Abstract
Human Epidermal Growth Factor Receptor 2-positive (HER2+) breast cancer (BC) is a highly aggressive disease commonly treated with chemotherapy and anti-HER2 drugs, including trastuzumab. There is currently no way to predict which HER2+ BC patients will benefit from these treatments. Previous prognostic signatures for HER2+ BC were developed irrespective of the subtype or the hierarchical organization of cancer in which only a fraction of cells, tumor-initiating cells (TICs), can sustain tumor growth. Here, we used serial dilution and single-cell transplantation assays to identify MMTV-Her2/Neu mouse mammary TICs as CD24+:JAG1− at a frequency of 2–4.5%. A 17-gene Her2-TIC-enriched signature (HTICS), generated on the basis of differentially expressed genes in TIC versus non-TIC fractions and trained on one HER2+ BC cohort, predicted clinical outcome on multiple independent HER2+ cohorts. HTICS included up-regulated genes involved in S/G2/M transition and down-regulated genes involved in immune response. Its prognostic power was independent of other predictors, stratified lymph node+ HER2+ BC into low and high-risk subgroups, and was specific for HER2+:estrogen receptor alpha-negative (ERα−) patients (10-y overall survival of 83.6% for HTICS− and 24.0% for HTICS+ tumors; hazard ratio = 5.57; P = 0.002). Whereas HTICS was specific to HER2+:ERα− tumors, a previously reported stroma-derived signature was predictive for HER2+:ERα+ BC. Retrospective analyses revealed that patients with HTICS+ HER2+:ERα− tumors resisted chemotherapy but responded to chemotherapy plus trastuzumab. HTICS is, therefore, a powerful prognostic signature for HER2+:ERα− BC that can be used to identify high risk patients that would benefit from anti-HER2 therapy.
Keywords: HER2+ breast cancer, cancer stem cells, prognostic signature, mouse models
Breast cancer (BC) represents multiple diseases, including Human Epidermal Growth Factor Receptor 2-positive (HER2+), estrogen receptor (ER)α+ (luminal A and B), and triple-negative (basal-like, Claudin-low) tumors. HER2+ BC is caused by overexpression/amplification of the HER2/ERBB2/NEU tyrosine kinase receptor and constitutes 15–20% of cases. Approximately 50% of these are ERα+ tumors, and 50% are ERα−. Current treatment of HER2+ BC involves chemotherapy plus trastuzumab (Herceptin; Genentech), a monoclonal antibody directed against HER2 (1–3). Despite improvement in disease-free survival (DFS) over a 4-y follow-up (4), the cost of trastuzumab, adverse effects such as cardiac failure, and emergence of drug-resistance metastases represent serious limitations for its use, particularly in low-income countries (5). A prognostic signature that can predict clinical outcome from tumor biopsies at time of presentation may help prioritize patients for anti-HER2 therapy.
Because BC consists of several different subtypes, each with distinct pathological features and clinical behaviors, predictive prognostic signatures may need to be developed for each subtype. In addition, many types of cancer exhibit hierarchical organization whereby only a fraction of cells, termed tumor-initiating cells (TICs), sustains growth, whereas the remaining tumor cells, which descend from TICs, have lost their tumorigenic potential (6). HER2/Neu drives asymmetrical cell division, increases the frequency of TICs relative to mammary stem cells (7), and its continuous expression is required to sustain tumorigenesis (8). One strategy to identify prognostic signatures would be to base it on gene expression in enriched TIC populations for specific BC subtype. However, so far, most prognostic signatures for BC were generated irrespective of TICs or BC subtype. As a result, these signatures are predictive for ERα+ tumors, which represent 60–70% of human BC, but not for HER2+:ERα− or triple-negative BC (9). Thus, Oncotype, a 21-gene recurrence signature (10), is highly predictive for ERα+ [hazard ratio (HR), 4.79] but not other subtypes such as HER2+ (HR, 1.0); the invasiveness gene signature (IGS) generated from CD44+/CD24-/low breast TICs (11) scores on ERα+ (HR, 2.12) but not HER2+ patients (HR, 0.96) (10) (this study); and a stroma-derived prognostic predictor (SDPP) (12) is shown herein to predict clinical outcome for HER2+:ERα+ but not for HER2+:ERα− BC.
We hypothesized that to be highly predictive, a prognostic signature for HER2+:ERα− BC should reflect gene expression in enriched TICs for this particular subtype. Here, we describe the development of a prognostic signature [Her2-TIC-enriched signature (HTICS)] for HER2+:ERα− BC based on transcriptional profiling of highly purified TICs from a mouse model for this subtype. HTICS identifies a subgroup of HER2+:ERα− patients that does not respond well to conventional chemotherapy but benefits from trastuzumab, and may therefore be used to identify and prioritize high-risk HER2+:ERα− patients for anti-HER2 therapy.
