Abstract
Salmonella Enteritidis is the major foodborne pathogen that is primarily transmitted by contaminated chicken meat and eggs. We recently demonstrated that Salmonella Enteritidis strains from poultry differ in their ability to invade human intestinal cells and cause disease in orally challenged mice. Here we hypothesized that the differential virulence of Salmonella Enteritidis strains is due to the differential fitness in the adverse environments that may be encountered during infection in the host. The responses of a panel of six Salmonella Enteritidis strains to acid stress, oxidative stress, survival in egg albumen, and the ability to cause infection in chickens were analyzed. This analysis allowed classification of strains into two categories, stress-sensitive and stress-resistant, with the former showing significantly (p<0.05) reduced survival in acidic (gastric phase of infection) and oxidative (intestinal and systemic phase of infection) stress. Stress-sensitive strains also showed impaired intestinal colonization and systemic dissemination in orally inoculated chickens and failed to survive/grow in egg albumen. Comparative genomic hybridization microarray analysis revealed no differences at the discriminatory level of the whole gene content between stress-sensitive and stress-resistant strains. However, sequencing of rpoS, a stress-regulatory gene, revealed that one of the three stress-sensitive strains carried an insertion mutation in the rpoS resulting in truncation of σS. Finding that one of the stress-sensitive strains carried an easily identifiable small polymorphism within a stress-response gene suggests that the other strains may also have small polymorphisms elsewhere in the genome, which likely impact regulation of stress or virulence associated genes in some manner.
Introduction
Salmonella enterica serovar Enteritidis is the most common serotype of Salmonella isolated from cases of foodborne gastroenteritis throughout the world (EFSA, 2007; WHO, 2008). Chickens are the single largest reservoir host for Salmonella Enteritidis, and source attribution studies have determined that contaminated poultry and poultry products are the major sources of human infection (Kimura et al., 2004; Patrick et al., 2004). In the chicken host, Salmonella Enteritidis is transmitted via a fecal-oral route. Young chickens <2 weeks of age often develop gastroenteritis and systemic disease with varying degrees of mortality. In contrast, most adult hens become colonized with Salmonella Enteritidis, but typically remain asymptomatic carriers with intermittent fecal shedding of Salmonella. During intestinal colonization, inflammation ensues and is characterized by an early influx of heterophils (avian counterpart to mammalian neutrophils) and macrophages that play an important role in host defense (Henderson et al., 1999; Chappell et al., 2009). Salmonella Enteritidis has the ability to survive within these cells (Henderson et al., 1999; Stabler et al., 1994; Okamura et al., 2005) and also penetrate the mucosal epithelium, resulting in dissemination to organs, including spleen, liver, and reproductive tract (Desmidt et al., 1997; Gantois et al., 2009). Due to the specialized ability of Salmonella Enteritidis to colonize the avian reproductive tract and to contaminate internal contents of eggs, contaminated eggs or egg products have been implicated in the majority of foodborne outbreaks worldwide (Kimura et al., 2004; Patrick et al., 2004).
The specialized ability of Salmonella Enteritidis to contaminate and survive within the egg is important from the public health perspective. The potential for Salmonella Enteritidis to be deposited in egg contents primarily depends on the “virulence” of a particular strain, i.e., the ability of the bacterium to invade avian intestinal epithelium with subsequent dissemination to internal tissues (Gast and Benson, 1996; Gast, 1994; Gast and Beard, 1990). It has been reported that naturally occurring Salmonella Enteritidis strains vary in their virulence in chickens in terms of organ invasiveness (Gast and Benson 1995, 1996; Dhillon et al., 1999), and the ability to contaminate and survive within the egg contents (Petter, 1993; Clavijo et al., 2006; Yim et al., 2010). These differences in virulence exist irrespective of the phage type (Poppe, et al., 1993) or clonal lineages (Yim et al., 2010; Olsen et al., 1999). The basis for this differential virulence of Salmonella Enteritidis in the chicken is somewhat understood, because phenotypic variation has been traced to the level of the single nucleotide polymorphism rather than to differences in whole gene content (Morales et al., 2007).
Avian innate defenses play an important role in the outcome of infection. For instance, the two major avian innate defenses that impose severe stress on an invading Salmonella Enteritidis include the acidic stress from gastric acidity (pH 2.6) (Joyner and Kokas, 1971; Carter and Collins, 1974) and oxidative stress from effects of hydrogen peroxide (H2O2) produced by infected avian heterophils and macrophages (Qureshi et al., 2000; Withanage et al., 2005; Kogut et al., 2002). Furthermore, egg albumen contains several antibacterial substances such as lysozyme, ovatransferrin, and avian B-defensins (Van Immerseel, 2010; Kang et al., 2006), thereby presenting another detrimental environment for the survival and growth of this bacterium. Assessing the ability of Salmonella Enteritidis strains to survive under different stress conditions encountered early during infection in chickens may facilitate better understanding of the pathogenesis of Salmonella Enteritidis. Nevertheless, limited studies have been conducted to determine the responses of Salmonella Enteritidis to various stress conditions (Yim et al., 2010; Humphrey et al., 1993, 1995). For instance, Humphrey et al. (1995) reported that Salmonella Enteritidis strains isolated from poultry had reduced tolerance to acid and oxidative stress as compared with the strains isolated from human clinical cases. In another study, Yim et al. (2010) reported that vast majority of Salmonella Enteritidis strains isolated from food sources showed diminished survival in egg albumen as compared with the clinical strains. In addition, Salmonella Enteritidis produces a specialized capsular-like lipopolysaccharide O-antigen that contributes to the survival of the pathogen in the egg, but its production varies between strains (Guard-Bouldin et al., 2004). We hypothesized that differential virulence of Salmonella Enteritidis strains is due to the differential fitness in the stressful environments encountered in the host during infection. To address this hypothesis, we tested the ability of several Salmonella Enteritidis strains to survive under various stressful conditions that may be encountered in the chicken host such as acidic-stress, oxidative-stress, growth in egg albumen, and in vivo virulence in orally inoculated chickens.
