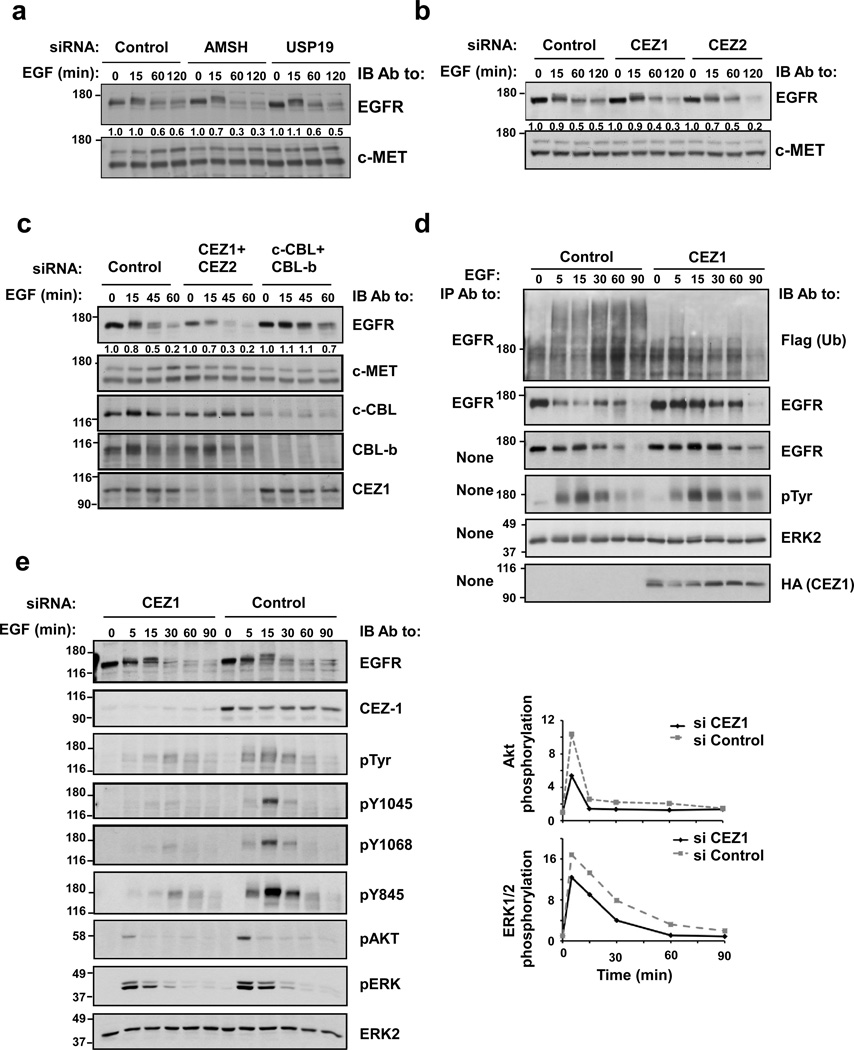
To identify EGFR-regulating enzymes, we performed siRNA screens of the majority of human DUBs. Because EGFR is expressed in epithelial cell types, we assumed that the putative DUB is co-expressed in epithelial tumor cells, such as KB oral cells. Hence, we transfected KB cells with siRNA duplexes against different DUBs, and stimulated the cells with EGF. This strategy was confirmed by transfecting KB cells with siRNA oligonucleotides specific to AMSH (as a positive control), or USP19, as a negative control. Ligand-induced degradation of EGFR was moderately enhanced upon AMSH knockdown, consistent with previous reports (Ma et al., 2007; McCullough et al., 2004) (Fig. 1a). In addition to AMSH, our screens of 95 human DUBs identified Cezanne-1, a DUB previously implicated in NFκB signaling (Enesa et al., 2008; Evans et al., 2003), and its family member, Cezanne-2, as candidate DUBs of EGFR (Fig. 1b). Accordingly, simultaneous knockdown of Cezanne-1 and Cezanne-2 by using two pools of four oligonucleotides (approximately 70% and 50%, respectively) moderately enhanced both basal and inducible degradation of EGFR, whereas the reciprocal silencing of c-CBL and CBL-b showed the expected opposite effect (Fig. 1c).
Figure 1. siRNA screens of human deubiquitinating enzymes identify Cezanne-1 and Cezanne-2 as EGFR-specific DUBs that enhance EGF signaling.
(a) KB cells were transfected with siRNAs against AMSH and USP19. Forty-eight hours later, cells were stimulated with EGF (20 ng/ml), as indicated. Thereafter, lysates were immunoblotted (IB) with antibodies against EGFR and c-MET. The EGFR signals were quantified, normalized and indicated. (b) KB cells were transfected with siRNAs targeting Cezanne-1 and Cezanne-2, and cells were treated and analyzed as in a. (c) HeLa cells were transfected with combinations of siRNAs against Cezanne-1/Cezanne-2 and c-CBL/CBL-b. Forty-eight hours following transfection, cells were treated with EGF and whole cell lysates analyzed. (d) HeLa cells were transfected with a plasmid encoding Cezanne-1, or an empty plasmid, alongside with a plasmid encoding a Flag-tagged ubiquitin. Forty-eight hours later, cells were incubated with EGF (20 ng/ml) for the indicated intervals. Cell lysates and EGFR immunoprecipitates were analyzed as indicated. (e) HeLa cells were transfected with siRNAs targeting Cezanne-1, or with control oligonucleotides. Forty-eight hours later, cells were stimulated with EGF (20 ng/ml) for the depicted intervals. Thereafter, whole cell extracts were immunoblotted with the indicated antibodies, including antibodies to the phosphorylated forms of AKT and ERK, as well as antibodies specific to specific phosphorylated tyrosines of EGFR. Signal intensities of phosphorylated AKT and ERK2 were quantified, normalized and graphically presented (right panels). All panels presented in this figure were repeated at least thrice.
Because Cezanne-1 is frequently overexpressed in cancer cells (see below), but Cezanne-2 displays no extensive alterations, we focused on Cezanne-1. To extend the effects beyond KB cells to other Cezanne-1 expressing cell, we employed the widely used HeLa cells, which were co-transfected with plasmids encoding Cezanne-1 and a Flag-tagged ubiquitin. The results presented in Figure 1d confirmed that Cezanne-1 overexpression associates with remarkably reduced ubiquitination of EGFR, as well as with stabilization the receptor and an increase in auto-phosphorylation. Application of a reciprocal approach in HeLa cells, which employed specific siRNAs, confirmed that knockdown of Cezanne-1 correlates with accelerated degradation and decreased receptor phosphorylation. Moreover, diminished phosphorylation of AKT and ERK1/2 was detected (Fig. 1e). Notably, silencing Cezanne-1 inactivated EGFR's kinase prior to receptor degradation, probably due to enhanced ubiquitination that either disabled the enzyme or occluded lysine acceptors of ubiquitin. In conclusion, our genetic screens and validation tests indicated that Cezanne-1 regulates EGFR levels, as well as downstream signals.
