Abstract
In mycobacteria, polyketide synthases and nonribosomal peptide synthetases (NRPSs) produce complex lipidic metabolites by using a thio-template mechanism of catalysis. In this study, we demonstrate that off-loading reductase (R) domain of mycobacterial NRPSs performs two consecutive [2 + 2]e- reductions to release thioester-bound lipopeptides as corresponding alcohols, using a nonprocessive mechanism of catalysis. The first crystal structure of an R domain from Mycobacterium tuberculosis NRPS provides strong support to this mechanistic model and suggests that the displacement of intermediate would be required for cofactor recycling. We show that 4e- reductases produce alcohols through a committed aldehyde intermediate, and the reduction of this intermediate is at least 10 times more efficient than the thioester-substrate. Structural and biochemical studies also provide evidence for the conformational changes associated with the reductive cycle. Further, we show that the large substrate-binding pocket with a hydrophobic platform accounts for the remarkable substrate promiscuity of these domains. Our studies present an elegant example of the recruitment of a canonical short-chain dehydrogenase/reductase family member as an off-loading domain in the context of assembly-line enzymology.
Keywords: chain release, glycopeptidolipid, NAD(P)H, tyrosine-dependent oxidoreductase
Polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs) are multifunctional proteins that are known to produce a variety of complex natural products (1). While most of these natural products can be classified as secondary metabolites, in mycobacteria PKSs and NRPSs synthesize lipidic metabolites that are important for their survival and pathogenesis (2). The biosynthetic mechanism involves assembly-line repetitive condensation of specific monomeric units. During this process, the intermediates remain covalently tethered to the proteins through the thiol group of phosphopantetheine (ppant) moiety that is posttranslationally added onto the carrier domains (3). The ppant-arm-bound substrate reaches out to the active centers of the various domains to facilitate successive catalytic steps. The chain-releasing domain then performs dual function of detaching the mature product and playing a role in determining the final structure of the metabolite.
The most well-studied chain-releasing domains are thioesterases (TE) that hydrolyze the thioester bond to release linear as well as macrocyclic products (4). Recently, a new mechanism of chain release catalyzed by the reductase (R) domains has been identified. These domains utilize NAD(P)H as cofactor to reductively release the final product as aldehyde or alcohol (Fig. S1A) (5). Interestingly, 2 R domain homologues are shown to perform cofactor-independent Dieckmann’s cyclization (referred to as R* domains) (6). Broadly, R domains show homology to the family of short-chain dehydrogenases/reductases (SDRs). The SDR family consists of tyrosine-dependent oxidoreductases that are known to share common sequence motifs and the overall mechanism of catalysis (7). Presently, no structural and mechanistic details are available regarding how R domains function in the context of multifunctional synthases.
Mycobacteria contain several homologues of NRPS and PKS proteins, some of which possess R domains at their carboxy-termini (C-termini). In this study, we have elucidated the biochemical, structural and mechanistic aspects of two such R domains: RGPL (Fig. 1A) from glycopeptidolipid (GPL) cluster of Mycobacterium smegmatis (Msmeg) and RNRP from NRPS of Mycobacterium tuberculosis (Mtb). Both RGPL and RNRP reductively release acyl chains as corresponding alcohol through a committed aldehyde intermediate. Interestingly, 4e- reductases catalyze the second reductive step (of converting aldehyde to alcohol) with approximately 15-fold greater efficiency. Our structural studies suggest that NAD(P)H cofactor exchange would involve major conformational changes and require displacement of aldehyde intermediate. We propose that 4e- reductases perform two reductive steps through a nonprocessive mechanism of catalysis.
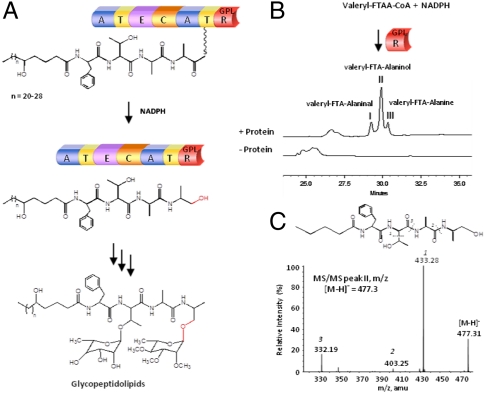
Fig. 1.
Mycobacterial glycopeptidolipid (GPL) biosynthesis involves thioester reduction to alcohol: (A) Proposed mechanism of chain release during GPL biosynthesis. (B) HPLC analysis of RGPL assay with valeryl-L-Phe-L-Thr-L-Ala-L-Ala-CoA (valeryl-FTAA-CoA) and NADPH shows valeryl-FTA-Alaninol (peak II) as the major product along with valeryl-FTA-Alaninal (peak I) and valeryl-FTA-Alanine (peak III). Control reaction in the absence of protein did not show any product. (C) Tandem mass spectrometric analysis of valeryl-FTA-Alaninol peak at m/z [M-H]- = 477.3.
Results
Off-Loading Reductase Domains from Mycobacteria.