Methods
Bacterial strains
A panel of six Salmonella Enteritidis strains (UK, G1, BC8, C19, C45, and G45) isolated from poultry or poultry-associated environments were analyzed in this study. Previous work from our laboratory showed that the UK (corresponds to the sequenced phage type-4 P125109 strain), G1 (phage type-4), and BC8 (phage type-8) strains differ from C19 (phage type-13), C45 (phage type-13), and G45 (phage type-13a) strains in terms of their invasiveness into human intestinal epithelial cells (Caco-2), virulence in orally challenged mice, motility, biofilm production, and phage types (Table 1) (Shah et al., 2011). Frozen stocks of cultures were grown on Luria-Bertani (LB) agar incubated at 37°C for overnight. For all the experiments, a single colony from the overnight culture was inoculated into LB broth and grown at 37°C or 42°C for 16 h with shaking at 200 rpm.
Table 1.
Phenotypic and Genotypic Characteristics of Poultry-Associated Salmonella Enteritidis Strains Used in This Study (Shah et al., 2011)
| Strain | Phage type | MLVA type | Caco-2 cell invasiveness | Survival in chicken macrophages | Motility | Biofilm |
|---|---|---|---|---|---|---|
| C19 | PT13 | 11 | Low | Low | Low | Negative |
| C45 | PT13 | 11 | Low | Low | Low | Negative |
| BC8 | PT8 | 9a | High | High | High | Mixed |
| G1 | PT4 | 13a | Medium | High | High | Positive |
| G45 | PT13a | 4a | Low | Low | Low | Negative |
| UK | PT4 | 13a | High | High | High | Positive |
MLVA, multi-locus variable-tandem repeat analysis.
Survival assays
The stationary phase (16 h) cultures of Salmonella Enteritidis strains were used to measure their survival rates in LB broth adjusted to pH 2.6±0.02 with 1 M HCl (Humphrey et al., 1993, 1995) and in normal saline (0.9% NaCl) after addition of H2O2 to a final concentration of 15 mM (Robbe-Saule et al., 2003). Controls included the cultures incubated in the same media but in the absence of stressors. Stationary phase cultures of Salmonella Enteritidis strains were used because this growth phase provides a reliable model to measure the differences in tolerance to acid and oxidative stress (Humphrey et al., 1995). All the media, tubes and reagents were prewarmed to avoid a sudden shift in temperatures that may otherwise markedly alter stress tolerance in Salmonella Enteritidis (Humphrey et al., 1993). For the acid stress survival assay, ∼1×108 colony forming units (CFU) of each Salmonella Enteritidis strain was inoculated in LB (pH 2.6) at 37°C or at 42°C followed by incubation at respective temperatures. For the oxidative stress survival assay, ∼1×108 CFU of the stationary phase cultures were resuspended in normal saline at either 37°C or 42°C. Subsequently, H2O2 was added to the final concentration of 15 mM, mixed thoroughly, and the suspension was incubated at 37°C or at 42°C. Aliquots from acid and oxidative stress cultures were collected at 10 min and 1 h post-inoculation. Serial 10-fold dilutions of each culture was prepared using maximum recovery diluent (MRD; Oxoid, Lenexa, KS) and plated on LB agar for enumeration of viable CFU. Each strain was tested in duplicate, and at least three independent experiments were completed.
Growth in egg albumen
The ability of Salmonella Enteritidis strains to survive in egg albumen was quantified as described previously with minor modifications (Lu et al., 2003). Briefly, organic, unfertilized, antibiotic-free eggs were purchased from Chino Valley Ranchers, CA and stored for less than 1 week at 4°C. For each experiment, two to three eggs were disinfected by immersion into 70% ethanol and aseptically broken to collect egg albumen into a sterile container. Egg albumen was pooled, and 1 mL aliquots were distributed into 96-well blocks (Qiagen, Valencia, CA). An overnight culture of each bacterial strain was added to egg albumen to a final concentration of ∼500 CFU/mL and thoroughly mixed and incubated at 25±2°C for 24 h. After incubation, the bacteria-albumen mixture was diluted in MRD and plated on LB agar to enumerate the viable bacterial counts. Uninoculated egg albumen served as a negative control. Five independent experiments were completed for each strain.
