Abstract
The half-a-tetratricopeptide repeat (HAT) motif is a helical repeat motif found in proteins that influence various aspects of RNA metabolism, including rRNA biogenesis, RNA splicing, and polyadenylation. This functional association with RNA suggested that HAT repeat tracts might bind RNA. However, RNA binding activity has not been reported for any HAT repeat tract, and recent literature has emphasized a protein binding role. In this study, we show that a chloroplast-localized HAT protein, HCF107, is a sequence-specific RNA binding protein. HCF107 consists of 11 tandem HAT repeats and short flanking regions that are also predicted to form helical hairpins. The minimal HCF107 binding site spans ∼11 nt, consistent with the possibility that HAT repeats bind RNA through a modular one repeat–1 nt mechanism. Binding of HCF107 to its native RNA ligand in the psbH 5′ UTR remodels local RNA structure and protects the adjacent RNA from exonucleases in vitro. These activities can account for the RNA stabilizing, RNA processing, and translational activation functions attributed to HCF107 based on genetic data. We suggest that analogous activities contribute to the functions of HAT domains found in ribonucleoprotein complexes in the nuclear–cytosolic compartment.
Keywords: helical repeat protein, plastid, pentatricopeptide repeat
The half-a-tetratricopeptide (HAT) motif is a helical repeat motif that has been functionally linked to RNA because of its presence exclusively in complexes that influence RNA metabolism (1). Examples of HAT domain proteins include Utp6, which is involved in nuclear pre-rRNA processing, Prp6, which is involved in nuclear pre-mRNA splicing, and CstF-77, which is involved in pre-mRNA cleavage and polyadenylation. The HAT motif is related to the tetratricopeptide repeat (TPR), a degenerate 34-aa motif that forms a pair of antiparallel α-helices (reviewed in ref. 2). TPR motifs are typically found in tandem arrays, which stack to form a broad surface that binds protein ligands. Crystal structures of CstF-77 confirmed that HAT repeat tracts adopt a TPR-like structure (3, 4). The possibility that HAT repeat tracts might bind RNA has been suggested (1, 5–7), but the notion that HAT repeat tracts serve as a scaffold for binding other proteins dominates recent literature (3, 8–10). This view is reflected by the annotation of the HAT motif at InterPro, which states only that “the repeats may be involved in protein–protein interactions” (http://www.ebi.ac.uk/interpro/IEntry?ac=IPR003107).
Although TPR, HEAT, and several other helical repeat motifs form protein binding surfaces, similar structures have been shown to bind nucleic acids. Puf and pentatricopeptide repeat (PPR) motifs have been shown to bind RNA (reviewed in refs. 11 and 12), and mTERF, ALK, and TAL repeat motifs have been shown to bind DNA (reviewed in refs. 13 and 14). Thus, it seemed quite plausible that the presence of HAT domains exclusively in ribonucleoprotein complexes reflects the fact that HAT repeat tracts interact directly with RNA. In this study, we show that the HAT domain protein HCF107 does, in fact, bind RNA and that it does so with high affinity and specificity for its genetically defined site of action. HCF107 consists almost entirely of HAT motifs and lacks other functional domains. Thus, findings obtained with HCF107 are likely to elucidate general mechanisms through which HAT domains act.
HCF107 is a nucleus-encoded protein that localizes to the chloroplasts of land plants. Mutations in Arabidopsis HCF107 cause defects in the translation and stabilization of mRNAs derived from the chloroplast psbH gene (6, 15). Our results show that HCF107 binds ssRNA with specificity for sequences at the 5′ end of the processed psbH mRNA isoforms that require HCF107 for their accumulation. In addition, we show that bound HCF107 (i) blocks a 5′ → 3′ exonuclease in vitro, accounting for its ability to stabilize psbH RNA in vivo, and (ii) remodels local RNA structure in a manner that can account for its ability to enhance psbH translation. We suggest that the RNA binding, RNA protection, and RNA remodeling activities observed for HCF107 are likely to be general features of long HAT repeat tracts, and that these properties are likely to contribute to the functions of HAT domains found in other ribonucleoprotein complexes throughout the cell.
Results
HCF107 Is a Sequence-Specific RNA Binding Protein.
HCF107 is targeted to chloroplasts by an N-terminal transit peptide (6). Mature HCF107 (i.e., lacking the cleaved transit peptide) has 11 consecutive HAT motifs flanked by ∼100 aa on each side (Fig. S1A). However, a structural model generated by I-TASSER (16) predicts that virtually the entire mature portion of HCF107 consists of helical hairpins (Fig. S1B), with approximately two additional helical hairpin units flanking each side of the canonical HAT repeats.
Mature HCF107 was expressed as a fusion to maltose binding protein in Escherichia coli and purified by successive amylose affinity and gel filtration chromatography (Fig. S2). Recombinant HCF107 (rHCF107) has a monomeric molecular mass of 67 kDa but eluted from a size exclusion column at a position corresponding to a globular protein of ∼150 kDa, indicating that it is an elongated monomer or forms homodimers.