Results
Enrichment of Her2/Neu TICs in CD24+:JAG1− Fraction.
In this study, we used a mouse model of HER2+ BC, MMTV-Her2/Neu, which sprouts mammary tumors with similar characteristics as the human disease (13). Previously, mammary TICs were identified in this model in the CD31−, CD45−, TER119− (lineage-depleted lin−), 7AAD− (live), CD24+ cell fraction at a frequency of ∼1/300 (14). The cell surface markers CD49f, Sca-1, CD29, CD90, CD18 and CD14 failed to subdivide the CD24+ cell population for enrichment of TICs (14, 15). We found that mechanical rather than enzymatic dissociation improved recovery of Her2/Neu TICs ∼3-fold to 1/84 (Fig. 1A). As levels of the Notch-ligand Jagged1 (JAG1) can predict outcome in BC (16), we tested for expression of Jagged1 and its receptor Notch1 in lin− CD24+ cells by flow cytometry. Interestingly, both Jagged1 and Notch1 independently subdivided the CD24+ fraction into two populations (Fig. 1B). Immunostaining of Neu tumors confirmed widespread expression of Jagged1 (Fig. 1C).
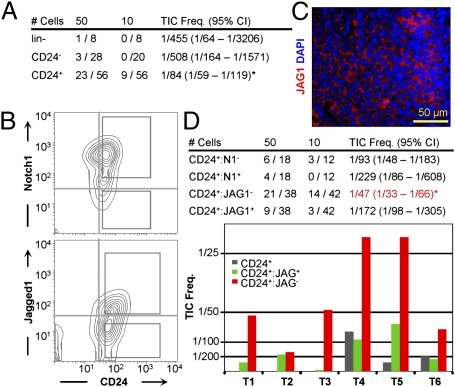
Fig. 1.
Identification of Her2/neu TICs as CD24+, JAG1−. (A) TIC frequency in CD24+, CD24−, and lineage-depleted (lin−) Her2/Neu tumor cells purified by mechanical dissociation and cell sorting. (B) Representative flow cytometric profiles of lin− PI− Her2/Neu tumor cells for CD24-Notch1 and CD24-Jagged1 and gating conditions used to sort cells for transplantation. (C) Immunofluorescent staining for Jagged1 in an MMTV-Neu tumor. DAPI was used to label nuclei. (D, Upper) Average TIC frequency and 95% CIs following serial-dilution transplantations of indicated fractions from six independent MMTV-Neu primary tumors. *P = 0.0005 against CD24+ (ANOVA). (D, Lower) Average TIC frequency for CD24+:JAG1+ and CD24+:JAG1− populations for six individual tumors. The CD24+ fraction was also analyzed in tumors 4–6.
To test whether these antigens could enrich TICs, tumor cells were sorted on the basis of CD24 plus JAG1 or Notch1 expression, serially diluted and transplanted into mammary glands of syngeneic mice. TIC frequency varied from tumor to tumor but was consistently higher in CD24+:JAG1− relative to CD24+:JAG1+ cells in six different tumors (Fig. 1D). Overall TIC frequency in the CD24+:JAG1− fraction was 1/47 (∼2%) compared with 1/172 in the CD24+:JAG1+ population (3.6-fold enrichment) and 1/455 in the lin− population (9.7-fold enrichment). TICs were also enriched 2.4-fold in the CD24+:Notch1− fraction relative to CD24+:Notch1+ (Fig. 1D, Upper). Flow cytometric profiles of secondary tumors arising after transplantation of either CD24+:JAG1− or CD24+:JAG1+ cells were similar to primary tumors (Fig. S1A), suggesting the presence of some contaminating CD24+:JAG1− TICs in the CD24+:JAG1+ fraction or that both fractions contained TICs, albeit at different frequencies, and that the JAG1− and JAG1+ states were interconnected.
The HER2/NEU and NOTCH1 pathways antagonize each other (17). However, we found that HER2/NEU expression in four independent tumors was not statistically different in CD24+:JAG1+ relative to CD24+:JAG1− cells (Fig. S1B), indicating that JAG1 does not significantly affect HER2/NEU expression in this mouse model. It was previously reported that the MMTV-Her2/Neu transgene, which encodes rat Her2/Neu (13, 18), elicits immunorejection or immunoediting response in transplanted mice, leading to silencing of the transgene in secondary tumors (19). However, secondary tumors expressed HER2 at comparable levels as primary tumors, and incidence of tumor formation was as high in isogenic immunocompetent recipient mice as in immunocompromised mice (Fig. S2), Thus, at least following transplantation of a small number of lin− cells (≤50) the rat Her2/Neu transgene does not induce overt immunorejection in mice.
Her2/Neu TICs Are Functionally Stable.