Comparative genomic hybridizations
The microarray was constructed by MYcroarray Inc. (Ann Arbor, MI) using the genomic information for the sequenced Salmonella Enteritidis PT4 P125109 (Thomson et al., 2008) which is referred to as the UK strain in this study. This microarray consists of a total of 16,320 features. Of these, 4,343 are control features (spike-in positive controls, empty spots, negative control probes, etc). The remaining 11,977 features (∼47-mers) represent 4,200 ORFs as follows: three specific probes/ORF (3,742 genes), two specific probes/ORF (293 genes), and one specific probe/ORF (165 genes). Three arrays per slide were generated by in situ synthesis using a proprietary light-directed oligonucleotide synthesis technology (www.mycroarray.com). Each strain was tested in triplicate (3 arrays/strain) using hybridization protocols as described previously (Call et al., 2003). Slides were scanned using a Genepix400B scanner (Axon Instruments, Inverurie, Scotland). After acquisition of signal intensity data, each spot was corrected by subtracting local background values and each “corrected” spot intensity was used to calculate median signal intensity for each array. Inferior spot intensities (empty spots or those where signal intensity was less than two times the background) were reset to an arbitrary number (50) and were not used in the median calculation. The array median intensity was then used for within array normalization. The data was log2 transformed and quantile normalized between arrays using SOLO (http://www-microarrays.u-strasbg.fr/Solo/index.html). Finally, the estimated probability of presence (EPP) for each probe was determined using a GACK-transformation as described previously (Kim et al., 2002). Microarray data and platform information have been submitted to GEO (under accession no. GSE33102).
rpoS gene sequencing
The total genomic DNA from each Salmonella Enteritidis strain was extracted using DNeasy Tissue kit (Qiagen) according to the manufacturer's protocol. The full length rpoS gene (993 bp) was amplified from all six strains using primers rpoS_F (5’-atgagtcagaatacgctgaaag-3’) and rpoS_R (5’-ttactcgcggaacagcgc-3’). PCR amplification included 28 cycles, each consisting of 30 s of denaturation at 96°C, 30 s of annealing at 50°C, and 45 s of extension at 72°C. PCR products were cloned into pGEM®-T Easy vector (Promega, Madison, WI) according to manufacturer's instructions. The rpoS gene insert was sequenced using universal reverse (5’-actttatgccggctcgtatgttgt-3’) and a universal forward (5’-atgtgtgcaaggcgattaagttggg-3’) primer at the Washington State University Genomics Core. The rpoS sequences were aligned against rpoS gene from a reference UK strain P125109 (GenBank accession no. AM933172) (Thomson et al., 2008). Sequence alignment and single nucleotide polymorphism were detected using Geneious Pro 5.3 (Biomatters Ltd., Auckland, New Zealand). The sequences of rpoS genes were deposited in the GenBank under the following accession numbers: JN588998 (UK), JN588996 (G1), JN588997 (BC8), JN588999 (C19), JN589000 (C45), and JN588995 (G45).
Chicken virulence assay
Twenty-one 1-day-old chickens were obtained from Belt Hatchery (Fresno, CA) and distributed in seven groups (n=3/group) in environmentally controlled isolation cages. Cloacal swabs were taken prior to placement of chicks in cages and examined for the presence of Salmonella by selective enrichment in tetrathionate broth (TTB; Difco, Franklin Lakes, NJ) followed by plating onto XLD agar (Dicfo). Antibiotic-free flock raiser diet (Purina, St. Louis, MO) and water was provided ad libitum throughout the experimental period. At 3 days of age, chickens were challenged orally with stationary phase cultures of Salmonella Enteritidis strains at a concentration of ∼107 CFU. One group served as the uninoculated negative control. The chickens in all groups were euthanized at 4 days post-infection, necropsied, and the small intestine, ceca, liver, and spleen were examined by selective enrichment in TTB followed by plating onto XLD-agar. The viable counts of Salmonella per gram in liver and spleen tissues from all the chickens were obtained by emulsification of weighed tissues in PBS followed by plating of serial dilutions of the ground tissues on XLD-agar plates. Presumptive Salmonella colonies were confirmed by slide agglutination test using serogroup D antiserum (Difco). The animal challenge experiments were conducted in accordance with the protocol approved by the Washington State University Institutional Animal Care and Use Committee (WSU IACUC).
Statistical analysis
Data was analyzed using one-factor analysis of variance (ANOVA) with Tukey's Kramer post-test in NCSS 2007 (NCSS, LLC, Kaysville, UT).
Results and Discussion
Because the ability of Salmonella to withstand stomach acidity is a recognized virulence attribute, we tested the survivability of stationary phase cultures of six Salmonella Enteritidis strains by substantially mimicking the avian stomach environment (pH 2.6) (Joyner and Kokas, 1971). While there were no differences in the survival of Salmonella Enteritidis strains in the absence of acid-stress (data not shown), significant differences in the tolerance of Salmonella Enteritidis strains to acid-stress were observed (Table 2). When 8 log CFU of Salmonella Enteritidis strains were exposed to pH 2.6 at 37°C for 10 min, the survival rate of UK, G1, and BC8 strains (>7 log CFU) was significantly higher (p<0.05, Tukey's Kramer test) as compared with the C19, C45, and G45 strains (<5 log CFU) (Table 2). At 1 h post-incubation, <1 log CFU of C19 and C45 and 1.2 log CFU of G45 strain survived, whereas >1.96 log CFU of the UK, G1, and BC8 strains survived this treatment. These differences were statistically significant (p<0.05). At 42°C (chicken body temperature), the counts of C19, C45, and G45 strains declined from an initial 8 log CFU to ≤1 log within 10 min. Thus, killing of stress-sensitive strains was more pronounced at the higher body temperature of the bird (Table 2). In contrast, >4 log CFU of the UK, G1, and BC8 strains survived this treatment, indicating that these strains were resistant to short-term acidic stress at 37°C and 42°C. We were not able to recover any of the Salmonella Enteritidis strains after 1 h of incubation at 42°C, suggesting that none of the strains survived prolonged acidic stress at high temperature. These results indicate that C19, C45, and G45 strains are acid-sensitive, whereas the UK, G1, and BC8 strains are relatively acid-resistant (Table 2).