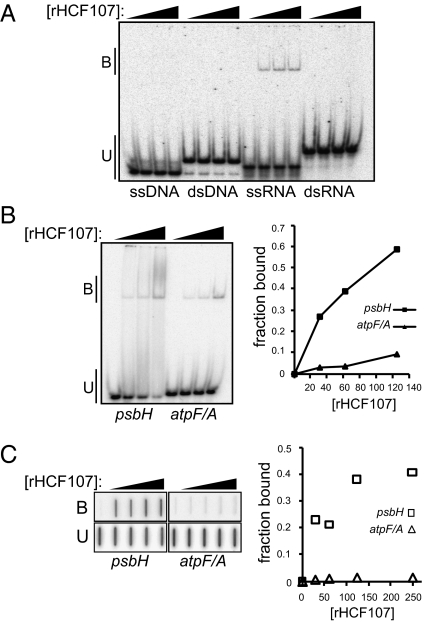
To test whether rHCF107 has the capacity to bind nonspecifically to nucleic acids, gel mobility shift assays were performed under low stringency binding conditions (i.e., in the absence of competitor RNA or heparin) using oligonucleotides of the same sequence but in the form of either ssRNA, dsRNA, ssDNA, or dsDNA (Fig. 1A). rHCF107 bound to the ssRNA but not the other nucleic acid forms.
Fig. 1.
Gel mobility shift assays showing the RNA binding capacity of rHCF107. (A) Synthetic 31-mers of the same sequence but in ssRNA, dsRNA, ssDNA, or dsDNA form were incubated with rHCF107 (0, 50, 100, and 200 nM) and resolved on a native polyacrylamide gel. Bound (B) and unbound (U) RNAs/DNAs are marked. (B) RNAs derived from the 5′ UTR of Arabidopsis psbH (26 nt) or the maize atpF/atpA intergenic region (27 nt) were incubated with rHCF107 (0, 31, 62, and 125 nM) under stringent conditions (150 mM NaCl, 0.5 mg/mL heparin). The complex formed between rHCF107 and the psbH RNA did not form a sharp band, but the loss of unbound RNA as protein concentration increases is easily visible. The total counts per lane and the counts in unbound RNA were quantified and used to calculate the fraction-bound RNA (Right). (C) Binding reactions performed as in B were assayed by filter binding: reactions were filtered through stacked nitrocellulose and nylon membranes such that RNA bound to protein was retained on the nitrocellulose, and unbound RNA passed through the nitrocellulose and was retained on the nylon membrane. The bound (B) and unbound (U) RNAs were quantified and used to generate the plot in Right.
HCF107 influences the metabolism of the polycistronic RNA derived from the chloroplast psbB-psbT-psbH-petB-petD gene cluster (15). Mutations in HCF107 cause the specific loss of processed RNAs with a 5′ end 44 nt upstream of the psbH start codon (Figs. 2B and 3A), and also decrease psbH translational efficiency (15). The PPR protein PPR10 has analogous activities but acts on different chloroplast mRNAs (17). Based on the mechanisms established for PPR10 (18), we hypothesized that the HCF107 binding site maps to the immediate 5′ end of those RNA isoforms that fail to accumulate in its absence and that bound HCF107 blocks 5′ → 3′ RNA degradation. As a first step toward testing this hypothesis, we determined whether rHCF107 binds with specificity to a 26-nt RNA with a 5′ end that maps 2 nt upstream of the HCF107-dependent psbH 5′ terminus. A 27-nt RNA matching a different intergenic region was used as a negative control, and binding reactions included 0.5 mg/mL heparin to reduce nonspecific interactions. rHCF107 bound with much higher affinity to the RNA from the psbH 5′ UTR than it bound the unrelated RNA, showing it to be a sequence-specific RNA binding protein (Fig. 1 B and C).
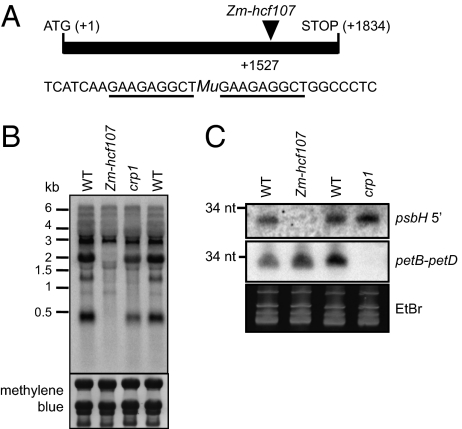
Fig. 2.