Cancer heterogeneity attributable to clonal evolution and functional instability of TICS can lead to occurrence of distinct secondary tumors (20), complicating generation of TIC-based prognostic signatures. To test for functional stability of CD24+:JAG1− Her2/Neu TICs, we performed single cell transplantation assays as depicted in Fig. 2A. Tumors (n = 4; N133, N181, N182, N202) were mechanically dissociated, lineage-depleted and sorted for single, live (PI negative), CD24+:JAG1− cells. Sorted cells were seeded, one cell per well, into Terasaki plates, which have a conical flat bottom, facilitating identification of wells with single cells (Fig. 2A, Inset). Content of each well containing a single cell was mixed with matrigel and injected into the no. 4 mammary gland of 4–5 wk old FvB female mouse. For each tumor, we performed 60 single-cell injections. The entire procedure, from tumor resection to transplantation took ∼12 h. Of 240 injections, 11 mice developed mammary tumors within 6 mo (average latency, 3.9 mo) with an overall frequency of 1/22 (∼4.6%) (Fig. 2A). TIC frequency for the four individual tumors was 1/30 for N133 (i.e., 2 tumors/60 single-cell injections), 1/30 for N181 (2/60), 1/20 for N182 (3/60), and 1/15 for N202 (4/60). As control, we injected female mice with 2,000 lin− tumor cells; all injected mice in this group developed tumors (termed lin−-derived tumors).
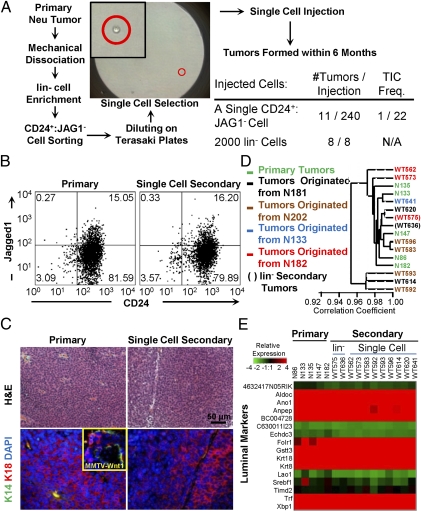
Fig. 2.
CD24+:JAG1− TICs are functionally stable. (A) Scheme for single cell transplantation assays. (B) Representative flow cytometric profiles for CD24 and Jagged1 of primary and single (CD24+:JAG1−) cell-derived Neu tumors. (C) Histology and marker analysis of primary and single cell-derived tumors: Keratin 14 (K14, green) and 18 (K18, red). (Inset) Positive staining of Keratin14 plus Keratin18 in MMTV-Wnt1 tumors. (D) Cluster analysis of primary, lin−-derived and single cell-derived Neu tumors showing close clustering with >0.95 correlation coefficient. (E) Heat map of representative luminal genes in indicated tumors.
We next determined whether individual secondary tumors were similar to the tumors from which they were derived, as well as to other primary and secondary tumors. With the exception of one single-cell derived tumor, WT614, all exhibited similar flow cytometry profiles for CD24 plus Sca1, CD49f or JAG1, as did primary and lin−-derived tumors (Fig. 2B and Fig. S3 A–C). Notably, single-cell-derived tumors gave rise not only to CD24+:JAG1− but also to CD24+:JAG1+ cells, indicating that they could expand and regenerate the cellular complexity found in Her2/Neu tumors. The outlier, WT614, showed an expanded CD24+:JAG1+ fraction, but similar profiles for CD24-Sca1 and CD24-CD49f (Fig. S3 A–C). The single cell-derived tumors, like primary Her2/Neu tumors and bulk lin−-derived tumors, exhibited a similar histology of poorly differentiated adenocarcinoma as well as similar expression of HER2 and the luminal marker cytokeratin 18, with no expression of K14 or vimentin (Fig. 2C and Fig. S3 D and E). Finally, transcriptional profiling and hierarchical clustering revealed that tumors derived from single cell injections (n = 9), injections of lin− cells (n = 2), as well as primary tumors (n = 5) clustered together with a correlation coefficient of over 0.95, indicating a high degree of similarity among samples (Fig. 2 D and E and Fig. S3 F–H). Only 16 genes, enriched in IFN-associated factors, were differentially expressed in single cell-derived tumors relative to primary lesions (Fig. S3 I–K). Thus, CD24+:Jag1− Her2/Neu TICs are functionally stable and their frequency is ∼2% (serial dilution) to 4.6% (single cell transplantation).
CD24+:JAG1− Her2/Neu TIC Fraction Is Enriched in Cell Division-Associated Pathways and Depleted for Differentiation Pathways.
To gain an insight into molecular regulations that determine Her2+ TIC function, we analyzed genes and pathways specifically expressed in TIC-enriched cell populations. Four independent MMTV-Neu primary mammary tumors (N250, N261, N283, N222/N229) were harvested, lin− cells were sorted according to CD24 and JAG1 expression and subjected to gene expression microarray analysis. With the exception of CD24, which, as expected, was elevated in TICs, expression of several luminal markers was high and similar in TIC and non-TIC fractions, whereas expression of basal markers was generally low in both fractions (Fig. 3A).