Table 2.
Survival Values for High- and Low-Pathogenic Salmonella Enteritidis Strains Exposed to Acidic (pH 2.6) and Oxidative (15 mM H2O2) Stress
| |
Number of survivors (mean log10 CFU±SE)a |
|||||
|---|---|---|---|---|---|---|
| |
Stress-resistant |
Stress-sensitive |
||||
| Conditions | UK | BC8 | G1 | C19 | G45 | C45 |
| pH 2.6 for 10 min (37°C) | 7.33±0.4 | 7.08±0.43 | 7.33±0.36 | 4.05±1.01b | 4.84±0.75b | 4.14±1.06b |
| pH 2.6 for 1 h (37°C) | 1.96±1.15 | 2.88±1.67 | 3.21±1.35 | <0.01b | 1.2±0.41b | 0.09±0.09b |
| pH 2.6 for 10 min (42°C) | 5.35±0.9 | 4.28±1.29 | 5.02±1.17 | 0.66±0.5b | 1.02±0.65b | 1.01±0.64b |
| pH 2.6 for 1 h (42°C) | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 mM H2O2 for 10 min (37°C) | 8.07±0.04 | 8.07±0.06 | 8.06±0.05 | 6.1±0.18b | 5.53±0.19b | 6.33±0.19b |
| 15 mM H2O2 for 1 h (37°C) | 7.95±0.05 | 7.81±0.24 | 7.92±0.12 | 1.54±1.04b | 0.4±0.4b | 1.47±0.6b |
| 15 mM H2O2 for 10 min (42°C) | 8.06±0.08 | 8.08±0.1 | 8.06±0.07 | 6.4±0.23b | 5.33±0.19b | 6.5±0.07b |
| 15 mM H2O2 for 1 h (42°C) | 7.94±0.09 | 7.43±0.19 | 7.85±0.01 | 0 | 0 | 0 |
An initial 8 log colony forming units (CFU) of each strain was used in each stress-survival assay and the mean log10 CFU±standard error (SE) were calculated from three independent experiments.
For each stressor, means followed by asterisk indicate a significant difference from stress-resistant strains exposed to same stressor (p<0.05, Tukey's Kramer test).
Hydrogen peroxide produced by avian macrophages and heterophils plays a crucial role in Salmonella killing because it can penetrate cell membranes and may act on intracellular targets (Desmidt et al., 1997; Qureshi et al., 2000; Withanage et al., 2005; Kogut et al., 2002). Therefore, we determined the ability of Salmonella Enteritidis strains to counteract the effects of hydrogen peroxide. Irrespective of the temperature (37°C or 42°C) or time (10 min or 1 h) of incubation, there was no significant decline in the counts of UK, G1, and BC8 strains (Table 2) indicating that these strains were highly resistant to the oxidative stress induced by hydrogen peroxide. In contrast, the counts of C19, C45, and G45 strains declined by 1.5–2.5 log within 10 min at 37°C or 42°C (Table 2). By 1 h, the counts further declined by 6.5–7.5 log, indicating that these strains were highly sensitive to the oxidative stress induced by hydrogen peroxide (Table 2). Overall, these results indicate that C19, C45, and G45 strains (stress-sensitive strains) show impaired ability to survive in both acid and oxidative stress when compared with the UK, G1, and BC8 strains (stress-resistant strains).
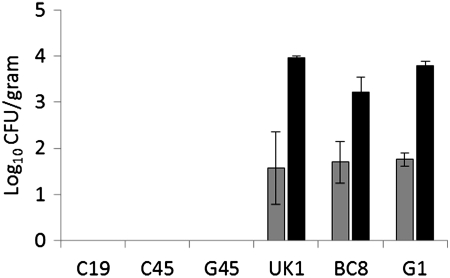
To assess the virulence (i.e., the ability to invade and systemically spread in the internal organs) of stress-sensitive and stress-resistant Salmonella Enteritidis strains in chickens, 3-day-old chickens were orally inoculated with ∼107 CFU of each strain and sacrificed at 4 days post-infection. For stress-resistant strains, the mean log10 CFU of Salmonella Enteritidis/gram of liver ranged from 1.57±0.79 (UK) to 1.75±0.14 (G1), whereas mean log10 CFU of Salmonella Enteritidis/gram of spleen ranged from 3.22±0.32 (BC8) to 3.96±0.05 (UK) (Fig. 1). In contrast, none of the stress-sensitive strains were recovered from either liver or spleen by either direct plating or by enrichment procedure, indicating that the acid-sensitive strains had impaired ability to invade the internal tissues. Similar to the results obtained in this study, Jorgensen et al., (2000) reported that two acid-sensitive Salmonella Typhimurium strains failed to invade chicken tissues, whereas Humphrey et al. (1996) reported that one acid and H2O2 sensitive Salmonella Enteritidis strain was significantly less invasive in chickens when compared with one acid and H2O2 resistant strain. It was also reported that the fecal carriage rates of stress-sensitive strains of Salmonella Tyhimurium were not significantly different from those of stress-resistant strains (Jorgensen et al., 2000; Williams et al., 1998). In this study, the intestinal samples collected from chickens challenged with stress-resistant strains were positive for Salmonella by selective enrichment procedure, indicating that intestinal carriage of these strains was not affected. Nevertheless, none of the intestinal samples collected from chickens infected with stress-sensitive strain were positive for Salmonella, indicating that stress-sensitive strains had an impaired ability to colonize the intestine. Studies on oral infection of Salmonella Enteritidis in mice have shown that only 1% of the inoculum survives the low pH during the passage through the stomach (Carter and Collins, 1974). About 80% of the bacteria that survive the passage through the stomach are passed with the feces within 6–10 h post-infection, whereas 15% remain localized in the lumen of cecum and large intestine, while only 5% manage to penetrate the intestinal wall of the small intestine and reach gut associated lymphoid tissues (Carter and Collins, 1974). Because the stress-sensitive strains tested in this study showed impaired ability to survive in gastric acidity and oxidative stress, these strains appear to have cleared from the intestine before the birds were sacrificed 4 days post-infection. Additional time-course studies to investigate changes in the intestinal carriage and systemic spread of stress-sensitive and stress-resistant strains may be required to fully understand the differences in their kinetics of infection in chickens.