A maize hcf107 mutant (Zm-hcf107) lacks an sRNA corresponding to the HCF107-dependent psbH 5′ end. (A) Position of a Mu1-transposon insertion in Zm-hcf107. The diagram shows its position within the spliced ORF. The sequence flanking the insertion is shown below, with the target site duplication underlined. (B) RNA gel blot showing loss of processed psbH mRNAs in the Zm-hcf107 mutant. The missing transcripts match those transcripts that are missing in Arabidopsis hcf107 mutants (15). (C) RNA gel blot showing chloroplast sRNAs from two intergenic regions. Total leaf RNA (15 μg) from a Zm-hcf107 mutant, a crp1 mutant, and a phenotypically normal sibling of each (WT) was fractionated in a polyacrylamide gel and electrophoretically transferred to nylon membrane. Replicate membranes were probed with oligodeoxynucleotides complementary to the sRNA in the psbH 5′ UTR (the putative HCF107 footprint) or the sRNA in the petB–petD intergenic region (the putative CRP1 footprint) (20). The position of a 34-nt RNA size marker is shown.
Fig. 3.
Defining the HCF107 binding site. (A) Diagram of the psbT–psbH intergenic region. These genes lie within the psbB-psbT-psbH-petB-petD transcription unit. The sequence shows the 5′ end of the RNAs that fail to accumulate in hcf107 mutants. The HCF107 binding site (as established through the assays shown in B) is underlined; dashes indicate nucleotides that likely contribute to binding affinity but that are not essential for binding. (B) Gel mobility shift assays used to define the HCF107 binding site. Binding reactions used rHCF107 (0, 50, 100, and 200 nM) under the same conditions as in Fig. 1B with 0.5 mg/mL heparin. The sequences of the RNAs are shown below. (C) Equilibrium Kd of rHCF107 for RNA 1 and RNA 10. Binding reactions were performed as in B. The raw data are shown in Fig. S3. (D) Multiple sequence alignment showing conservation of the HCF107 binding site among angiosperms. At, A. thaliana; Hv, Hordeum vulgare; Nt, Nicotiana tabacum; Os, Oryza sativa; Pa, Populus alba; Zm, Zea mays.
Additional evidence that the native binding site of HCF107 resides within the 5′-most 21 nt of HCF107-dependent psbH mRNA isoforms comes from the observation that this region is represented by abundant small RNAs (sRNAs) in Arabidopsis, rice, barley, and maize (19, 20). The binding sites of several PPR proteins accumulate as sRNAs in vivo, and there is evidence that these sRNAs are in vivo protein footprints that accumulate because they are protected by the cognate protein from endogenous ribonucleases (17, 19, 20). To evaluate whether the sRNA mapping to the 5′ end of processed psbH RNA is the HCF107 footprint, we determined whether it fails to accumulate in a maize hcf107 (Zm-hcf107) mutant (Fig. 2A). Zm-hcf107 is allelic to the photosystem II mutant hcf3 (21), and maize and Arabidopsis hcf107 mutants lack the same subset of psbH RNAs (Fig. 2B). The psbH sRNA was absent in the Zm-hcf107 mutant but accumulated normally in a crp1 mutant (Fig. 2C), which lacks a PPR protein that stabilizes a different RNA segment in the same transcription unit (22, 23). These in vivo results in conjunction with the in vitro RNA binding data provide strong evidence that the sRNA derived from the psbH 5′ UTR harbors the native binding site of HCF107.
Mapping the Minimal HCF107 Binding Site.
To define the minimal RNA segment required for a high-affinity interaction with HCF107, we assayed binding to truncated RNAs derived from the psbH 5′ UTR (Fig. 3). Truncations from the 3′ direction through residue −28 (with respect to the psbH start codon) caused little effect on binding (compare RNAs 1–5 and note the decrease in unbound RNA as protein concentration increases). However, removal of the C and A residues at positions −29 and −30 eliminated binding (compare RNA 5 with RNA 6). These results place the 3′ end of the minimal HCF107 binding site at −29 or −30.
Truncation from the 5′ direction through residue −44 did not affect binding significantly (compare RNAs 1–3), whereas truncation through the A residue at position −39 abolished binding (compare RNA 10 with RNA 11). Qualitative comparison of the data for RNAs 5, 7, and 10 suggested that one or more residues between −40 and −43 also contribute to binding affinity. These results place the 5′ boundary of the minimal HCF107 binding site at position −39 and imply incremental contributions by several additional nucleotides upstream. A quantitative comparison of binding to RNAs 1 and 10 confirmed that nucleotides outside of the minimal binding site increase binding affinity, and showed that rHCF107 binds the intact site with a Kd in the low nanomolar range (Fig. 3C).
Taken together, these experiments showed that the HCF107 binding site spans at least 11 and no more than 15 nt. This size correlates well with the number of helical hairpin modules in HCF107; 11 canonical HAT repeats are flanked by several predicted helical hairpins that do not conform to the HAT consensus. In light of the modular mechanism of RNA recognition established for Puf domains (24), it seems reasonable to propose that HAT repeats likewise bind RNA through a modular one repeat–1 nt mechanism.