Cezanne-1 stabilizes and physically interacts with EGFR
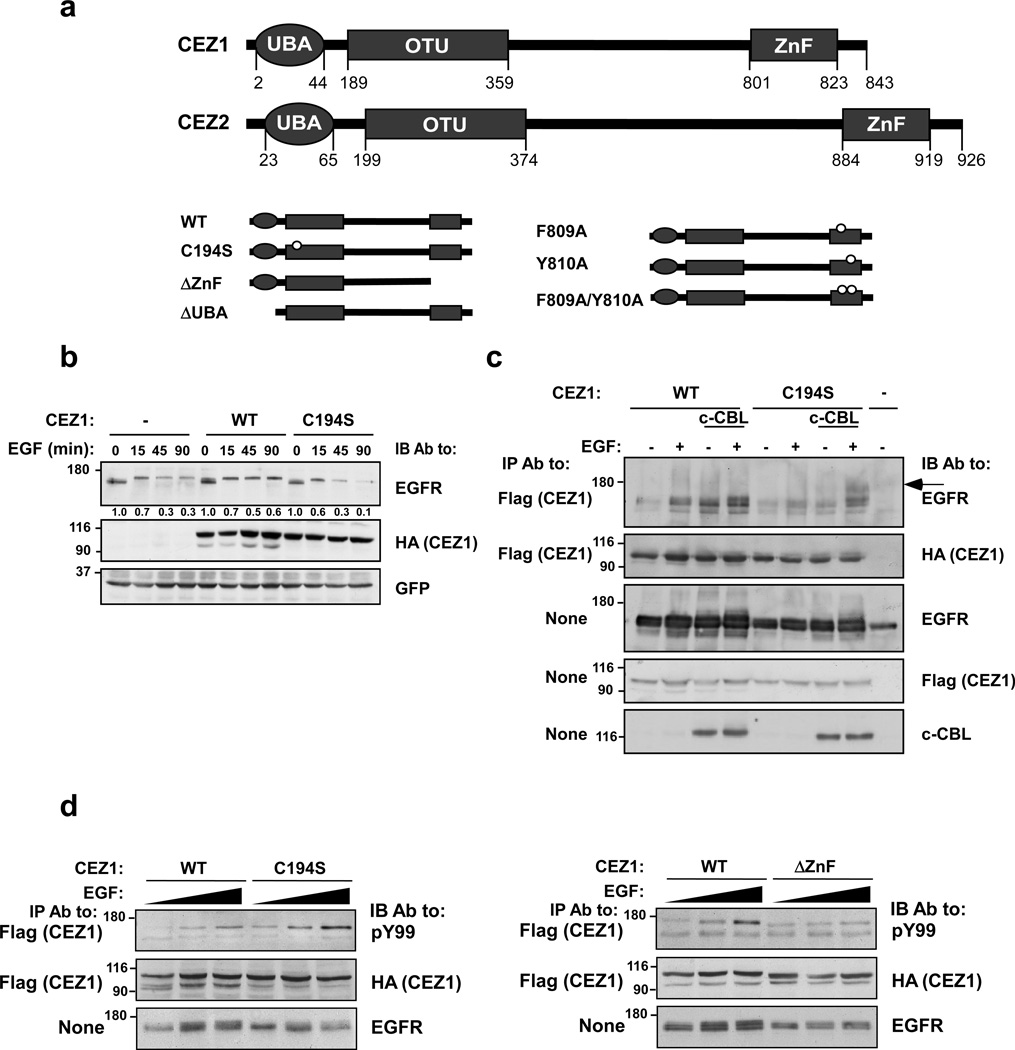
Cezanne-1 and Cezanne-2 belong to the family of Otubains, which harbor a catalytically active deubiquitinating domain (OTU) and two ubiquitin-binding domains (UBDs): a ubiquitin-associated domain (UBA) and an A20-like Zinc Finger (ZnF; Fig. 2a). To study the functions of Cezanne-1, we generated a set of mutants: The catalytic cysteine of OTU was replaced by a serine, generating a catalytically inactive mutant (C194S). Likewise, we deleted the UBA and the ZnF (ΔUBA and ΔZnF). Previous studies defined the A20-like ZnF of Rabex as a UBD that employs a diaromatic patch to recognize a polar region centered on Asp58 of ubiquitin (Lee et al., 2006). Hence, we also mutated the respective diaromatic patch of Cezanne-1 (mutants denoted F809A and Y810A). Comparison of Cezanne-1 WT and C194S confirmed that overexpression of WT caused stabilization of EGFR following stimulation with EGF, but C194S accelerated EGFR degradation, in line with functionality of the catalytic domain (Fig. 2b).
Figure 2. Cezanne-1 physically binds with and stabilizes EGFR and undergoes EGF-induced phosphorylation.
(a) Schematic representations of the domain structures of Cezanne proteins. The following domains are depicted: Ubiquitin associated domain (UBA), otubain (OTU) and the Zinc Finger (ZnF). The mutants of Cezanne-1 used in this study (lower panel) include a point mutant in the OTU domain, a deletion mutant lacking the ZnF, a deletion of the UBA domain, and single and double point mutants within the ZnF. (b) HEK-293T cells were transfected with EGFR- and GFP-encoding plasmids, along with plasmids encoding Cezanne-1, either WT or C194S. After forty-eight hours, cells were incubated at 37°C with EGF (20 ng/ml). Cell lysates were immunoblotted with the indicated antibodies. This experiment was repeated twice. (c) HEK-293T cells were transfected with plasmids encoding EGFR, c-CBL or an empty plasmid. In addition, cells were transfected with the WT Cezanne-1 or C194S (Flag and HA tagged). Forty-eight hours later, cells were either stimulated with EGF (100 ng/ml) for 1 hour at 4°C, or left untreated. Thereafter, cleared cell extracts were subjected to immunoprecipitation (IP) with the indicated antibodies, or directly electrophoresed. An arrow indicates a form of EGFR whose mobility is retarded. This experiment was repeated thrice. (d) The indicated forms of HA-Flag-tagged Cezanne-1 were co-expressed together with EGFR in HEK-293T cells. Forty-eight hours after transfection, cells were treated for 10 minutes with increasing concentrations of EGF (0, 10 and 50 ng/ml) and cell lysates subjected to immunoprecipitation (IP) and immunoblotting (IB) using the indicated antibodies.
Next, we sought to determine if Cezanne-1 and EGFR maintain bi-directional enzyme-substrate interactions. As a first step, we overexpressed WT and C194S, along with EGFR, in HEK-293T cells. In addition, to enhance ubiquitination, we also transfected c-CBL, and stimulated the cells under conditions that partly inhibit EGFR degradation. This experiment suggested that both forms of Cezanne-1 can co-immunoprecipitate with the un-stimulated EGFR molecule, and this interaction can be enhanced upon EGF stimulation (Fig. 2c). Co-expression of c-CBL, which leads to increased receptor ubiquitination, further strengthened the interaction. Interestingly, C194S displayed slightly weaker co-precipitation. Nevertheless, when combined with c-CBL, this mutant precipitated a high molecular weight form of EGFR, probably the ubiquitinated version (marked by an arrow; Fig. 2c). These observations implied that Cezanne-1 pre-assembles with EGFR molecules. Upon activation by EGF, these interactions are enhanced, probably due to receptor ubiquitination. In line with this model, when we co-expressed EGFR and Cezanne-1 under conditions that inhibit receptor degradation due to overexpression, we observed EGF-induced tyrosine phosphorylation of Cezanne-1 (Fig. 2d). Importantly, a mutant lacking the ZnF lost the ability to undergo inducible phosphorylation, but the catalytically-defective C194S mutant retained tyrosine phosphorylation. These observations predicted rapid onset of EGFR-Cezanne complexes, followed by dissociation due to deubiquitination. Indeed, in a coimmunoprecipitation experiment of the endogenous EGFR and Cezanne-1, the peak time of complex formation was reached 5 minutes after stimulation, and this was followed by a rapid decay (Fig. 3a). In conclusion, EGF-induced and CBL-mediated ubiquitination of EGFR permit phosphorylation and recruitment of Cezanne-1 to multiple ubiquitins, but subsequent deubiquitination by Cezanne-1 dissociates the complex.