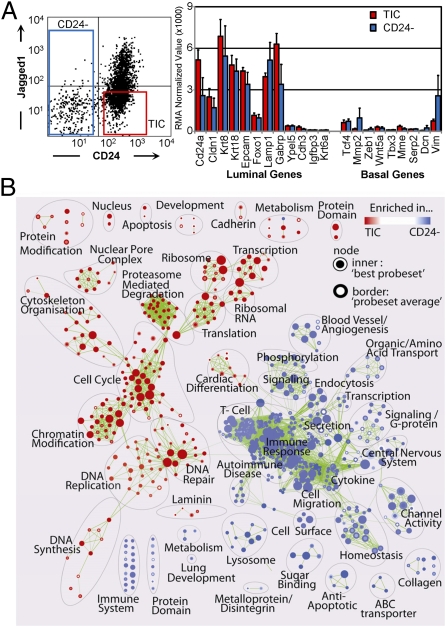
Fig. 3.
The Her2/Neu CD24+:JAG1− TIC fraction is enriched in genes associated with dividing but not differentiating cells. (A) Left, gating conditions used to sort lin− MMTV-Her2/neu tumor cells. Right, expression of luminal and basal genes in CD24+:JAG1− TICs versus non-TICs. (B) Functional enrichment map for TIC/CD24+:JAG1− versus non-TIC/CD24− fractions revealing distinct pathways in each group. Nodes (circles) represent significantly enriched pathways; red or blue color indicates gene sets enriched in TIC or non-TIC fractions, respectively.
Using Gene Set Enrichment Analysis (GSEA) software (21) and “Functional Enrichment Maps” to visualize the results (22), we identified marked differences in pathway activity in CD24+:JAG1−/TIC versus CD24−/non-TIC populations, with 262 up-regulated and 492 down-regulated gene-sets (Fig. 3B). Gene sets enriched in the TIC fraction included pathways associated with cell division; the non-TIC fraction was enriched in pathways associated with differentiation as well as immune response and angiogenesis.
Generation of a HTICS.
To generate a Her2/Neu TIC-enriched prognostic signature, we analyzed publicly available cohorts with clinical outcomes and microarray expression data from RNA extracted from fresh tumor biopsies (Dataset S1A). Because HER2 status, as determined by immunostaining, was not available for most cohorts, we used twofold increase in expression of three or more of five genes on the HER2 amplicon (HER2/ErbB2, Stard3, Perld1, Grb7, C17orf37) as the basis to collate HER2+ patients. This criterion, previously used to generate HDPP (23), selected 69.5% of HER2 patients, as determined by IHC (Fig. S4).
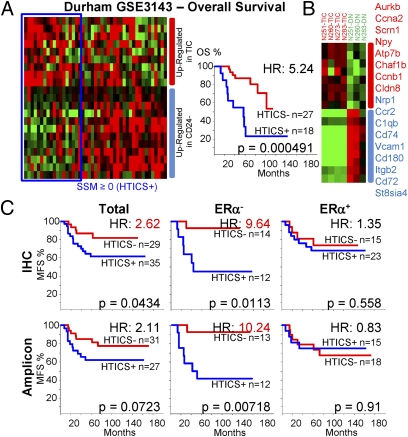
We first identified differentially expressed genes (329) that showed twofold or greater increase or decrease in enriched TIC versus non-TIC fractions with a significant P value (≤ 0.05). A total of 284 of these genes were found on a human overall survival (OS) cohort (GSE3143), which we used to train the signature. We classified patients using a “Score for Signature Match (SSM)” algorithm, modified from Ref. 9 (SI Material and Methods). Without training, the 284 genes could separate HER2+ patients in the GSE3143 cohort into poor and good prognosis groups with a HR of 2.54 (P = 0.072; Fig. S5A). After testing for association and significance of each gene to patient outcome, a 40-gene signature was derived which could stratify patients with HR of 3.53 (P = 0.00742; Fig. S5A). Further optimization resulted in a 17 Her2 TIC-enriched Signature, which we termed HTICS. Gene expression heat map of 45 HER2+ patients with descending SSM scores in the training cohort using HTICS is shown (Fig. 4A, Left). A SSM > 0 cutoff was selected to evaluate its predictive power by Kaplan-Meier analysis. On this training cohort, tumors expressing HTICS had a reduced OS relative to tumors that did not express the signature (HR, 5.24; P = 0.000491; Fig. 4B). HTICS was specific to HER2+ tumors; its predictive power for all BC subtypes or for HER2-negative tumors was statistically insignificant (Fig. S5B).
Fig. 4.