FIG. 1.
The mean log10 colony forming units (CFU)±standard error (SE) of stress-sensitive and stress-resistant Salmonella Enteritidis strains recovered from liver (gray bars) and spleen (black bars) of chicks. Chicks in each group (n=3) were orally inoculated at number (#) of days of age with ∼1×107 CFU of Salmonella Enteritidis strains, and the challenge strains were recovered from the internal organs at 4 days post-infection.
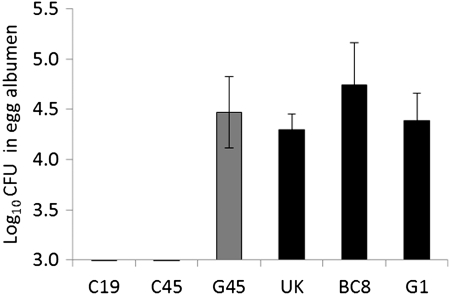
It has been hypothesized that stress-induced survival mechanisms may enable certain Salmonella Enteritidis strains to cope with the antimicrobial compounds present in the egg albumen (Van Immerseel, 2010). Consequently, we tested all six Salmonella Enteritidis strains for their ability to survive and/or grow in egg albumen. With the starting inoculum of ∼500 CFU, all the stress-resistant strains grew in egg albumen up to 4–5 log after 24 h of incubation at 25°C (Fig. 2). In contrast, two stress-sensitive strains (C19 and C45) did not survive or grow in egg albumen, whereas one strain (G45) grew as well as the stress-resistant strains. These results indicate that some stress-sensitive strains may also have an impaired ability to respond to the hostile environment of egg albumen—an environment that includes iron restriction, high pH, and other enzymatic activities. Further research is required to see if additional factors, such as expression of an O-antigen capsule, also contribute to the survival and growth of some strains Salmonella Enteritidis in albumen.
FIG. 2.
The mean log10 colony forming units (CFU)±standard error (SE) of stress-sensitive (gray bars) and stress-resistant (black bars) Salmonella Enteritidis strains in egg albumen. An overnight culture of each bacterial strain was added to egg albumen to a final concentration of ∼500 CFU/mL and incubated at 25±2°C for 24 h.
To determine if the differential stress response and virulence observed in this study could be due to the differences in the gene content between different strains, we compared the genomes of all the strains using CGH microarray designed based on the genomic information available for the sequenced strain. The CGH analysis revealed no consistent differences in the genomes between the six strains tested in this study. These results corroborate previous reports indicating that Salmonella Enteritidis strains are relatively genetically homogeneous, despite geographical, temporal, and source differences between the different strains (Porwollik et al., 2005; Morales et al., 2005; Olson et al., 2007; Betancor et al., 2009). It is important to point out that the microarray used in this study contained genes representing a single Salmonella Enteritidis strain and therefore only genes that are present on the array can be assayed. Some strains tested in this study may contain additional genes (e.g., phage-related genes) that were not represented on the microarray. Overall, these results indicate that the differential stress-response and virulence is less likely due to the variation in the whole gene content, but are more likely due to small polymorphisms that may alter regulation or expression of stress or virulence associated genes. To determine if the phenotypic differences were related to the rpoS gene, we sequenced the full length rpoS from all the six Salmonella Enteritidis strains. Several studies have shown that σS, encoded by the rpoS gene, controls the general stress response of bacteria, in particular the ability to resist hydrogen peroxide and acid stress during stationary phase (Hengge-Aronis, 2002). In addition, it has been reported that RpoS contributes to the virulence of Salmonella (Jorgensen et al., 2000; Nickerson and Curtiss, 1997). The rpoS gene in two stress-resistant strains (UK and G1) and one stress-sensitive strain (C19) was 100% identical in DNA sequence to the rpoS gene of Salmonella Enteritidis P125109 (GenBank accession no. AM933172). Strains BC8 contained a synonymous substitution from G to A at nucleotide 375 and a non-synonymous mutation at codon 36 (AGT [Ser] to AGG [Arg]). Strain C45 contained a non-synonymous mutation at codon 126 (GGG [Gly] to GAG [Glu]). Strain G45 contained two mutations, a synonymous substitution from T to C at nucleotide 605 and an insertion of A at nucleotide position 666. The insertion resulted in the introduction of an internal stop codon (TAA). Thus, one of the three stress-sensitive strains analyzed had a discernible mutation that interrupted the ORF of rpoS, consistent with the hypothesis that differential survivorship between strains is related to the single nucleotide polymorphisms in the chromosome. It is possible that the other strains may also have small polymorphisms elsewhere in the genome, which likely impact regulation of stress or virulence associated genes in some manner.