HCF107 Blocks 5′ → 3′ and 3′ → 5′ Exoribonucleases in Vitro.
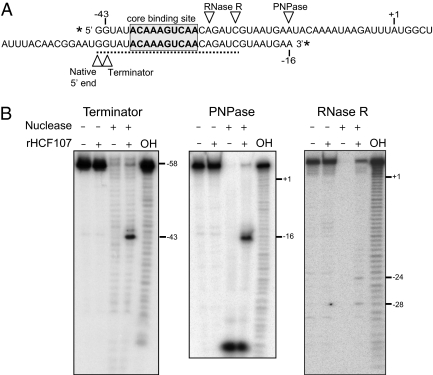
The in vitro RNA binding data in conjunction with prior genetic data show that HCF107 is required for the accumulation of processed mRNAs with a 5′ end several nucleotides upstream of its binding site (Figs. 2 and 3) (15). We hypothesized that HCF107 positions this 5′ end and stabilizes the RNA downstream by serving as a barrier to 5′ → 3′ exoribonucleases. To test this hypothesis, we performed in vitro assays using rHCF107, a 3′ end-labeled RNA spanning the HCF107 binding site (Fig. 4A), and Terminator Exonuclease, a commercially available 5′ → 3′ exoribonuclease. In the absence of rHCF107, Terminator Exonuclease fully degraded the RNA (Fig. 4B). Addition of rHCF107 blocked the exonuclease, yielding a major product with a 5′ end that maps to position −43. The 5′ end that is stabilized by HCF107 in vivo was not mapped to single-nucleotide resolution in Arabidopsis (15), but it is likely to map to −44 based on data from barley and tobacco (20, 25). Thus, these results strongly support the view that HCF107 promotes the accumulation of 5′-processed psbH mRNAs by blocking an endogenous 5′ → 3′ exoribonuclease.
Fig. 4.
HCF107 impedes exonucleases and defines the position of the processed psbH 5′ end. (A) The RNA shown above was 5′ end-labeled and used in the PNPase and RNase R assays. The RNA shown below was 3′ end-labeled and used in the Terminator assay. The HCF107-dependent sRNA is marked by the dashed line. (B) The labeled RNAs were incubated with Terminator, PNPase, or RNase R in the presence or absence of rHCF107. A partial alkaline hydrolysis ladder (OH) of the labeled RNA was included in each gel, and was used to infer the positions at which the exonucleases were blocked by rHCF107.
Analogous experiments were performed with a 5′ end-labeled RNA (Fig. 4A) and the 3′ → 5′ exonucleases polynucleotide phosphorylase (PNPase) or RNase R (Fig. 4B). rHCF107 blocked PNPase-mediated RNA degradation ∼14 nt downstream of the minimal HCF107 binding site, whereas it blocked RNase R at two positions, mapping 2 and 6 nt downstream of the minimal HCF107 binding site. This difference in RNase R and PNPase behavior likely reflects a difference in the structure of these enzymes: PNPase stalls ∼9 nt downstream from the base of stable RNA hairpins because of steric interference with the enzyme's narrow neck (26, 27). Thus, the positions at which rHCF107 blocks PNPase and RNase R correlate well with the 3′ boundary of HCF107's binding site. The boundaries of HCF107's footprint in the exonuclease protection assays are similar to the boundaries of the HCF107-dependent sRNA (Fig. 4A), providing additional evidence that this sRNA is HCF107's in vivo footprint.
HCF107 Remodels RNA Structure in a Manner That Accounts for Its Ability to Enhance Translation.
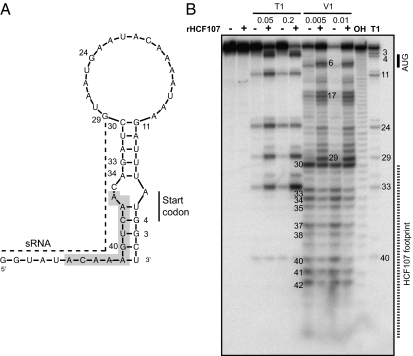
In addition to stabilizing processed psbH mRNAs, HCF107 increases the efficiency of psbH translation (15). Translation initiation in chloroplasts involves mechanisms similar to those in bacteria: internal binding of ribosomes to unstructured RNA near the start codon (sometimes with the aid of a Shine–Dalgarno interaction) (reviewed in refs. 28 and 29). It had been proposed that HCF107 enhances psbH translation by increasing the abundance of 5′-processed psbH RNAs, which were predicted to have reduced potential for inhibitory RNA–RNA pairing in the ribosome binding region (compared with their precursors) (15). However, when RNA secondary structure is predicted by MFold (30) based on a temperature of 30 °C (closer to physiological temperatures than the 35 °C used in the prior study), the psbH start codon and flanking nucleotides are predicted to form a duplex with the HCF107 binding site (Fig. 5A). Thus, we hypothesized that HCF107 enhances psbH translation by sequestering the RNA to which it is bound, which would otherwise form an inhibitory duplex with the translation initiation region. An analogous mechanism was shown for the PPR protein PPR10, which binds in the chloroplast atpH 5′ UTR and enhances atpH translation (17, 18).