Generation of a Her2/Neu TIC-enriched prognostic signature (HTICS). (A, Left) Gene expression heat map of 45 HER2+ patients with descending “Score for Signature Match” (SSM) using GSE3143 as a training cohort, with patients who match (blue line) or do not match (red line) HTICS. (A, Right) SSM > 0 cutoff was selected to evaluate predictive power by Kaplan–Meier analysis. (B) HTICS differentiates TIC versus non-TIC mammary tumor fractions. The 17-gene HTICS is shown on the right. (C) MFS curves and HRs using HTICS for HER2+, HER2+:ERα−, and HER2+:ERα+ patients collated from GSE2034 and GSE2603 on the basis of IHC (Upper) or 5-gene HER2 amplicon (Lower).
HTICS consists of eight up-regulated (Aurkb, Ccna2, Scrn1, Npy, Atp7b, Chaf1b, Ccnb1, Cldn8) and nine down-regulated genes (Nrp1, Ccr2, C1qb, Cd74, Vcam1, Cd180, Itgb2, Cd72, St8sia4; Fig. 4B and Fig. S5C). The up-regulated subset includes genes associated with passage through the S/G2/M phase of the cell cycle (Aurkb, cyclinB1, Cyclin A2; Chaf1b). Down-regulated in HTICS are genes involved in cell adhesion, angiogenesis and immune-response.
HTICS Predicts Clinical Outcome for HER2+:ERα− BC Patients Treated with Chemotherapy.
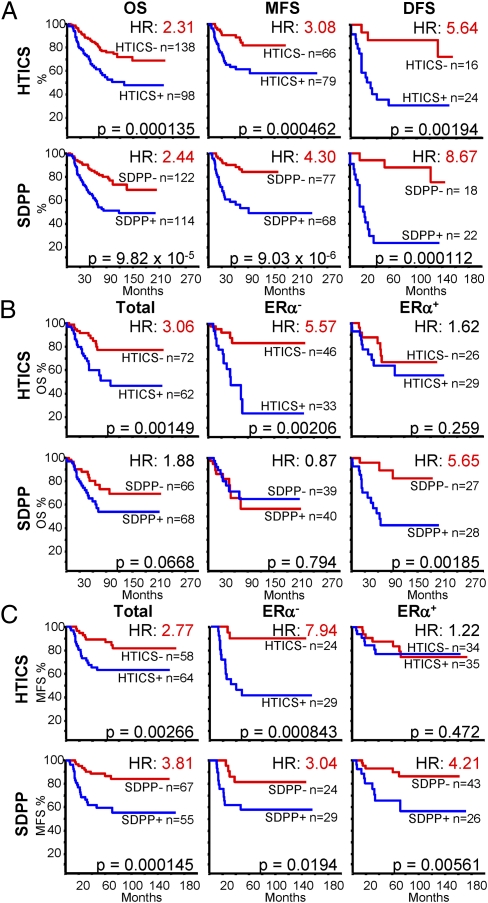
We initially evaluated the prognostic power of HTICS using two metastasis-free survival (MFS) cohorts (n = 64) with annotated HER2 expression data determined by IHC. HTICS+ patients exhibited poor MFS with HR of 2.62 relative to the HTICS-negative group (P = 0.043; Fig. 4C). As MMTV-Neu tumors are ERα-negative, we determined the effect of ERα-expression on the predictive power of HTICS. Remarkably, HTICS predicted 10-y MFS of 83.6% for signature-negative and 46.3% for signature-positive HER2+:ERα− patients (HR, 9.64; P = 0.01), and was not predictive for the HER2+:ERα+ group (Fig. 4C, Upper). Importantly, similar results were obtained when patients from these cohorts were collated on the basis of the five-gene HER2 amplicon, with MFS of 83.6% and 41.7% for HTICS− and HTICS+ HER2+:ERα− groups, respectively (HR, 10.24; P = 0.007; Fig. 4C, Lower).
Next, we extended our analysis to other MFS, OS and DFS cohorts (excluding the training cohort) using the five-gene HER2 amplicon to identify HER2+ patients. HTICS predicted OS, MFS and DFS for HER2+ patients with HR of 2.1, 3, and 5.6, respectively (P < 0.002; Fig. 5A). Moreover, for cohorts with available ERα data, HTICS predicted clinical outcomes for HER2+:ERα− patients with a 10-y OS of 83.6% versus 24.0% (HR, 5.57; P = 0.002) and MFS of 90.9% versus 47.2% (H = 7.94; P = 0.00084) (Fig. 5 B and C).
Fig. 5.
HTICS predicts clinical outcome for HER2+:ERα− BC patients treated with chemotherapy; SDPP predicts clinical outcome for HER2+:ERα+ patients. (A) Kaplan–Meier analyses of combined OS, MFS, or DFS using HTICS or SDPP. (B and C) Kaplan–Meier analyses on OS (B) and MFS (C) cohorts with known ERα status.