Mycobacterial genome mining using the known R domain protein sequences led to the identification of several multifunctional proteins that possess R domains at their C-termini (Fig. S1B). We decided to examine R domains from Mps2 protein, which is implicated in the biosynthesis of GPLs in Msmeg (8, 9) and from Nrp protein with unknown function from Mtb (10). Pairwise sequence alignments of RGPL and RNRP with other R domains revealed overall homology in the range of 45–50% with other aldehyde/alcohol-forming R domains and also with the Dieckmann’s cyclization-catalyzing R* domains (Fig. S1C). In order to identify the biochemical function of RGPL and RNRP, we decided to perform in vitro reconstitution of purified recombinant proteins. These domains are integral components of multifunctional proteins and have been suggested to show poor stability when expressed as independent proteins (11). We therefore decided to express RGPL and RNRP as independent domains as well as bifunctional proteins (T-RGPL and T-RNRP) along with the upstream thiolation (T) domains. The NRPS-PKS tool was used to identify the junction between the T and R domains (12). The RNRP, RGPL domain and T-RNRP didomain constructs showed good soluble expression in Escherichia coli (E. coli). The T-RGPL didomain construct showed poor expression in soluble form. The recombinant proteins were expressed with hexa-histidine tags and could be purified to homogeneity.
Glycopeptidolipid RGPL Is an Alcohol-Producing Reductase Domain.
Retrobiosynthetic approach suggests that the substrate for RGPL would be an acyl-tetrapeptide (Fig. 1A). The formation of alaninol could be a two-stage process, wherein either the RGPL would first produce alaninal and then another unknown protein could convert this to alaninol, or the RGPL could directly synthesize alaninol by 4e- reduction. Initially, the assays for RGPL and T-RGPL were carried out with smaller analogues of this acyl-tetrapeptide-ppant moiety. Because L-Ala is at the (n-1) position from the scissile bond, Boc-L-Alanyl-CoA (tert-butyloxycarbonyl-L-Alanyl Coenzyme A) was synthesized (Fig. S2A). However, no activity could be detected for T-RGPL (in the presence of surfactin synthetase-activating enzyme, 4’ phosphopantetheinyl synthetase) (13) or for RGPL under varying experimental conditions. We then proceeded to synthesize a tetrapeptide substrate with a smaller fatty-acyl (C5, valeryl) chain. The valeryl group was used to facilitate the solubility of the substrate in aqueous buffer. The synthetic lipopeptide-CoA was purified on HPLC (Fig. S2B), and the identity was confirmed by using tandem mass spectrometry (MS) analysis (Fig. S2 C and D). Enzymatic assays of RGPL showed robust activity with both valeryl-D-Phe-D-Thr-D-Ala-L-Ala-CoA and valeryl-L-Phe-L-Thr-L-Ala-L-Ala-CoA (valeryl-FTAA-CoA). The products were resolved on HPLC and further analyzed on ESI-MS. Three closely eluting peaks at 220 nm could be separated at 48% acetonitrile/water (0.1% TFA) (Fig. 1B). The most prominent peak (II) at 29.8 min showed m/z mass of [M-H]- = 477.3, which is consistent with the molecular formula C24H37N4O6 corresponding to valeryl-FTA-Alaninol. Tandem mass spectrometric analysis of molecular ion peak at m/z = 477.3 clearly showed several fragments, including the loss of Thr side chain from molecular ion and cleavage of the -CO-NH- peptide bonds (Fig. 1C). The smaller peak (I) at 29.2 min corresponded to valeryl-FTA-Alaninal with [M-H]- = 475.2 (Fig. S3A). The molecular ion for peak III corresponded to the free peptide (Fig. S3B) and this probably is due to the noncatalytic hydrolysis of the substrate. Our studies thus demonstrate that RGPL is a 4e- reductase which can convert p-pant-tethered lipopeptide to lipopeptide alcohol during GPL biosynthesis.
RNRP also Performs 4e- Reductions to Synthesize Lipopeptide Alcohol.