To determine whether any of the mutations altered the production of σS, we measured the activity of rpoS indirectly by analysis of hydrogen peroxidase II (HPII) activity using a semiqualitative catalase test as described previously (Taylor and Achanzar, 1972). Catalase test is based on principle that when 3% H2O2 is added to the bacterial colonies grown on LB plates, HPII breaks down H2O2 with the concomitant release of O2 resulting in bubbling of the colony. Strains producing RpoS bubble vigorously (+++), whereas strains bearing null alleles either do not form bubbles (-) or bubble only slightly (+) (Taylor and Achanzar 1972). We found that UK, G1, BC8, C19 and C45 strains formed vigorous bubbles (+++), suggesting that rpoS mutations in strains BC8 and C45 did not affect the σS function. In contrast, G45 strain with truncated rpoS did not form bubbles (-), indicating impaired σS function and a possibility of impaired KatN, an rpoS regulated non-hem catalase expression (Robbe-Saule, et al., 2001). It is important to note that the G45 strain is impaired in motility and also lacks a large virulence plasmid (Shah et al., 2011) and Salmonella plasmid virulence (spv) genes whose expression is controlled by rpoS during systemic infection (Jorgensen et al., 2000; Nickerson and Curtiss, 1997). Therefore it is likely that the impaired motility or regulation of virulence genes in this particular strain may have led to the reduced pathogenicity in chickens. Interestingly, the growth of G45 strain in the egg albumen was not affected indicating that σS may not be required for the survival of Salmonella Enteritidis in egg albumen (Fig. 2). Previous studies have reported that genes involved in bacterial cell wall structure and function, amino acid and nucleic acid metabolism, motility and stress responses may contribute to the survival of Salmonella Enteritidis in egg albumen (Clavijo et al., 2006; Lu et al., 2003; Gantois et al., 2008).
Conclusion
The results of this study indicate that wild-type strains of Salmonella Enteritidis show differential response to the acid stress, oxidative stress and survival in the egg albumen. Stress-sensitive strains may be less virulent as compared to the stress-resistant strains and therefore differential virulence of wild-type strains may be attributed to the underlying differences in their response to common stressful conditions encountered during infection. The differences in the regulation of virulence or stress-associated genes contribute to, but do not fully explain, the phenotypic differences observed in this study. Comparative transcriptomic and proteome profiling of these well-characterized stress-sensitive and stress-resistant strains may provide further insights into the molecular basis of differential stress response and virulence of Salmonella Enteritidis strains.
Acknowledgments
We gratefully acknowledge the technical assistance of Lisa Orfe for conducting CGH arrays. This project was funded in part with the federal funds from the NIAID, National Institutes of Health (under contract no. N01-A1-30055) and by the Agricultural Animal Health Program, College of Veterinary Medicine, Washington State University. Quincy Hawley was supported in part by the National Institutes of Health (grant RR07049).
Disclosure Statement
No competing financial interests exist.
References
- Betancor L. Yim L. Fookes M, et al. Genomic and phenotypic variation in epidemic-spanning Salmonella enterica serovar Enteritidis isolates. BMC Microbiol. 2009;9:237. doi: 10.1186/1471-2180-9-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call DR. Borucki MK. Besser TE. Mixed-genome microarrays reveal multiple serotype and lineage-specific differences among strains of Listeria monocytogenes. J Clin Microbiol. 2003;41:632–639. doi: 10.1128/JCM.41.2.632-639.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter PB. Collins FM. The route of enteric infection in normal mice. J Exp Med. 1974;139:1189–203. doi: 10.1084/jem.139.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell L. Kaiser P. Barrow P, et al. The immunobiology of avian systemic salmonellosis. Vet Immunol Immunopathol. 2009;128:53–59. doi: 10.1016/j.vetimm.2008.10.295. [DOI] [PubMed] [Google Scholar]
- Clavijo RI. Loui C. Andersen GL, et al. Identification of genes associated with survival of Salmonella enterica serovar Enteritidis in chicken egg albumen. Appl Environ Microbiol. 2006;72:1055–1064. doi: 10.1128/AEM.72.2.1055-1064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmidt M. Ducatelle R. Haesebrouck F. Pathogenesis of Salmonella enteritidis phage type four after experimental infection of young chickens. Vet Microbiol. 1997;56:99–109. doi: 10.1016/S0378-1135(96)01350-8. [DOI] [PubMed] [Google Scholar]
- Dhillon AS. Alisantosa B. Shivaprasad HL, et al. Pathogenicity of Salmonella enteritidis phage types 4, 8, and 23 in broiler chicks. Avian Dis. 1999;43:506–515. [PubMed] [Google Scholar]
- [EFSA] European Food Safety Authority. The community summary report on trends and sources of zoonoses, zoonotic agents, antimicrobial resistance and foodborne outbreaks in the European Union in 2006. EFSA J. 2007;130:34–117. [Google Scholar]