Fig. 5.
HCF107 remodels the structure of the psbH 5′ UTR to expose the translation initiation region. (A) Secondary structure of the 5′ UTR of processed psbH mRNA was calculated with MFold assuming a temperature of 30 °C. The core HCF107 binding site is shaded, and the sRNA sequence (the putative in vivo footprint of HCF107) is indicated by a dashed line. (B) The RNA shown in A was radiolabeled at its 5′ end and incubated with RNase T1 or V1 in the presence or absence of rHCF107. Each ribonuclease was used at two different concentrations. An alkaline hydrolysis ladder (OH) and the RNase T1 cleavage pattern of the denatured RNA (T1) provide size markers.
To test this hypothesis, the effect of rHCF107 on the structure of the psbH 5′ UTR was assayed by probing with ribonucleases T1 and V1 (Fig. 5B). RNase T1 cleaves after guanosines in a single-stranded context, whereas RNase V1 cleaves regions in which the bases are stacked, as when they are present in an RNA duplex. The cleavage patterns in the absence of rHCF107 support the structure predicted by MFold: residues in the 5′ portion of the predicted RNA duplex were digested by RNase V1, and the pair of G residues within the predicted duplex (residues 3 and 4) were highly resistant to cleavage by RNase T1. The addition of rHCF107 caused a dramatic increase in RNase T1 digestion at G3 and G4, which are within and adjacent to the start codon. rHCF107 also enhanced RNase T1 cleavage at positions 11, 24, 29, and 33. Therefore, the binding of rHCF107 shifts the equilibrium structure near the psbH start codon from largely duplex in nature to a predominantly single-stranded structure. As expected, rHCF107 also caused a generalized protection of residues within and adjacent to its binding site from RNase V1.
These results support the view that the binding of HCF107 reduces the formation of a secondary structure that would otherwise interfere with ribosome binding. In contrast with the previous proposal that HCF107 enhances psbH translation by stimulating accumulation of the processed psbH isoform (15), our results argue that it is the presence of HCF107 on the psbH 5′ UTR that enhances translation.
Discussion
In this study, we explored the basis for the effects of the HAT protein HCF107 on chloroplast gene expression. We showed that HCF107, which consists almost entirely of HAT repeats, is a sequence-specific RNA binding protein that binds in the psbT–psbH intercistronic region. After it is bound, HCF107 blocks the progression of 5′ → 3′ exoribonucleases, which defines the 5′ end of processed psbH transcripts and also stabilizes the downstream RNA segment. In addition, HCF107 binding remodels the structure of the psbH 5′ UTR in a way that can account for its ability to enhance psbH translation. These findings explain the molecular defects in hcf107 mutants (15) and are likely to be relevant to the similar phenotypes observed for mutations in Mbb1, the HCF107 ortholog in Chlamydomonas (31). Furthermore, the position of the HCF107 binding site matches the position of the intercistronic expression element, a cis-element reported to generate stable, translatable mRNAs in tobacco chloroplasts (25); this finding strongly suggests that the intercistronic expression element acts by recruiting the tobacco HCF107 ortholog.
RNA Binding Activity of HAT Repeats.
When the HAT motif was first recognized to be a variant of the TPR motif (1), it was noted that all characterized HAT proteins are found in complexes that interact with RNA. This theme has been maintained with the characterization of additional HAT proteins. Structural analyses have confirmed that HAT repeats adopt a TPR-like α-solenoid structure (3, 4, 10), but little is known about the biochemical functions of HAT repeats. Early reports speculated that HAT repeats might bind RNA (1, 5, 7). However, a lack of evidence for RNA binding and the fact that some HAT repeats can bind peptides (8, 9) have led recent literature to assume that HAT motifs serve only to support protein–protein interactions (3, 9, 10). Our demonstration that the HAT protein HCF107 binds RNA with high affinity and sequence specificity together with the exclusive association of HAT proteins with RNA-containing complexes strongly suggest that RNA binding activity contributes to the biological functions of most HAT domains.
Functional Parallels Between HAT and PPRs.