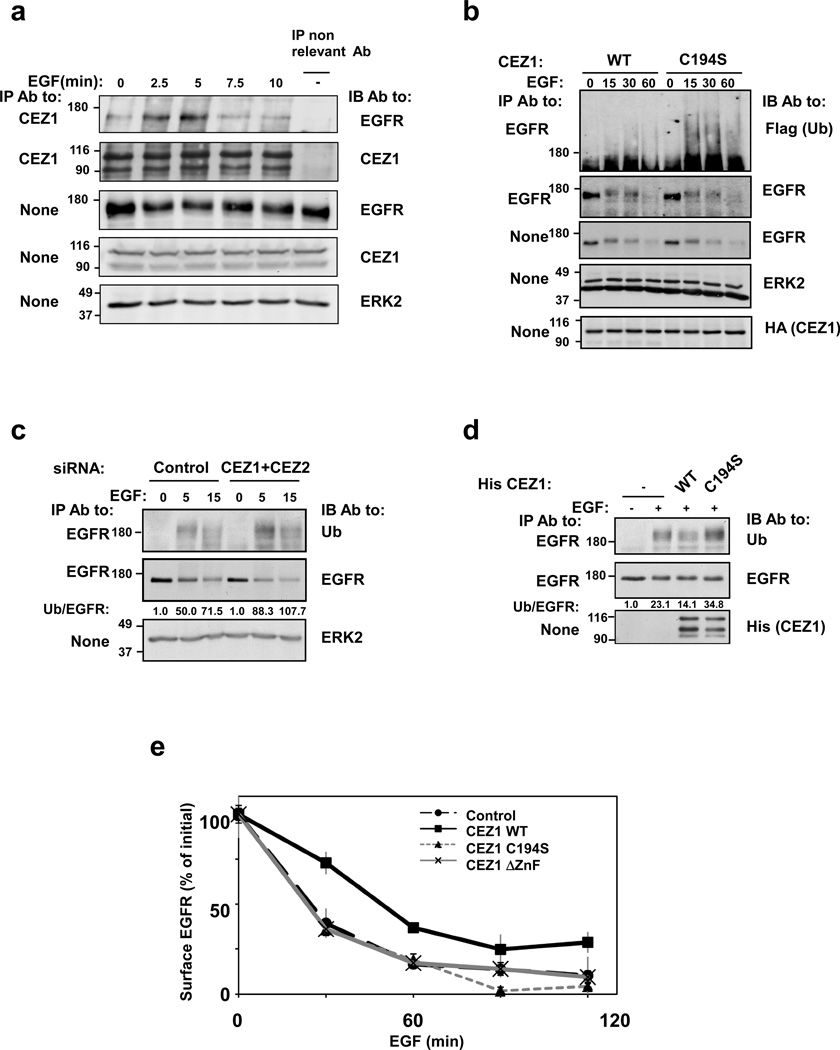
Figure 3. The catalytic and ubiquitin-binding functions of Cezanne-1 are essential for deubiquitination of EGFR and for inhibition of ligand-induced receptor downregulation.
(a) MDA-MB-468 cells were stimulated with EGF (20 ng/ml) for the indicated time points. Whole cell lysates were subjected to immunoprecipitation with anti-Cezanne-1 antibodies, or with a non-relevant antibody. Immunoprecipitates were immunoblotted with antibodies against EGFR and Cezanne-1. Whole cell extracts were probed with antibodies to EGFR, Cezanne-1, and ERK2. (b) HeLa cells were transfected with plasmids encoding Cezanne-1, either WT or C194S, together with a Flag-tagged ubiquitin. Thereafter, cells were stimulated with EGF (20 ng/ml). Cell extracts were subjected to immunoprecipitation (IP) and/or immunoblotting (IB) with the indicated antibodies. (c) HeLa cells were transfected with a mixture of siRNAs against Cezanne-1 and Cezanne-2, or with control oligonucleotides. Forty-eight hours after transfection, cells were incubated with EGF (20 ng/ml) and lysates were subjected to immunoprecipitation and immunoblotting as indicated. Signals were quantified, normalized and the ubiquitin/EGFR ratio indicated. (d) HeLa cells were stimulated for 15 minutes with EGF (20 ng/ml). Subsequently, cells were lysed and their extracts subjected to immunoprecipitation with anti-EGFR antibodies. Thereafter, EGFR immunoprecipitates were incubated for 1 hour at 37°C with the WT or C194S forms of a His-tagged Cezanne-1, previously purified from bacteria. The immunoprecipitates were subsequently immunoblotted with antibodies against ubiquitin and EGFR; signals were quantified, normalized and their ratio presented. (e) HeLa cells were transfected with plasmids encoding the indicated forms of Cezanne-1, or with an empty vector. Thereafter, cells were stimulated with EGF (2 ng/ml) for the indicated time intervals. Next, EGFR downregulation was assayed using a radioactive EGF. Averages of triplicates and S.D. values (bars) are presented. All experiments presented in this figure were repeated at least thrice.
Deubiquitination of EGFR by Cezanne-1 inhibits receptor downregulation
To ascertain the effect of Cezanne-1 on EGFR ubiquitination, HeLa cells were co-transfected with plasmids encoding Cezanne-1, either WT or a catalytically inactive mutant, and a Flag-tagged ubiquitin. The results presented in Figure 3b confirmed the necessity of the catalytic function of Cezanne-1 for deubiquitination of EGFR. The reciprocal experiment, in which we knocked-down both Cezanne-1 and Cezanne-2 detected increased ubiquitin/receptor ratios and enhanced rates of receptor degradation (Fig. 3c). In summary, the combination of gain- and loss-of-function approaches lent support to the conclusion that Cezanne family proteins regulate the level of EGFR ubiquitination.
To validate enzyme-substrate relations, we performed in vitro deubiquitination experiments. HeLa cells were stimulated with EGF, and EGFR was immunoprecipitated. In parallel, recombinant forms of a His-tagged Cezanne-1 (WT and C194S) were purified from bacteria, and incubated with the immunoprecipitated receptor. Blotting for ubiquitinated receptors confirmed that Cezanne-1 WT can deubiquitinate EGFR in vitro. In contrast, C194S showed no evidence of activity (Fig. 3d). Next we analyzed the effects of Cezanne-1-mediated deubiquitination on receptor endocytosis. HeLa cells were stimulated for up to 2 hours with physiological concentrations of EGF (2 ng/ml), and receptors remaining on the cell surface were quantified with a radioactive EGF. Overexpression of Cezanne-1 WT inhibited receptor downregulation at all time intervals. In contrast, both C194S and ΔZnF exerted no inhibitory effects and a third mutant, ΔUBA, resembled the phenotype induced by WT (Fig. 3e and data not shown). In conclusion, the deubiquitinating and ubiquitin-binding function of Cezanne-1 enable rescue of EGFR from intracellular degradation.