- Gantois I. Ducatelle R. Pasmans F, et al. Mechanisms of egg contamination by Salmonella Enteritidis. FEMS Microbiol Rev. 2009;33:718–738. doi: 10.1111/j.1574-6976.2008.00161.x. [DOI] [PubMed] [Google Scholar]
- Gantois I. Ducatelle R. Pasmans F, et al. Salmonella enterica serovar Enteritidis genes induced during oviduct colonization and egg contamination in laying hens. Appl Environ Microbiol. 2008;74:6616–2662. doi: 10.1128/AEM.01087-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gast RK. Beard CW. Isolation of Salmonella Enteritidis from internal organs of experimentally infected hens. Avian Dis. 1990;34:991–993. [PubMed] [Google Scholar]
- Gast RK. Benson ST. Intestinal colonization and organ invasion in chicks experimentally infected with Salmonella Enteritidis phage type 4 and other phage types isolated from poultry in the United States. Avian Dis. 1996;40:853–857. [PubMed] [Google Scholar]
- Gast RK. Benson ST. The comparative virulence for chicks of Salmonella Enteritidis phage type 4 isolates and isolates of phage types commonly found in poultry in the United States. Avian Dis. 1995;39:567–774. [PubMed] [Google Scholar]
- Gast RK. Understanding Salmonella Enteritidis in laying chickens: the contributions of experimental infections. Int J Food Microbiol. 1994;21:107–116. doi: 10.1016/0168-1605(94)90204-6. [DOI] [PubMed] [Google Scholar]
- Guard-Bouldin J. Gast RK. Humphrey TJ, et al. Subpopulation characteristics of egg-contaminating Salmonella enterica serovar Enteritidis as defined by the lipopolysaccharide O chain. Appl Environ Microbiol. 2004;70:2756–2763. doi: 10.1128/AEM.70.5.2756-2763.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson SC. Bounous DI. Lee MD. Early events in the pathogenesis of avian salmonellosis. Infect Immun. 1999;67:3580–3586. doi: 10.1128/iai.67.7.3580-3586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge-Aronis R. Recent insights into the general stress response regulatory network in Escherichia coli. J Mol Microbiol Biotechnol. 2002;4:341–346. [PubMed] [Google Scholar]
- Hengge-Aronis R. Stationary phase gene regulation: What makes an Escherichia coli promoter sigmaS-selective? Curr Opin Microbiol. 2002;5:591–595. doi: 10.1016/s1369-5274(02)00372-7. [DOI] [PubMed] [Google Scholar]
- Humphrey TJ. Richardson NP. Statton KM, et al. Effects of temperature shift on acid and heat tolerance in Salmonella Enteritidis phage type 4. Appl Environ Microbiol. 1993;59:3120–3122. doi: 10.1128/aem.59.9.3120-3122.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey TJ. Slater E. McAlpine K, et al. Salmonella Enteritidis phage type 4 isolates more tolerant of heat, acid, or hydrogen peroxide also survive longer on surfaces. Appl Environ Microbiol. 1995;61:3161–3164. doi: 10.1128/aem.61.8.3161-3164.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey TJ. Williams A. McAlpine K, et al. Isolates of Salmonella enterica Enteritidis PT4 with enhanced heat and acid tolerance are more virulent in mice and more invasive in chickens. Epidemiol Infect. 1996;117:79–88. doi: 10.1017/s0950268800001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen F. Leach S. Wilde SJ, et al. Invasiveness in chickens, stress resistance and RpoS status of wild-type Salmonella enterica subsp. enterica serovar Typhimurium definitive type 104 and serovar Enteritidis phage type 4 strains. Microbiology. 2000;146:3227–3235. doi: 10.1099/00221287-146-12-3227. [DOI] [PubMed] [Google Scholar]
- Joyner WL. Kokas E. Action of serotonin on gastric (proventriculus) secretion in chickens. Comp Gen Pharmacol. 1971;2:145–150. doi: 10.1016/0010-4035(71)90004-8. [DOI] [PubMed] [Google Scholar]
- Kang H. Loui C. Clavijo RI, et al. Survival characteristics of Salmonella enterica serovar Enteritidis in chicken egg albumen. Epidemiol Infect. 2006;134:967–976. doi: 10.1017/S0950268806006054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CC. Joyce EA. Chan K, et al. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 2002;3:RESEARCH0065. doi: 10.1186/gb-2002-3-11-research0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura AC. Reddy V. Marcus R, et al. Chicken consumption is a newly identified risk factor for sporadic Salmonella enterica serotype Enteritidis infections in the United States: A case-control study in FoodNet sites. Clin Infect Dis. 2004;38:S244–S252. doi: 10.1086/381576. [DOI] [PubMed] [Google Scholar]
- Kogut M. Rothwell L. Kaiser P. Differential effects of age on chicken heterophil functional activation by recombinant chicken interleukin-2. Dev Comp Immunol. 2002;26:817–830. doi: 10.1016/s0145-305x(02)00040-x. [DOI] [PubMed] [Google Scholar]
- Lu S. Killoran PB. Riley LW. Association of Salmonella enterica serovar Enteritidis yafD with resistance to chicken egg albumen. Infect Immun. 2003;71:6734–6741. doi: 10.1128/IAI.71.12.6734-6741.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales CA. Musgrove M. Humphrey TJ, et al. Pathotyping of Salmonella enterica by analysis of single-nucleotide polymorphisms in cyaA and flanking 23S ribosomal sequences. Environ Microbiol. 2007;9:1047–1059. doi: 10.1111/j.1462-2920.2006.01233.x. [DOI] [PubMed] [Google Scholar]