The finding that HAT repeats can bind RNA was foreshadowed by the discovery of two other helical repeat motifs with RNA binding activity: the Puf and PPR motifs. Puf domains consist of eight repeating units of three α-helices that stack to form a curved solenoid (32). The basis for sequence-specific RNA binding by Puf domains has been characterized in depth (24, 33). Several amino acids in each Puf repeat mediate the recognition of specific nucleotides, and consecutive repeats generally bind consecutive nucleotides. PPR proteins comprise a large protein family whose members localize to mitochondria and chloroplasts, where they influence various aspects of organellar RNA metabolism (reviewed in ref. 12). Like the HAT motif, the PPR motif is related to the TPR motif (34), and both HAT and PPR tracts are variable in length (typically, 10–15 repeats). Several PPR proteins have been shown to bind specifically in vitro to their genetically defined RNA targets (17, 20, 35–37). Although structures are not available for PPR/RNA complexes, current data support a modular 1–repeat 1 nucleotide recognition mechanism that is reminiscent of the Puf/RNA interaction (18, 38). The correspondence between the length of the minimal HCF107 binding site and the length of its HAT repeat tract suggests the same to be true for HAT repeats.
The parallels between HAT and PPR proteins extend to their biological functions. The HAT protein HCF107 stabilizes psbH mRNA, defines the processed psbH 5′ end, and enhances psbH translation in vivo (6, 15). Analogous activities have been attributed to several members of the PPR family (reviewed in ref. 12), the most thoroughly characterized of which is PPR10 (17, 18). PPR10 stabilizes several processed mRNA segments and defines their termini by impeding 5′ and 3′ exonucleases. At the same time, PPR10 enhances the translation of an ORF mapping downstream from one of its binding sites by remodeling RNA structure to increase the single-stranded character of the ribosome binding region. These potent activities result simply from sequestering an extended RNA segment (∼25 nt), preventing that RNA from interacting with other macromolecules. This mechanism is analogous to the mechanism of many sRNAs in bacteria, which by sequestering an RNA segment in a duplex, influence translation or RNA stability. HCF107's activities in vivo and in vitro are strikingly similar to those of PPR10, supporting the view that HCF107's fundamental activity is, likewise, to prevent the RNA to which it is bound from interacting with other proteins (i.e., exonucleases) or RNA (i.e., the downstream ribosome binding region). Genetic analysis of a second chloroplast HAT protein implies a similar mechanism (39). It seems likely that any HAT repeat tract of sufficient length would have analogous effects on the RNA to which it binds and that these activities contribute to the functions of HAT proteins that participate in nuclear rRNA biogenesis, pre-mRNA splicing, and polyadenylation.
In contrast to the striking functional and biochemical similarities between HCF107 and PPR proteins, the evolutionary histories of the HAT and PPR families are very different. The PPR protein family expanded to >400 members during the evolution of land plants, whereas the HAT family has remained small (∼10–15 members) and highly conserved across the eukaryotes. Almost all PPR proteins localize to organelles, whereas almost all HAT proteins localize to the nucleus. It will be interesting to discover the basis for this apparent difference in the evolutionary malleability of these two otherwise very similar protein classes.
Materials and Methods
Preparation of Recombinant HCF107.
A cDNA encoding mature HCF107 (i.e., lacking the transit peptide) was amplified by PCR from Arabidopsis cDNA in two steps. First, two overlapping fragments were amplified with primer pairs (i) k116 (5′-GGGGggatccGCAGTCGTGGACAGGTCTTCTTCAGG) and k119 (5′-AACGTCCAGCTTCAAAGGATCTAACGACAGC) and (ii) k117 (5′-GGGGgtcgacTCAAGCACCATTTATTCTTCCTCTAG) and k118 (5′-GCTGTCGTTAGATCCTTTGAAGC). Second, the two fragments were joined by amplification with primers k116 and k117. The product was digested with Sal I and Bam HI and cloned into pMAL-TEV. maltose binding protein-HCF107 was expressed in E. coli, purified by amylose affinity chromatography, and cleaved with TEV protease, and the untagged protein was then purified on a gel filtration column as described previously for PPR5 (36), except that the lysis and dialysis buffers contained 400 mM NaCl and did not include CHAPS detergent.
RNA Binding Assays.
Synthetic RNAs (Integrated DNA Technologies) were 5′ end-labeled with [γ-32P]-ATP and T4 polynucleotide kinase and then purified on a denaturing polyacrylamide gel. Except where otherwise indicated, binding reactions contained 150 mM NaCl, 50 mM Tris⋅HCl, pH 7.5, 4 mM DTT, 0.04 mg/mL BSA, 0.5 mg/mL heparin, 10% (vol/vol) glycerol, 10 units RNAsin (Promega), and 15 pM radiolabeled RNA. Reactions were incubated for 30 min at 25 °C and resolved on 5% native polyacrylamide gels (gel mobility shift assays) or filtered through stacked nitrocellulose and nylon membranes (filter binding assays). Results were visualized on a Storm phosphorimager. Data quantification was performed with ImageQuant (Molecular Dynamics). The binding curve in Fig. 3 was graphed, and the apparent Kd value was calculated with KaleidaGraph software as described (40). The 31-mers used to test sequence nonspecific binding to various nucleic acid forms (Fig. 1A) were of the same sequence and prepared in the same manner as those used previously (36). The nonspecific oligonucleotide used in Fig. 1 B and C came from the maize chloroplast atpF–atpA intergenic region and had the following sequence: 5′-UUAGAAUUUAGGCAUUAUUUUUCCCUU.