The Zinc Finger of Cezanne-1 interacts with the non-canonical ubiquitin face centered on aspartate 58
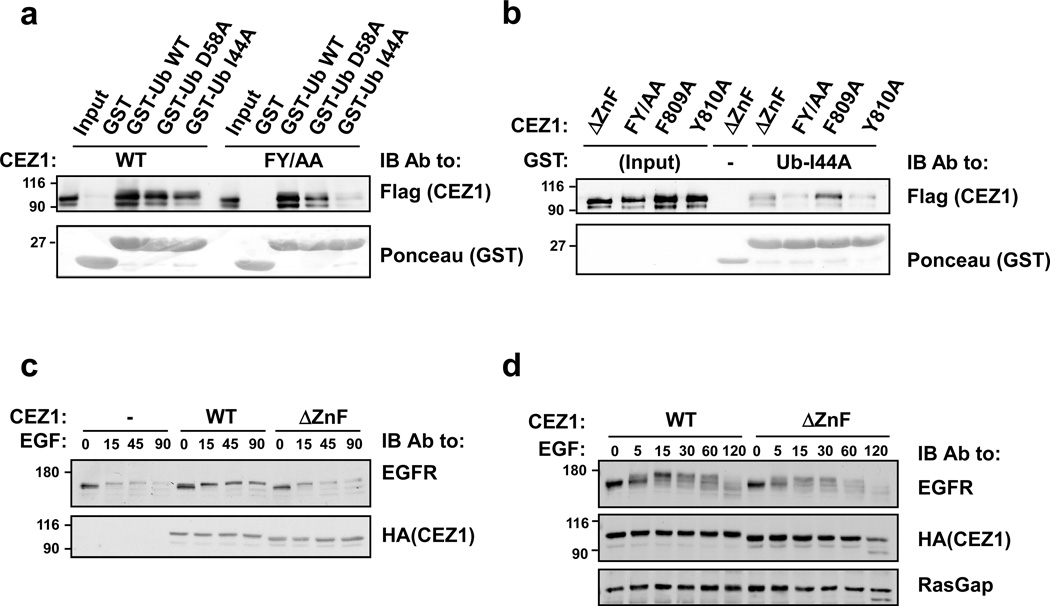
While many UBDs recognize the canonical surface of ubiquitin centered on Ile-44, ubiquitin may also be recognized via an Asp-58-centered interface (Lee et al., 2006; Penengo et al., 2006). To determine the specificity of the ZnF, we employed mutants of the diaromatic motif of the ZnF of Cezanne-1, the putative Asp-58 interfacing site, as well as ubiquitins mutated at Ile44 and Asp-58. As a first step, we incubated cell extracts containing Cezanne-1 (WT or FY/AA) with bacterially expressed GST fusion proteins of ubiquitin (Ub; WT, D58A, or I44A). The results of this experiment indicated that the WT form of Cezanne-1 specifically binds with either surface of ubiquitin, presumably because the UBA recognizes Ile-44 whereas the ZnF binds with Asp-58. This model was supported by the observed inability of FY/AA to recognize Ub-I44A (Fig. 4a). Further support was provided by an experiment that tested individual ZnF mutants: F809A (which bound Ub-I44A) and Y810A, which practically lost the ability to recognize Ub-I44A (Fig. 4b). To determine the affinity of monoubiquitin binding to the ZnF, we used Surface Plasmon Resonance. The GST-ZnF was immobilized on a CM-5 chip, and a flow of HA-monoubiquitin was injected at different concentrations, essentially as described before (Crosetto et al., 2008). The calculated dissociation constant (KD), 205 µM, for the ZnF-Ub complex (Supplementary Fig. S1) is well within the low affinity range of interactions compared to other UBDs (Dikic et al., 2009).
Figure 4. The Zinc Finger of Cezanne-1 binds with the Asp58-centered surface of ubiquitin.
(a) HEK-293T cells were transfected with plasmids encoding Flag-tagged Cezanne-1 (WT and FY/AA). Forty-eight hours later, lysates were incubated for 1 hour at 4°C with immobilized GST-ubiquitin, either WT, I44A or D58A. Pull-downs were stained or probed with an antibody to Flag. The Input lane represents 10% of the amount of cell extract used per lane. This experiment was repeated twice. (b) Lysates of HEK-293T cells transiently expressing Cezanne-1 were incubated for 1 hour at 4°C with an immobilized GST-I44A-Ub. Pull-downs were stained or probed with an antibody to Flag. The input extracts were electrophoresed for reference. This experiment was repeated twice. (c) HEK-293T cells were transfected with plasmids encoding EGFR and the indicated forms of Cezanne-1. Forty-eight hours later, cells were stimulated with EGF (20 ng/ml). Cleared extracts were probed as indicated. (d) HeLa cells were transfected with the indicated forms of Cezanne-1. After 48 hours cells were stimulated with EGF (20 ng/ml). Whole extracts were probed as indicated.
To evaluate the biological significance of the recognition of ubiquitin by the ZnF, we co-transfected cells with EGFR, along with either WT or ΔZnF Cezanne-1. As expected, the WT form of Cezanne-1 inhibited ligand-dependent degradation of EGFR. However, ΔZnF failed to inhibit receptor degradation (Fig. 4c), in line with a critical role for ubiquitin binding. A similar analysis, in which we applied a more detailed time course, proposed that the ZnF is essential for Cezanne-1 mediated retardation of EGFR degradation (Fig. 4d), possibly by binding to the Asp-58 surface of ubiquitins decorating internalized EGFRs. In summary, our data indicate that the A20-like ZnF of Cezanne-1 contacts ubiquitin via Asp-58, and that the interfacing residue involves Tyr-810 of Cezanne-1. Moreover, the Asp-58/Tyr-810 interaction is necessary for the stabilizing effect of Cezanne-1 toward EGFR.
Cezanne-1 physically interacts with HECT domain ubiquitin ligases involved in EGFR endocytosis
Nedd4 and AIP4 are WW domain ubiquitin ligases, which share catalytic HECT domains (see a scheme of Nedd4 in Supplementary Fig. S2a) and regulate EGFR endocytosis by coupling to CBL (Courbard et al., 2002; Magnifico et al., 2003). To investigate possible interactions with Cezanne-1, we performed coimmunoprecipitation experiments by co-overexpressing these proteins in HEK-293T cells. Cezanne-1 physically interacted with AIP4, as well as with both the intact and a catalytically disabled mutant of Nedd4 (Supplementary Figs. S2b and S2c). However, deletion of the WW domains of Nedd4 abolished interactions. These results are consistent with findings reported by a previous mass-spectrometry study (Sowa et al., 2009), and they reinforce the notion that Cezanne-1 regulates receptor endocytosis.
Cezanne-1 associates with cellular proliferation, migration and malignancy of breast lesions
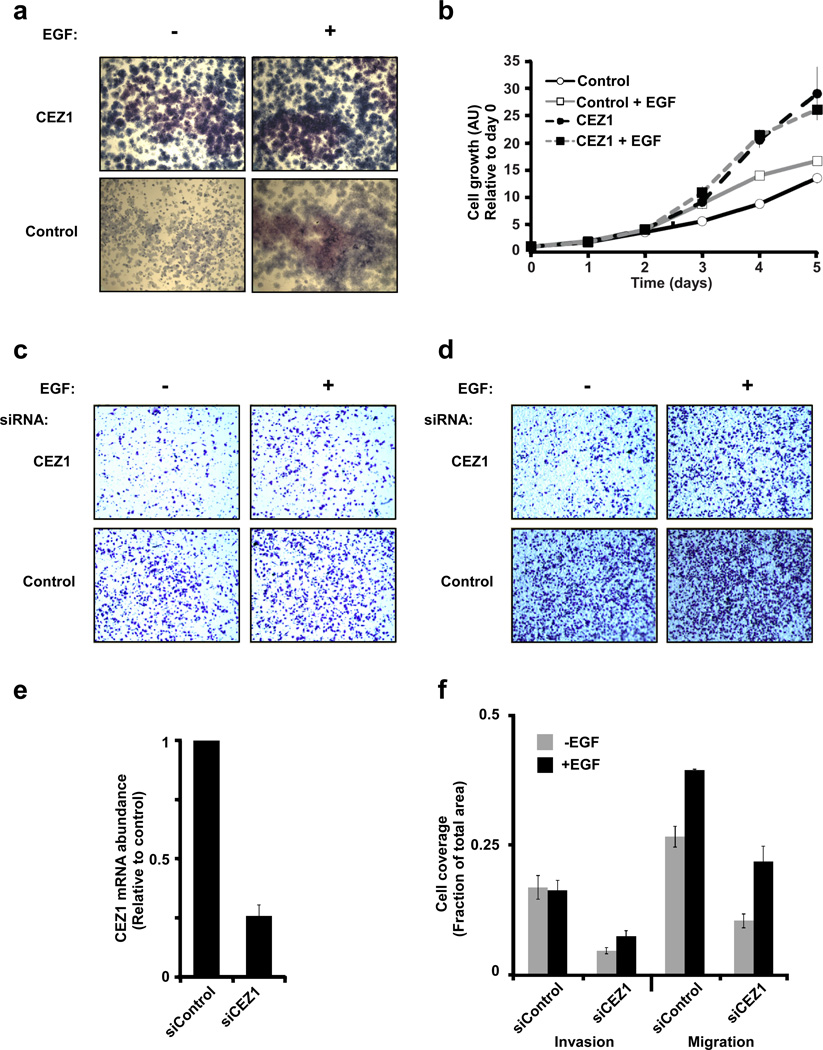
To address the consequences of Cezanne-1-mediated stabilization of active EGFRs, we established HeLa clones stably overexpressing the enzyme, and performed a clonogenic assay under anchorage-dependent conditions. As expected, overexpression of the DUB resulted in more abundant and larger colonies (Fig. 5a). The reason for these differences was provided by a cell growth assay: whereas control cells reached saturation, overexpression of Cezanne-1 released cells from density-dependent arrest (Fig. 5b). Interestingly, stimulation with EGF exerted relatively minor effects on cell growth under the conditions we examined. Hence, we tested the ability of EGF and Cezanne-1 to increase an alternative outcome, namely migration and invasion of mammary tumor cells. MDA-MB-231 cells were first transfected with siRNA oligonucleotides and 24 hours later they were plated on porous filters, which were either uncoated (migration assay) or pre-coated with an extracellular matrix (Matrigel; invasion assay). Cells that migrated through the filter toward EGF in the lower compartment were stained and photographed (Figs. 5c and 5d). Importantly, pretreatment with a pool of oligonucleotides directed at Cezanne-1 effectively reduced expression of the enzyme as determined by PCR (Fig. 5e), as well as reduced both invasion and migration (Fig. 5f), but once again EGF exerted only limited effects.