- Morales CA. Porwollik S. Frye JG, et al. Correlation of phenotype with the genotype of egg-contaminating Salmonella enterica serovar Enteritidis. Appl Environ Microbiol. 2005;71:4388–4399. doi: 10.1128/AEM.71.8.4388-4399.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson CA. Curtiss R., 3rd Role of sigma factor RpoS in initial stages of Salmonella typhimurium infection. Infect Immun. 1997;65:1814–1823. doi: 10.1128/iai.65.5.1814-1823.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura M. Lillehoj HS. Raybourne RB, et al. Differential responses of macrophages to Salmonella enterica serovars Enteritidis and Typhimurium. Vet Immunol Immunopathol. 2005;107:327–335. doi: 10.1016/j.vetimm.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Olsen JE. Tiainen T. Brown DJ. Levels of virulence are not determined by genomic lineage of Salmonella enterica serotype Enteritidis strains. Epidemiol Infect. 1999;123:423–430. doi: 10.1017/s0950268899003155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson AB. Andrysiak AK. Tracz DM, et al. Limited genetic diversity in Salmonella enterica serovar Enteritidis PT13. BMC Microbiol. 2007;7:87. doi: 10.1186/1471-2180-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick ME. Adcock PM. Gomez TM, et al. Salmonella Enteritidis infections, United States, 1985–1999. Emerg Infect Dis. 2004;10:1–7. doi: 10.3201/eid1001.020572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petter JG. Detection of two smooth colony phenotypes in a Salmonella Enteritidis isolate which vary in their ability to contaminate eggs. Appl Environ Microbiol. 1993;59:2884–2890. doi: 10.1128/aem.59.9.2884-2890.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppe C. Demczuk W. McFadden K, et al. Virulence of Salmonella Enteritidis phagetypes 4, 8 and 13 and other Salmonella spp. for day-old chicks, hens and mice. Can J Vet Res. 1993;57:281–287. [PMC free article] [PubMed] [Google Scholar]
- Porwollik S. Santiviago CA. Cheng P, et al. Differences in gene content between Salmonella enterica serovar Enteritidis isolates and comparison to closely related serovars Gallinarum and Dublin. J Bacteriol. 2005;187:6545–6555. doi: 10.1128/JB.187.18.6545-6555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi MA. Heggen CL. Hussain I. Avian macrophage: Effector functions in health and disease. Dev Comp Immunol. 2000;24:103–119. doi: 10.1016/s0145-305x(99)00067-1. [DOI] [PubMed] [Google Scholar]
- Robbe-Saule V. Algorta G. Rouilhac I, et al. Characterization of the RpoS status of clinical isolates of Salmonella enterica. Appl Environ Microbiol. 2003;69:4352–4358. doi: 10.1128/AEM.69.8.4352-4358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbe-Saule V. Coynault C. Ibanez-Ruiz M. Hermant D. Norel F. Identification of a non-haem catalase in Salmonella and its regulation by RpoS (sigmaS) Mol Microbiol. 2001;39:1533–1545. doi: 10.1046/j.1365-2958.2001.02340.x. [DOI] [PubMed] [Google Scholar]
- Shah DH. Zhou X. Addwebi T, et al. Cell invasion of poultry-associated Salmonella enterica serovar Enteritidis isolates is associated with pathogenicity, motility and proteins secreted by the type III secretion system. Microbiology. 2011;157:1428–1445. doi: 10.1099/mic.0.044461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabler JG. McCormick TW. Powell KC, et al. Avian heterophils and monocytes: Phagocytic and bactericidal activities against Salmonella enteritidis. Vet Microbiol. 1994;38:293–305. doi: 10.1016/0378-1135(94)90148-1. [DOI] [PubMed] [Google Scholar]
- Taylor WI. Achanzar D. Catalase test as an aid to the identification of Enterobacteriaceae. Appl Microbiol. 1972;24:58–61. doi: 10.1128/am.24.1.58-61.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson NR. Clayton DJ. Windhorst D, et al. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 2008;18:1624–1637. doi: 10.1101/gr.077404.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Immerseel F. Stress-induced survival strategies enable Salmonella Enteritidis to persistently colonize the chicken oviduct tissue and cope with antimicrobial factors in egg white: A hypothesis to explain a pandemic. Gut Pathog. 2010;2:23. doi: 10.1186/1757-4749-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [WHO] World Health Organization. 2008. http://www.who.int/gfn/activities/CDB_poster_Sept09.pdf. [Dec 15;2011 ]. http://www.who.int/gfn/activities/CDB_poster_Sept09.pdf
- Williams A. Davies AC. Wilson J, et al. Contamination of the contents of intact eggs by Salmonella Typhimurium DT104. Vet Rec. 1998;143:562–563. [PubMed] [Google Scholar]
- Withanage GS. Mastroeni P. Brooks HJ, et al. Oxidative and nitrosative responses of the chicken macrophage cell line MQ-NCSU to experimental Salmonella infection. Br Poult Sci. 2005;46:261–267. doi: 10.1080/00071660500098608. [DOI] [PubMed] [Google Scholar]
- Yim L. Betancor L. Martinez A, et al. Differential phenotypic diversity among epidemic-spanning Salmonella enterica serovar Enteritidis isolates from humans or animals. Appl Environ Microbiol. 2010;76:6812–6820. doi: 10.1128/AEM.00497-10. [DOI] [PMC free article] [PubMed] [Google Scholar]