RNA Gel Blot Analysis.
RNA gel blots used for detection of sRNAs used polyacrylamide gels and electrophoretic transfer to a charged nylon membrane as previously described (20). The following DNA oligonucleotides were used as probes: petB–petD (5′-AGCAATGAAATACCACAACCTACCCGATATG-3′) and psbH 5′ UTR (5′-AATCATTGGTGTTGACTTTGTATACT-3′). The RNA gel blot showing psbH mRNAs in Zm-hcf107 mutants was performed similarly, except that the RNAs were resolved on a 1% agarose gel. This blot was probed with the same oligonucleotide used to detect the psbH sRNA.
Exonuclease Protection and RNA Structure Probing Assays.
The exonuclease protection assays were performed as described previously (18) with the following minor modifications. For the PNPase assay, rHCF107 (400 nM) was bound to the 5′-radiolabeled RNA shown in Fig. 4A and incubated with 2 units Synechocystis PNPase (Sigma-Aldrich) at 25 °C for 30 min. For the Terminator assay, the RNA oligonucleotide shown in Fig. 4A was labeled at its 3′ end, bound to rHCF107 (400 nM), and incubated with 2 units Terminator exonuclease (Epicentre Biotechnologies) for 1 h. For the RNase R assay, the binding reaction was performed in 20 mM Tris⋅HCl (pH 8.0), 100 mM KCl, and 1 mM MgCl2, and 1 unit RNase R (Epicentre Biotechnologies) was used for digestion (30 min). RNA structure probing was also performed as described previously (18) with the following minor modifications. The synthetic RNA diagrammed in Fig. 5 (120 pM) was radiolabeled at its 5′ end and incubated in the absence or presence of 400 nM rHCF107 at 25 °C in reaction buffer containing no BSA and only 50 ng/μL heparin. RNase T1 (0.05 or 0.2 units; Ambion) or RNase V1 (0.01 or 0.005 units; Ambion) was added, and reactions were incubated at 25 °C for 10 min. Samples were analyzed on a 12% polyacrylamide gel containing 8 M urea and 1× Tris/borate/EDTA.
Identification of Maize hcf107 Insertion Mutant.
The Zm-hcf107 mutant was initially detected in the laboratory of Don Miles (University of Missouri, Columbia, MO) as a Mu-transposon–induced mutant lacking photosystem II. Complementation crosses showed this mutant to be allelic to the photosystem II mutant hcf3 (21). Southern blot display of Mu-elements identified a 4.4-kb Hind III fragment harboring a Mu1-element that cosegregated with the mutant phenotype. This fragment was cloned by generating a size-enriched λ-library followed by plaque hybridization using a Mu1-probe. The insertion site was confirmed by PCR amplification of flanking sequences with Mu- and gene-specific primers followed by sequencing of the PCR products.
Supplementary Material
Acknowledgments
We thank Margarita Rojas at the University of Oregon for assistance with the purification of rHCF107; Don Miles at the University of Missouri, in whose laboratory the Mu-tagged allele of hcf3 was initially recovered; and Melissa Scott, Hortensia Rolan, and Sijiong Mou at Midwestern State University for characterization of the maize cDNA. This work was supported by a European Molecular Biology Organization Long-Term Fellowship (to K.H.) and National Science Foundation Grant MCB-0940979 (to A.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200318109/-/DCSupplemental.