Figure 5. Cezanne-1 associates with cell proliferation and migration.
(a and b) HeLa cells stably overexpressing Cezanne-1 (or an empty plasmid) were plated, allowed to adhere overnight, and stimulated with EGF (20 ng/ml). Subsequently, the cells were allowed to grow for 7 days, fixed and stained with Giemsa prior to image capturing (a). Alternatively, cells were stimulated with EGF (20 ng/ml) and grown for the indicated intervals prior to an assay that determined cell growth (b). The results represent average±S.D. values of quadruplets. (c and d) MDA-MB-231 cells were transfected with the indicated siRNA oligonucleotides and 24 hours later they were seeded in the upper compartment of an Invasion (c) or a Transwell (d) chamber. The lower compartment of each chamber was filled with either serum-containing medium or the same medium supplemented with EGF (10 ng/ml). Eighteen hours later, cells that migrated to the lower side of the filter were fixed, permeabilized and stained. (e) MDA-MB-231 cells were transfected with siRNA oligonucleotides as in c and d, and expression levels of Cezanne-1 were determined using a preparation of mRNA and PCR. (f) Quantification of cell invasion and migration (panels c and d) by measuring the fraction of the lower face of the intervening filter which was covered by migrating cells. Bars show the means and S.D. values of three determinations.
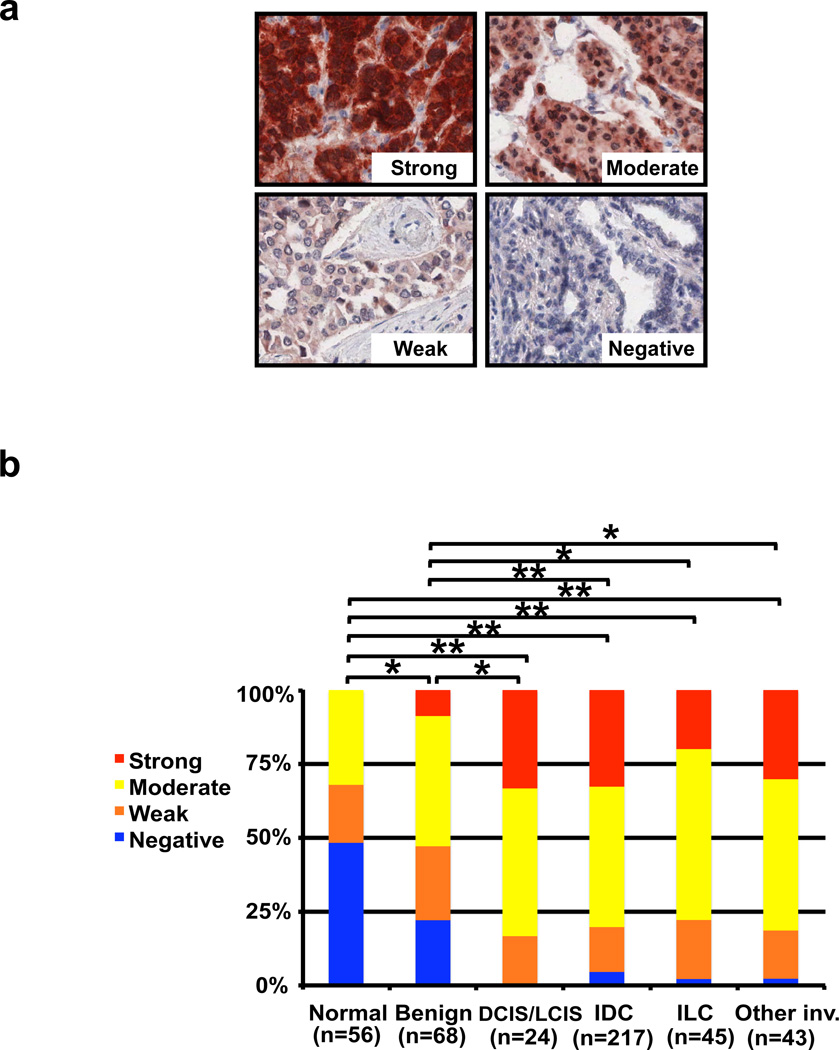
Because these observations raised the possibility that Cezanne-1 harbors attributes of an oncoprotein, we examined potential associations with markers of disease aggressiveness. To enable analyses of clinical specimens, we generated a monoclonal antibody by immunizing mice with a recombinant human Cezanne-1. Thereafter, we retrieved a breast tissue microarray, which included normal breast, fibrocystic mastopathy, atypical ductal hyperplasia (ADH), ductal and lobular carcinoma in situ (DCIS and LCIS) and several types of invasive carcinoma. In addition to hematoxylin-eosin stains, immunohistochemical analyses were carried out using our anti-Cezanne-1 monoclonal antibody. Cezanne-1 expression was scored as strong (3), moderate (2), weak (1) or negative (Fig. 6a). The scores resulting from our analyses revealed consistently higher expression of Cezanne-1 in malignant mammary tissues, as compared to normal breast or benign disease (Fig. 6b). Notably, this increase in Cezanne-1 expression could be observed already in early forms of the disease, such as DCIS and LCIS, but we cannot exclude association of the DUB with signaling pathways other than EGFR. Taken together with our biochemical and cell growth analyses, these results strongly implicate Cezanne-1 in breast cancer progression.
Figure 6. Cezanne-1 associates with advanced malignancy of breast lesions.
(a) Photomicrographs presenting scores of Cezanne-1 expression in breast cancer specimens, qualified as strong, moderate, weak and negative. (b) The abundance of Cezanne-1 is compared to the following disease subgroups (the respective numbers of cases are indicated): normal breast, benign disease, ductal and lobular carcinoma in situ (DCIS/LCIS), invasive ductal carcinoma (IDC), invasive lobular carcinoma (ILC), and other invasive carcinomas. Statistical p values were calculated (Chi square test): * represents p <0.01, and ** represents p <0.001.