HTICS may identify patients with poor prognosis or poor response to chemotherapy. To address this question, we performed a retrospective analysis on cohorts of patients treated or not with conventional chemotherapy (Fig. S5D). There was a tendency of HER2+ and HER2+:ERα+, but not HER2+:ERα− patients, to benefit from chemotherapy. HTICS− HER2+:ERα− survived better than the HTICS+ HER2+:ERα− patients both in treated and untreated settings. We note that retrospective analyses may miss subtle benefits that can only be observed in prospective studies. Nonetheless, our signature clearly identifies high-risk HER2+:ERα− patients with bad prognosis and poor response to conventional chemotherapy.
The tumor suppressor p53 is a transcriptional activator of ERα; mutation in p53 correlates with reduced ERα expression and bad prognosis (24). In a patient cohort with available p53 status (n = 32), the predictive power of HTICS was elevated in the p53 mutant arm (HR, 5.78; P = 0.0136) compared with the whole population (HR, 3.4; P = 0.028) or the p53 wild-type arm (HR, 2.34; P = 0.414; Fig. S5E), suggesting that this signature can discriminate clinical outcome for HER2+ patients depending not only on ERα but also on p53 status.
HTICS Predicts Clinical Outcome for HER2+:ERα− Patients: SDPP for HER2+:ERα+.
Next, we compared HTICS predictive power to other signatures (Dataset S1 C–N). The SDPP (12) was highly predictive for HER2+ BC patients (Fig. 5A). This predictive power was proportional to the ERα+ to ERα− ratio in these cohorts. Indeed, SDPP was not (OS, P = 0.794) or only moderately informative (MFS) (HR, 3.0; P < 0.02) for HER2+:ERα− patients but was highly predictive for HER2+:ERα+ patients with a HR of 5.65 for OS (P ≤ 0.002) and HR, 4.21 (P < 0.01) for MFS (Fig. 5 B and C). Thus, together, HTICS and SDPP can be used to predict clinical outcome for the two HER2+ BC subtypes. For HER2+:ERα+ patients, a HER2-derived prognostic predictor (HDPP) (23) was also predictive for MFS better than HTICS with HR of 3.47 (P < 0.007; Fig. S6). In contrast, a 70-gene/mammaPrint (9), IGS (11), and BC proliferation signatures (25) performed poorly on both HER2+:ERα+ and HER2+:ERα− patients (Fig. S6).
HTICS Predicts Clinical Outcome Independently of Other Predictors Including Node Status.
Next, we performed bi- and multivariate analyses of HER2+ and HER2+:ERα− patients to determine the effect, if any, of chemotherapy, tumor grade, tumor size, age at detection and lymph node involvement on the prediction power of HTICS. HTICS was highly predictive independently of these other variables (Fig. S7). The other most potent predictor was lymph node status with HRs of 3.28 and 8.29 in bi- and multivariate analysis of HER2+:ERα− patients, respectively. In the bivariate analysis, HTICS could further subdivide node+ tumors into high and low risk groups with HR of 5.2 or compounded HR of 3.28 × 5.2 = 17.0.
HTICS Predicts Clinical Outcome for HER2+:ERα− BC Patients Treated with Neoadjuvant Chemotherapy Plus Trastuzumab.
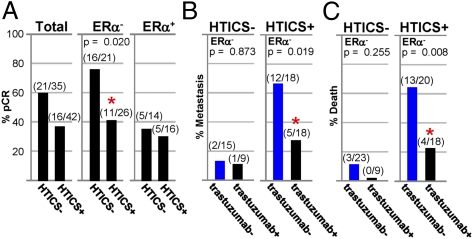
The aforementioned results indicate that HTICS+ patients do not respond well to conventional chemotherapy. We next sought to determine their response to trastuzumab. Only one patient cohort (n = 27) of neoadjuvant chemotherapy plus trastuzumab with microarray data and pathological complete response (pCR) is publicly available [GSE22358 (26)]. We combined it with a new dataset with clinical data (pCR, MFS, and OS) from 50 HER2+ patients who were treated with neoadjuvant chemotherapy (fluorouracil/epirubicin or Adriamycin/cyclophosphamide-taxol) plus trastuzumab at the MD Anderson Cancer Center and monitored in the past 7.5 y. This group of HER2+ patients included 32 ERα− and 18 ERα+ tumors. HTICS+ HER2+:ERα− patients exhibited significantly worse pCR (11/26 = 42%) relative to the HTICS− group (16/21 = 76%; P = 0.0195; Fig. 6 and Fig. S8A).
Fig. 6.
HTICS predicts response of HER2+:ERα− BC patients to trastuzumab. (A) pCR data for HER2+, HER2+:ERα−, and HER2+:ERα+ patients treated with chemotherapy/trastuzumab. (B and C) Fractions of patients that developed metastasis (B) or died (C) 4-y postsurgery in trastuzumab-untreated patients (trastuzumab−) selected from publicly available cohorts (Fig. 5C) versus patients treated with neoadjuvant chemotherapy plus trastuzumab (trastuzumab+).