References
- 1.Preker PJ, Keller W. The HAT helix, a repetitive motif implicated in RNA processing. Trends Biochem Sci. 1998;23:15–16. doi: 10.1016/s0968-0004(97)01156-0. [DOI] [PubMed] [Google Scholar]
- 2.Allan RK, Ratajczak T. Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones. 2011;16:353–367. doi: 10.1007/s12192-010-0248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai Y, et al. Crystal structure of murine CstF-77: Dimeric association and implications for polyadenylation of mRNA precursors. Mol Cell. 2007;25:863–875. doi: 10.1016/j.molcel.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 4.Legrand P, Pinaud N, Minvielle-Sébastia L, Fribourg S. The structure of the CstF-77 homodimer provides insights into CstF assembly. Nucleic Acids Res. 2007;35:4515–4522. doi: 10.1093/nar/gkm458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung S, McLean MR, Rymond BC. Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA. 1999;5:1042–1054. doi: 10.1017/s1355838299990635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sane AP, Stein B, Westhoff P. The nuclear gene HCF107 encodes a membrane-associated R-TPR (RNA tetratricopeptide repeat)-containing protein involved in expression of the plastidial psbH gene in Arabidopsis. Plant J. 2005;42:720–730. doi: 10.1111/j.1365-313X.2005.02409.x. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Yehuda S, et al. Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics. 2000;156:1503–1517. doi: 10.1093/genetics/156.4.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu S, Rauhut R, Vornlocher HP, Lührmann R. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. RNA. 2006;12:1418–1430. doi: 10.1261/rna.55406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Champion EA, Lane BH, Jackrel ME, Regan L, Baserga SJ. A direct interaction between the Utp6 half-a-tetratricopeptide repeat domain and a specific peptide in Utp21 is essential for efficient pre-rRNA processing. Mol Cell Biol. 2008;28:6547–6556. doi: 10.1128/MCB.00906-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Champion EA, Kundrat L, Regan L, Baserga SJ. A structural model for the HAT domain of Utp6 incorporating bioinformatics and genetics. Protein Eng Des Sel. 2009;22:431–439. doi: 10.1093/protein/gzp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3'UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 12.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Rubinson EH, Eichman BF. Nucleic acid recognition by tandem helical repeats. Curr Opin Struct Biol. 2012;22:101–109. doi: 10.1016/j.sbi.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: Finding plant genes for disease and defense. Curr Opin Plant Biol. 2010;13:394–401. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Felder S, et al. The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell. 2001;13:2127–2141. doi: 10.1105/TPC.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfalz J, Bayraktar OA, Prikryl J, Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009;28:2042–2052. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc Natl Acad Sci USA. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruwe H, Schmitz-Linneweber C. Short non-coding RNA fragments accumulating in chloroplasts: Footprints of RNA binding proteins? Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr1138. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhelyazkova P, et al. Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr1137. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leto KJ, Miles D. Characterization of three photosystem II mutants in Zea mays L. lacking a 32,000 dalton lamellar polypeptide. Plant Physiol. 1980;66:18–24. doi: 10.1104/pp.66.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barkan A, Walker M, Nolasco M, Johnson D. A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 1994;13:3170–3181. doi: 10.1002/j.1460-2075.1994.tb06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fisk DG, Walker MB, Barkan A. Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 1999;18:2621–2630. doi: 10.1093/emboj/18.9.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 25.Zhou F, Karcher D, Bock R. Identification of a plastid intercistronic expression element (IEE) facilitating the expression of stable translatable monocistronic mRNAs from operons. Plant J. 2007;52:961–972. doi: 10.1111/j.1365-313X.2007.03261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorentzen E, Dziembowski A, Lindner D, Seraphin B, Conti E. RNA channelling by the archaeal exosome. EMBO Rep. 2007;8:470–476. doi: 10.1038/sj.embor.7400945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spickler C, Mackie GA. Action of RNase II and polynucleotide phosphorylase against RNAs containing stem-loops of defined structure. J Bacteriol. 2000;182:2422–2427. doi: 10.1128/jb.182.9.2422-2427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barkan A. Expression of plastid genes: Organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 2011;155:1520–1532. doi: 10.1104/pp.110.171231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scharff LB, Childs L, Walther D, Bock R. Local absence of secondary structure permits translation of mRNAs that lack ribosome-binding sites. PLoS Genet. 2011;7:e1002155. doi: 10.1371/journal.pgen.1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaistij FE, Boudreau E, Lemaire SD, Goldschmidt-Clermont M, Rochaix JD. Characterization of Mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2000;97:14813–14818. doi: 10.1073/pnas.97.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell. 2001;7:855–865. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- 33.Koh YY, et al. Stacking interactions in PUF-RNA complexes. RNA. 2011;17:718–727. doi: 10.1261/rna.2540311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Small ID, Peeters N. The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 35.Okuda K, Nakamura T, Sugita M, Shimizu T, Shikanai T. A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J Biol Chem. 2006;281:37661–37667. doi: 10.1074/jbc.M608184200. [DOI] [PubMed] [Google Scholar]
- 36.Williams-Carrier R, Kroeger T, Barkan A. Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA. 2008;14:1930–1941. doi: 10.1261/rna.1077708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hattori M, Sugita M. A moss pentatricopeptide repeat protein binds to the 3′ end of plastid clpP pre-mRNA and assists with mRNA maturation. FEBS J. 2009;276:5860–5869. doi: 10.1111/j.1742-4658.2009.07267.x. [DOI] [PubMed] [Google Scholar]
- 38.Fujii S, Bond CS, Small ID. Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc Natl Acad Sci USA. 2011;108:1723–1728. doi: 10.1073/pnas.1007667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boudreau E, Nickelsen J, Lemaire SD, Ossenbühl F, Rochaix JD. The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J. 2000;19:3366–3376. doi: 10.1093/emboj/19.13.3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warf MB, Diegel JV, von Hippel PH, Berglund JA. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc Natl Acad Sci USA. 2009;106:9203–9208. doi: 10.1073/pnas.0900342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.