The gene encoding Cezanne-1 is frequently amplified in mammary tumors, and enhanced levels of the respective mRNA predict shorter patient survival
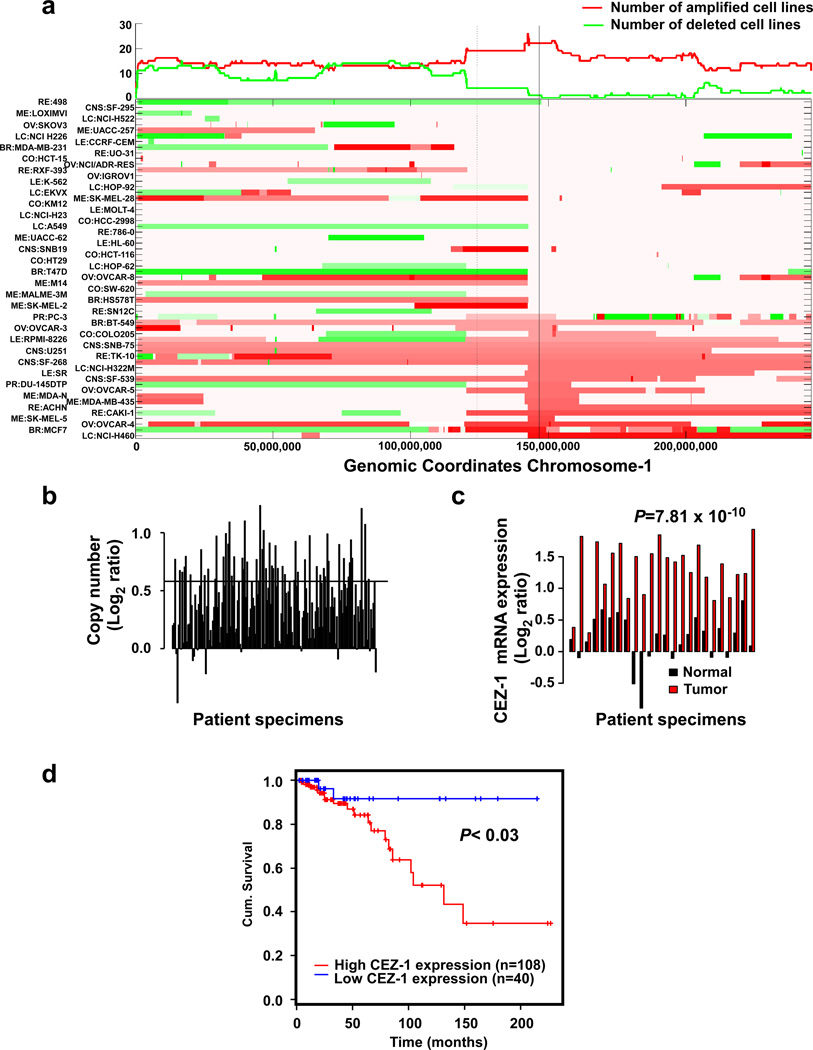
The remarkably variable abundance of Cezanne-1 in breast tissues (Fig. 6b) prompted us to analyze gene copy numbers in a collection of 60 human tumor cell lines (NCI-60). The corresponding array Comparative Genomic Hybridization (aCGH) data indicated that the gene, which is localized to 1q21.2, is amplified in one third of the cell lines (Fig. 7a and Supplementary Fig. S3a). It is notable that approximately 50% of breast tumors present gains at 1q, whereas the short arm presents mostly losses (Courjal and Theillet, 1997; Hawthorn et al., 2010; Orsetti et al., 2006). Indeed, immunoblotting of a set of tumor and normal mammary cell lines detected variable abundance of the characteristic bands of Cezanne-1 (Supplementary Figs. S3b and S3c). Hence, in the next step we analyzed a cohort of 173 breast tumors reported by The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov), and found that 32% presented a DNA amplification corresponding to the CEZ1 gene (Fig. 7b). We also analyzed the same cohort for correlation between mRNA levels and copy number of CEZ1, and found positive correlation (Pearson correlation coefficient r=0.46, p=7.75e-11). Hence, we compared mRNA abundance in tumors versus the adjacent normal tissues. The results we obtained (Fig. 7c) clearly indicated that Cezanne-1 is overexpressed in tumors compared to paired adjacent normal tissues. Importantly, analysis of 148 breast cancer patients from the same cohort, for whom clinical data was available, revealed that high abundance of the corresponding mRNA predicts shorter patient survival time (Fig. 7d). Collectively, the results we obtained using clinical data offer a model attributing to receptor deubiquitination and escape from degradation critical roles in promoting tumor aggressiveness.
Figure 7. Genomic amplification and high mRNA levels of Cezanne-1 in mammary tumors, and their association with shorter patient survival time.
(a) Heatmap representation of chromosome 1 aberrations, based on the NCI60 set of cancer cell lines. The x-axis represents genomic coordinates, including the centromer (dashed vertical line) and CEZ1 (solid line). The top panel summarizes the number of amplified (or deleted) cell lines. Each row in the bottom panel represents a cell line. The left axis lists all lines, sorted according to CEZ1 copy number. Amplifications are marked in red, deletions in green. The scale of colors is based on the log ratio between the respective cell line and a normal sample (log ratio of zero is shown in white). A CNV (at 149,400,000) deleted in 26 samples was not included. (b) DNA copy number of CEZ1 in tumors from breast cancer patients. The x-axis represents 173 patients, 56 of them have gained at least 1 extra copy of CEZ1. The y-axis represents log2 ratio of SNP intensity using paired blood-derived normal cells as control. The horizontal line represents log2 ratio value for 3 copies. (c) mRNA abundance of Cezanne-1 in pairs of a breast tumor and adjacent normal tissues from 24 breast cancer patients. The statistical p value, determined by using the paired two-sample T test, is indicated. (d) Kaplan-Meyer survival analysis was performed on 148 breast cancer patients. All patients were divided into high and low CEZ1 expression groups. The cutoff was optimized to achieve low p-value.
DISCUSSION
Early feedback loops controlling EGFR depend on covalent modifications, whereas late processes rely on newly synthesized proteins, and aberrant variants of both are implicated in cancer (Avraham and Yarden, 2010). Concentrating on the early loops, we previously identified DEP-1, a tyrosine phosphatase often deleted in carcinomas, as an enzyme that inactivates EGFR (Tarcic et al., 2009). Because phosphorylation and ubiquitination of EGFR are coupled, we assumed that the tumor suppressor function of DEP-1 would be mirrored by oncogenic functions of EGFR-specific DUBs. In line with this prediction, overexpression of the DUB we identified, Cezanne-1, is associated with a relatively aggressive course of breast cancers. Like DEP-1, Cezanne-1 assembles with EGFR prior to stimulation, and this physical interaction is enhanced by EGF (Fig. 3a), probably due to interactions of the two UBDs of Cezanne-1 with ubiquitins that decorate active EGFRs. Interestingly, our results attribute different binding specificities to the two UBDs of Cezanne-1: whereas the UBA binds with the canonical isoleucine-44 aspect of ubiquitin, the ZnF binds with aspartate-58. It is worth noting that A20, a negative regulator of the NFκB pathway, shares with Cezanne-1 both a ZnF and a catalytic OTU domain. This multifunctional enzyme acts as an editing enzyme: the enzyme first removes Lys-63-linked polyubiquitins from its substrate, RIP, and then it attaches to RIP Lys-48-linked polyubiquitins via its ZnF (Wertz et al., 2004). Whether Cezanne-1 similarly edits ubiquitinated EGFRs remains an intriguing question. Our results propose that this DUB rapidly associates with active receptors, once they undergo ubiquitination. Subsequently, Cezanne-1 d e-ubiquitinates the receptors, thereby delaying their degradation and prolonging growth and motility signals.