Whereas none of the HTICS− HER2+:ERα− patients died during this 7.5-y period, all five patients who died had HTICS+ tumors. However, because of the small size of the group and relatively short follow-up, the results were not statistically significant (P = 0.08; Fig. S8B). A similar trend of poor prognosis was observed for MFS (Fig. S8B).
To begin to assess benefits from trastuzumab, we retrospectively determined the fraction of HER2+:ERα− patients that developed metastasis within 48 mo postsurgery in the publicly available trastuzumab-untreated (n = 33; Fig. 5C) versus neoadjuvant chemotherapy/trastuzumab-treated patients (n = 27; Fig. 6A). For HTICS− patients, trastuzumab did not have a significant effect with 2/15 (13%) developing metastasis in the trastuzumab-negative group versus 1/9 (11%) in the chemotherapy plus trastuzumab group (P = 0.873; Fig. 6B). In contrast, for HTICS+ patients, 12/18 (66%) relapsed in the chemotherapy group compared with 5/18 (27%) in the chemotherapy/trastuzumab group (Fig. 6B). Despite the caveat of comparing independent patient cohorts, the results were highly significant (P = 0.019), indicating that trastuzumab reduced metastasis in HTICS+ HER2+:ERα− patients 2.4-fold (66%/27%). Similarly, no statistically significant difference in OS was found in trastuzumab-untreated versus chemotherapy/trastuzumab-treated HTICS− HER2+:ERα− patients (P = 0.255; Fig. 6C). In contrast, for HTICS+ patients, 13/20 (65%) died in the untreated compared with 4/18 (22%) in the chemotherapy/trastuzumab-treated cohorts (2.95-fold increase in OS; P = 0.008). Together these results suggest that HTICS+ HER2+:ERα− patients benefit from trastuzumab and should be prioritized for anti-HER2 therapy.
Discussion
We report on the generation of a prognostic signature (HTICS) that can identify a high-risk HER2+:ERα− BC subgroup, which, in retrospective analysis, resists chemotherapy but responds to chemotherapy plus trastuzumab. The signature was generated based on highly enriched TICs from a mouse model of HER2+:ERα− BC. A critical step was to demonstrate by single cell transplantation assays that Her2+ TICs are similar and stable, hence analysis of a few primary tumors is broadly informative. Because the TIC-enriched fraction contains only a small percentage of TICS (2–4.6%), most cells in this fraction represent early progenitors with expression patterns that likely reflect the proportion of TICs in the tumor. Genes associated with the ability of a single cell to self renew and form a tumor at an otherwise normal site following transplantation into recipient mice, may also endow it with the ability to form micrometastases at distal sites and acquire the necessary genetic changes needed for full macrometastases in human (27). Indeed, four of the eight up-regulated genes in HTICS are directly involved in cell cycle progression, DNA replication, and mitosis. In addition, HTICS includes down-regulated genes involved in immune response; this may allow TICs to evade immune surveillance, enhancing dissemination and metastasis.
In our retrospective analysis, chemotherapy plus trastuzumab treatment improved MFS 2.4-fold and OS 2.9-fold compared with trastuzumab-untreated therapy. Thus, HTICS can identify high-risk patients that may be prioritized for chemotherapy/trastuzumab therapy. This may be particularly relevant to low-income countries where trastuzumab therapy is not publicly available; HTICS+ can provide an objective criterion and an incentive for signature-positive patients to undergo trastuzumab therapy. In contrast, the effect of trastuzumab on HTICS− patients was insignificant for both MFS and OS cohorts over a 4-y period. We stress that our results do not justify, at this stage, withholding trastuzumab therapy from HTICS− patients. A prospective analysis of large cohorts is urgently needed to assess small benefits, if any, of trastuzumab in this group. Clearly, the lack of large, prospective cohorts with clinical outcome and microarray data from RNA derived from fresh tumor biopsies is a major limitation. Large cohorts of formalin-fixed, paraffin-embedded BC tissues with clinical outcome are available. NanoString technology can be used to assess gene expression on paraffin-embedded specimens (28). The application of this technology to large prospective cohorts of HER2+:ERα− patients is now required to bring HTICS to the clinic.
Materials and Methods
Animal, histology, TIC analysis, the generation of HTICS, and Kaplan–Meier analysis were performed as described previously (14, 29) and SI Materials and Methods. TIC frequency was calculated using L-Calc software (StemCell Technologies). Microarray data were normalized using RMA method via Partek software. Score for Signature Match (SSM) was calculated using:
Where I is gene index; 1 for up-regulated genes in TICs; −1 for down-regulated genes; X is log 2-transformed and median-centered gene expression value of the patient; n indicates signature gene number. SSM ≥0 was considered a match. Hazard ratios were calculated using COX proportional hazards survival regression. Heat maps and dendrograms were generated by JAVA tree-view.