Certain genomic amplicons have been associated with vesicle recycling in tumors (Mills et al., 2009). For example, 8p11-12 is frequently amplified in breast cancer. This region encodes the RAB-coupling protein, RCP, which confers oncogenic properties by shunting integrins and other receptors to a recycling route (Zhang et al., 2009). Likewise, RAB23 is encoded by the 6p11 amplicon and contributes to invasiveness of gastric cancer (Hou et al., 2008). The long arm of chromosome 1 is involved in quantitative anomalies in at least 50% of breast tumors. 1q22 encodes RAB25, an oncogenic partner of RCP, which is overexpressed in breast and in ovarian cancer (Cheng et al., 2004). A family member of RAB25, called RAB11A, is overexpressed in DCIS (Palmieri et al., 2006), and yet another component of the recycling route, RAB4A, is encoded by the 1q42 amplicon (Orsetti et al., 2006). The identification of Cezanne-1 as a regulator of EGFR trafficking adds a deubiquitination enzyme to the list of amplicon-encoded proteins that inhibit receptor degradation. Conceivably, amplicon-driven overexpression of Cezanne-1 results in deubiquitination of surface-bound or internalized EGFRs, thereby enhancing their firing in tumors. In addition, because recycling mechanisms are shared by several growth factor receptors and by adhesion molecules like integrins, overexpression of Cezanne-1 is expected to enhance recycling of multiple endosomal cargos, especially in the leading edge of tumor cells, thereby promoting cellular motility and invasion (Caswell et al., 2008). Validation of these predictions is expected to motivate pharmacological strategies to intercept Cezanne-1 in tumors.
MATERIALS AND METHODS
Reagents and antibodies
Unless indicated, siRNA oligonucleotides were acquired from Dharmacon (Lafayette, CO, USA), antibodies from Santa Cruz Biotechnology (Santa Cruz, CA, USA) and radioactive materials from IZOTOP (Budapest, Hungary).
DNA constructs and mutagenesis
The following plasmids were kind gifts: pHM6 encoding Cezanne-1 (from Paul Evans; Imperial College, London, UK), pIND encoding Nedd4 (from Marius Sudol; Danville, PA, USA), and pRK5 encoding AIP4 (from Daniel Birnbaum; Marseille, France). Mutations were introduced using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, Agilent Technologies, La Jolla, CA, USA). To clone GST fusion proteins, the relevant DNA sequences were cloned into a pGEX 4T1 vector (GE Healthcare, Uppsala, Sweden).
Cell lines and transfections
HeLa and KB cells were transfected using JetPEI (Polyplus transfection, Illkirch, France) or HiPerFect (Qiagen, Hilden, Germany). HEK-293T cells were transfected using calcium phosphate. To establish stable sub-lines, cells were co-transfected with a pLenti6 vector (Invitrogen, Carlsbad, CA, USA) harboring Cezanne-1 (Sigma, St Louis, MO, USA), together with a mixture of packaging plasmids. Following an overnight incubation, the medium was refreshed, and 48 hours after transfection, virus-containing media were collected. Subsequently, target cells were infected with lentiviruses in the presence of hexadimethrine bromide (8 µg/ml). Twenty-four hours later, cells were subjected to drug selection for approximately 10 days.
siRNA screen for deubiquitinating enzymes
KB cells were seeded in 6 well plates and transfected with oligonucleotides of siRNA oligonucleotides (5 nM) targeting the majority of human deubiquitinating enzymes, or with control oligonucleotides. Forty-eight hours later, cells were stimulated with EGF (20 ng/ml) for up to 120 minutes. Whole cell lysates were immunoblotted with anti-EGFR antibodies to measure the degradation of EGFR.
Immunoprecipitation and immunoblotting
Cells were solubilized in lysis buffer (50 mM Hepes pH 7.5, 150mM NaCl, 10% glycerol, 1% Triton X-100, 1mM EDTA, 1mM EGTA, 10mM NaF and 30 mM beta-glycerol phosphate) with freshly added 0.2 mM Na3VO4 and a protease inhibitor cocktail. Electroblotted proteins were detected with antibodies linked to horseradish peroxidase (HRP), and visualized using reagents from Amersham (Buckinghamshire, UK).
Receptor downregulation assays
Forty-eight hours after transfection, cells were starved for 4 hours and stimulated with EGF (2 ng/ml) at 37°C. Subsequently, the cells were placed on ice, rinsed with binding buffer (DME medium containing 20mM Hepes buffered at pH 7.5 and 1% albumin), and subjected to mild acid wash (0.2 M Na-acetate buffer pH 4.5, 0.5 M NaCl). Thereafter, cells were incubated at 4°C with a radioactive EGF for 1.5 hours. The control well was incubated with a radioactive EGF and an excess (50X) of unlabeled EGF. Finally, cells were lysed in 1M NaOH, and radioactivity was determined.
Colony formation and cell viability assays
For colony formation assays, cells (25 × 103) were plated in 10-cm plates and grown for 7 days. Subsequently, cells were fixed and stained with Giemsa (0.2% in saline). For viability assays, cells were seeded in 96-well plates and cell proliferation was assayed every 24 hours. In brief, 20 µl of MTT (3-(4,5-dimethylthiazolyl-2)-2.5-diphenyltetrazolium bromide) in PBS was added to each well. Cells were incubated for two additional hours at 37°C, 0.180 ml of solubilization solution (isopropanol in 40mM HCl) was added, and O.D.595 was determined. The experiments were performed in quadruplets.
Surface plasmon resonance measurements
Binding of the ZnF to ubiquitin was analyzed at 25°C, using a Biacore X100 system (GE Healthcare). Activation of the CM5 chip was achieved with 1:1 N-hydroxysuccinimide/1-ethyl-3-(3-dimethylaminopropyl) carbodiimide. GST and a fusion protein comprising the ZnF of Cezanne-1 (8000 response units of each) were immobilized on the sensor chip by covalent linkage to the N-terminus of GST. GST-fused proteins were passed over separate flow cells in 10 mM Na-acetate buffer (at pH 5.0), followed by a blocking step performed with ethanolamine (1M; pH 8.5). We measured the binding of mono-ubiquitin to the ZnF of Cezanne-1 by injecting it over the chip at different concentrations. Surface regeneration between subsequent injections of ubiquitin was done with 10 mM NaOH.
Immunohistochemical analyses
Prior to staining of tissue microarrays, antigen retrieval was achieved by pretreatment with citrate buffer for all the antibodies, except for Cezanne-1 staining, for which slides were pretreated with Pronase E. Staining was performed using a DAKO Tech Mate Horizon automated immunostainer.
Statistical analysis of clinical data
The gene expression data of tumors was obtained from analyses performed at the University of North Carolina, using Agilent 244K Custom Gene Expression G4502A-07-3. Data on copy number variations in breast cancer samples was derived from analyses performed at the Broad Institute (Cambridge, MA, USA) using Affymetrix Genome-Wide Human SNP Array 6.0. Other Analyses used the SPSS statistics 17.0 package. Statistical tests used a two-tailed significance level of p<0.05.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Tissue Bank of the National Center for Tumor Diseases, Heidelberg University Hospital (Heidelberg, Germany). Our research is supported by grants from the National Cancer Institute (5R37CA072981, CCSG and P30 CA16672), the European Commission, the German-Israeli Project Cooperation, the Israel Cancer Research Fund, the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation and the M.D. Moross Institute for Cancer Research. Y.Y. is the incumbent of the Harold and Zelda Goldenberg Professorial Chair.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary information is available at Oncogene’s web site (http://www.nature.com/onc).