Supplementary Material
Acknowledgments
We thank Dongyu Wang, Kolja Eppert, Shaheena Bashir, and Chao Lu for advice on statistical and microarray analysis; Miriam Zacksenhouse and Morag Park for discussions; and Kolja Eppert and John Dick for critical comments on the manuscript. J.C.L. was supported, in part, by a fellowship from the Canadian Breast Cancer Foundation. This study was conducted with the support of the Ontario Institute for Cancer Research through funding provided by the Government of Ontario, the Canadian Breast Cancer Foundation, and the Canadian Breast Cancer Research Alliance (E.Z.). The study was also supported by the Breast Cancer Research Foundation (F.J.E., L.P.); and the Komen Foundation for the Cure (S.E.E. and E.Z.).
Footnotes
Conflict of interest statement: A patent application has been filed on subject matter disclosed in this manuscript.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE29590 and GSE29616).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201105109/-/DCSupplemental.
References
- 1.Slamon DJ, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 2.Abramson V, Arteaga CL. New strategies in HER2-overexpressing breast cancer: Many combinations of targeted drugs available. Clin Cancer Res. 2011;17:952–958. doi: 10.1158/1078-0432.CCR-09-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dean-Colomb W, Esteva FJ. Her2-positive breast cancer: Herceptin and beyond. Eur J Cancer. 2008;44:2806–2812. doi: 10.1016/j.ejca.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Gianni L, et al. Herceptin Adjuvant (HERA) Trial Study Team Treatment with trastuzumab for 1 year after adjuvant chemotherapy in patients with HER2-positive early breast cancer: A 4-year follow-up of a randomised controlled trial. Lancet Oncol. 2011;12:236–244. doi: 10.1016/S1470-2045(11)70033-X. [DOI] [PubMed] [Google Scholar]
- 5.Martín M, et al. Minimizing cardiotoxicity while optimizing treatment efficacy with trastuzumab: Review and expert recommendations. Oncologist. 2009;14:1–11. doi: 10.1634/theoncologist.2008-0137. [DOI] [PubMed] [Google Scholar]
- 6.O'Brien CA, Kreso A, Dick JE. Cancer stem cells in solid tumors: An overview. Semin Radiat Oncol. 2009;19:71–77. doi: 10.1016/j.semradonc.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Cicalese A, et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 8.Korkaya H, Paulson A, Iovino F, Wicha MS. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008;27:6120–6130. doi: 10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desmedt C, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14:5158–5165. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- 10.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 11.Liu R, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 12.Finak G, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 13.Guy CT, et al. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu JC, Deng T, Lehal RS, Kim J, Zacksenhaus E. Identification of tumorsphere- and tumor-initiating cells in HER2/Neu-induced mammary tumors. Cancer Res. 2007;67:8671–8681. doi: 10.1158/0008-5472.CAN-07-1486. [DOI] [PubMed] [Google Scholar]
- 15.Vaillant F, et al. The mammary progenitor marker CD61/beta3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 2008;68:7711–7717. doi: 10.1158/0008-5472.CAN-08-1949. [DOI] [PubMed] [Google Scholar]
- 16.Reedijk M, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 17.Osipo C, et al. ErbB-2 inhibition activates Notch-1 and sensitizes breast cancer cells to a gamma-secretase inhibitor. Oncogene. 2008;27:5019–5032. doi: 10.1038/onc.2008.149. [DOI] [PubMed] [Google Scholar]
- 18.Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54:105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- 19.Kmieciak M, Knutson KL, Dumur CI, Manjili MH. HER-2/neu antigen loss and relapse of mammary carcinoma are actively induced by T cell-mediated anti-tumor immune responses. Eur J Immunol. 2007;37:675–685. doi: 10.1002/eji.200636639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Notta F, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–367. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 21.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS ONE. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staaf J, et al. Identification of subtypes in human epidermal growth factor receptor 2—positive breast cancer reveals a gene signature prognostic of outcome. J Clin Oncol. 2010;28:1813–1820. doi: 10.1200/JCO.2009.22.8775. [DOI] [PubMed] [Google Scholar]
- 24.Shirley SH, et al. Transcriptional regulation of estrogen receptor-alpha by p53 in human breast cancer cells. Cancer Res. 2009;69:3405–3414. doi: 10.1158/0008-5472.CAN-08-3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 26.Glück S, et al. TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine ± trastuzumab. Breast Cancer Res Treat. 2011 doi: 10.1007/s10549-011-1412-7. [DOI] [PubMed] [Google Scholar]
- 27.Valastyan S, Weinberg RA. Tumor metastasis: Molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 29.Jiang Z, et al. Rb deletion in mouse mammary progenitors induces luminal-B or basal-like/EMT tumor subtypes depending on p53 status. J Clin Invest. 2010;120:3296–3309. doi: 10.1172/JCI41490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.