REFERENCES
- Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol. 2010;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- Bowers K, Piper SC, Edeling MA, Gray SR, Owen DJ, Lehner PJ, et al. Degradation of endocytosed epidermal growth factor and virally ubiquitinated major histocompatibility complex class I is independent of mammalian ESCRTII. J Biol Chem. 2006;281:5094–5105. doi: 10.1074/jbc.M508632200. [DOI] [PubMed] [Google Scholar]
- Caswell PT, Chan M, Lindsay AJ, McCaffrey MW, Boettiger D, Norman JC. Rab-coupling protein coordinates recycling of alpha5beta1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J Cell Biol. 2008;183:143–155. doi: 10.1083/jcb.200804140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng KW, Lahad JP, Kuo WL, Lapuk A, Yamada K, Auersperg N, et al. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med. 2004;10:1251–1256. doi: 10.1038/nm1125. [DOI] [PubMed] [Google Scholar]
- Courbard JR, Fiore F, Adelaide J, Borg JP, Birnbaum D, Ollendorff V. Interaction between two ubiquitin-protein isopeptide ligases of different classes, CBLC and AIP4/ITCH. J Biol Chem. 2002;277:45267–45275. doi: 10.1074/jbc.M206460200. [DOI] [PubMed] [Google Scholar]
- Courjal F, Theillet C. Comparative genomic hybridization analysis of breast tumors with predetermined profiles of DNA amplification. Cancer Res. 1997;57:4368–4377. [PubMed] [Google Scholar]
- Crosetto N, Bienko M, Hibbert RG, Perica T, Ambrogio C, Kensche T, et al. Human Wrnip1 is localized in replication factories in a ubiquitin-binding zinc finger-dependent manner. J Biol Chem. 2008;283:35173–35185. doi: 10.1074/jbc.M803219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex JE, Sorkin A. RNA interference screen identifies Usp18 as a regulator of epidermal growth factor receptor synthesis. Mol Biol Cell. 2009;20:1833–1844. doi: 10.1091/mbc.E08-08-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enesa K, Zakkar M, Chaudhury H, Luong le A, Rawlinson L, Mason JC, et al. NF-kappaB suppression by the deubiquitinating enzyme Cezanne: a novel negative feedback loop in pro-inflammatory signaling. J Biol Chem. 2008;283:7036–7045. doi: 10.1074/jbc.M708690200. [DOI] [PubMed] [Google Scholar]
- Evans PC, Smith TS, Lai MJ, Williams MG, Burke DF, Heyninck K, et al. A novel type of deubiquitinating enzyme. J Biol Chem. 2003;278:23180–23186. doi: 10.1074/jbc.M301863200. [DOI] [PubMed] [Google Scholar]
- Fehrenbacher N, Bar-Sagi D, Philips M. Ras/MAPK signaling from endomembranes. Mol Oncol. 2009;3:297–307. doi: 10.1016/j.molonc.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosi Y, Anastasi S, Ballaro C, Varsano G, Castellani L, Maspero E, et al. A two-tiered mechanism of EGFR inhibition by RALT/MIG6 via kinase suppression and receptor degradation. J Cell Biol. 2010;189:557–571. doi: 10.1083/jcb.201002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh LK, Huang F, Kim W, Gygi S, Sorkin A. Multiple mechanisms collectively regulate clathrin-mediated endocytosis of the epidermal growth factor receptor. J Cell Biol. 189:871–883. doi: 10.1083/jcb.201001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh LK, Huang F, Kim W, Gygi S, Sorkin A. Multiple mechanisms collectively regulate clathrin-mediated endocytosis of the epidermal growth factor receptor. J Cell Biol. 2010;189:871–883. doi: 10.1083/jcb.201001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol. 2003;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- Hawthorn L, Luce J, Stein L, Rothschild J. Integration of transcript expression, copy number and LOH analysis of infiltrating ductal carcinoma of the breast. BMC Cancer. 2010;10:460. doi: 10.1186/1471-2407-10-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Q, Wu YH, Grabsch H, Zhu Y, Leong SH, Ganesan K, et al. Integrative genomics identifies RAB23 as an invasion mediator gene in diffuse-type gastric cancer. Cancer Res. 2008;68:4623–4630. doi: 10.1158/0008-5472.CAN-07-5870. [DOI] [PubMed] [Google Scholar]
- Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell. 2006;21:737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Katz M, Shtiegman K, Tal-Or P, Yakir L, Mosesson Y, Harari D, et al. Ligand-independent degradation of epidermal growth factor receptor involves receptor ubiquitylation and Hgs, an adaptor whose ubiquitin-interacting motif targets ubiquitylation by Nedd4. Traffic. 2002;3:740–751. doi: 10.1034/j.1600-0854.2002.31006.x. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Lee S, Tsai YC, Mattera R, Smith WJ, Kostelansky MS, Weissman AM, et al. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat Struct Mol Biol. 2006;13:264–271. doi: 10.1038/nsmb1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- Ma YM, Boucrot E, Villen J, Affar el B, Gygi SP, Gottlinger HG, et al. Targeting of AMSH to endosomes is required for epidermal growth factor receptor degradation. J Biol Chem. 2007;282:9805–9812. doi: 10.1074/jbc.M611635200. [DOI] [PubMed] [Google Scholar]
- Magnifico A, Ettenberg S, Yang C, Mariano J, Tiwari S, Fang S, et al. WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. J Biol Chem. 2003;278:43169–43177. doi: 10.1074/jbc.M308009200. [DOI] [PubMed] [Google Scholar]
- McCullough J, Clague MJ, Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills GB, Jurisica I, Yarden Y, Norman JC. Genomic amplicons target vesicle recycling in breast cancer. J Clin Invest. 2009;119:2123–2127. doi: 10.1172/JCI40256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosesson Y, Shtiegman K, Katz M, Zwang Y, Vereb G, Szollosi J, et al. Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J Biol Chem. 2003;278:21323–21326. doi: 10.1074/jbc.C300096200. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Tanaka N, Kitamura N, Komada M. Clathrin anchors deubiquitinating enzymes, AMSH and AMSH-like protein, on early endosomes. Genes Cells. 2006;11:593–606. doi: 10.1111/j.1365-2443.2006.00963.x. [DOI] [PubMed] [Google Scholar]
- Orsetti B, Nugoli M, Cervera N, Lasorsa L, Chuchana P, Rouge C, et al. Genetic profiling of chromosome 1 in breast cancer: mapping of regions of gains and losses and identification of candidate genes on 1q. Br J Cancer. 2006;95:1439–1447. doi: 10.1038/sj.bjc.6603433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri D, Bouadis A, Ronchetti R, Merino MJ, Steeg PS. Rab11a differentially modulates epidermal growth factor-induced proliferation and motility in immortal breast cells. Breast Cancer Res Treat. 2006;100:127–137. doi: 10.1007/s10549-006-9244-6. [DOI] [PubMed] [Google Scholar]
- Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, et al. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183–1195. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, et al. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416:451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- Row PE, Prior IA, McCullough J, Clague MJ, Urbe S. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J Biol Chem. 2006;281:12618–12624. doi: 10.1074/jbc.M512615200. [DOI] [PubMed] [Google Scholar]
- Sacco JJ, Coulson JM, Clague MJ, Urbe S. Emerging roles of deubiquitinases in cancer-associated pathways. IUBMB Life. 2010;62:140–157. doi: 10.1002/iub.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol. 2009;10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarcic G, Boguslavsky SK, Wakim J, Kiuchi T, Liu A, Reinitz F, et al. An unbiased screen identifies DEP-1 tumor suppressor as a phosphatase controlling EGFR endocytosis. Curr Biol. 2009;19:1788–1798. doi: 10.1016/j.cub.2009.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira AV, Lamaze C, Schmid SL. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, et al. Deubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu X, Datta A, Govindarajan K, Tam WL, Han J, et al. RCP is a human breast cancer-promoting gene with Ras-activating function. J Clin Invest. 2009;119:2171–2183. doi: 10.1172/JCI37622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.