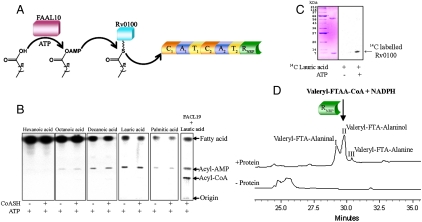
Bioinformatic analysis indicated a very high sequence identity between the RGPL and RNRP. We therefore proceeded to investigate whether RNRP protein would also show similar biochemical activity. Nrp is a bimodular protein with seven catalytic domains (12), which include two condensation domains (C), two adenylation domains (A), and 2 T domains and a RNRP domain organized as C1-A1-T1-C2-A2-T2-RNRP (Fig. 2A). Because the metabolite produced by the Nrp protein is unknown, the retrobiosynthetic approach cannot be used to design substrates for the RNRP domain. The NRPS-PKS tool does not predict substrate specificity of A1, but suggests A2 to be specific for phenylalanine. However, RNRP and T-RNRP assays using Boc-Phenylalanyl-CoA (Fig. S3C) did not result in formation of products. Because genes involved in the biosynthesis of nonribosomal peptides are clustered together in the genomes of various microorganisms, we analyzed the neighboring genes of nrp in the Mtb genome to ascertain possible chemical nature of the substrates. Interestingly, a fatty acyl-adenylate ligase (FAAL) homologue (Rv0099/FAAL10) and a T domain homologue (Rv0100) are positioned adjacent to nrp (Rv0101) in the genome. FAAL proteins in Mtb have been demonstrated to activate fatty acids as acyl adenylates, which are then transferred onto a thiolation domain to synthesize complex metabolites (14, 15) (Fig. 2A). We therefore decided to investigate the specificity of FAAL10 protein. Both the FAAL10 and Rv0100 proteins were cloned and expressed in E. coli with a C-terminal hexa-histidine tag and purified to homogeneity by using affinity chromatography (Fig. S3D). Enzymatic assays showed that FAAL10 could catalyze conversion of several medium-chain fatty acids to corresponding acyl-adenylates (Fig. 2B). The addition of coenzyme A did not result in the formation of acyl-CoA, confirming that this protein indeed belongs to the newly discovered FAAL family of proteins (14). Addition of posttranslationally modified Rv0100 protein to the assay showed facile transfer of acyl group on the ppant arm of Rv0100, thiolation protein (Fig. 2C). This acyl chain is expected to be utilized by the Nrp protein to perform the first round of condensation by the first module (C1-A1-T1). Based on these studies, we argued that the substrate of RNRP would resemble a lipopeptide. Because RGPL and RNRP share a very high sequence identity (66%), and also the fact that a lipotetrapeptidyl-CoA was synthesized for RGPL assays, we decided to examine whether valeryl-FTAA-CoA could be a substrate for RNRP. Strikingly, RNRP showed significant activity with valeryl-FTAA-CoA, and the product formed could be readily characterized by a combination of HPLC and mass spectrometric analysis (Fig. 2D). Analogous to RGPL assays, both aldehyde and alcohol products could be observed. The major product obtained was valeryl-FTA-Alaninol, thus confirming that RNRP is also a 4e- reducing off-loading domain.
Fig. 2.
FAAL10-nrp genetic cluster of Mtb: (A) Scheme showing the predicted function of FAAL10 and Rv0100 in the synthesis of a novel metabolite by this genetic cluster. (B) FAAL10 converts octanoic (C8) acid, decanoic (C10) acid, lauric (C12) acid and palmitic (C16) acid, but not hexanoic (C6) acid to corresponding acyladenylates. No acyl-CoA is formed when Coenzyme A (CoASH) is added to the reaction. Control reaction was set up using FACL1914 and lauric acid. (C) Coupled assays of FAAL10 with Rv0100, the left box shows proteins FAAL10 (approximately 60 kDa) and Rv0100 (approximately 10 kDa) used in the assay. The corresponding autoradiogram in the right box shows that in the presence of ATP and 14C lauric acid, the acyl chain gets transferred to the ppant arm of Rv0100 (shown with an arrow). (D) HPLC analysis of RNRP assay with valeryl-FTAA-CoA and NADPH showed valeryl-FTA-Alaninol (peak II) as the major product along with valeryl-FTA-Alaninal (peak I) and valeryl-FTA-Alanine (peak III).
Structure of the RNRP Domain.
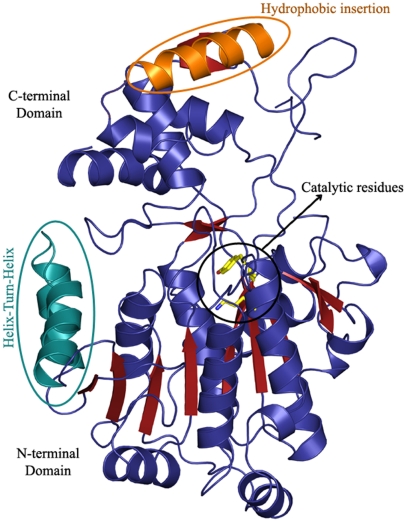
In order to gain better understanding of the catalytic mechanism and substrate promiscuity of R domains, we attempted to crystallize RNRP and RGPL proteins. While RGPL showed propensity for small needle-shaped crystals, good-quality RNRP crystals could be obtained in the space group P3121. Attempts to solve structure by using either molecular replacement or isomorphous replacement were unsuccessful due to poor sequence identity (less than 20% for RNRP and RGPL domains with other SDRs) and nonisomorphous crystals, respectively. The structure for RNRP was determined by using selenomethionine-substituted protein crystals by single wavelength anomalous dispersion (SAD) method to a resolution of 2.3 Å (Table S1). Overall, the structure comprises two intertwined domains: an N-terminal domain composed of 282 amino acids and a C-terminal domain with 144 amino acids (Fig. 3). The N-terminal domain has an extended NAD(P)H-binding α/β Rossmann fold with seven parallel beta strands (β2-6 and β8-10). These strands in the beta sheet are arranged in the order 4-3-2-5-6-8-10, which are flanked by five helices (α2, α5 and 6, α8, α10) (Fig. S4A). This feature is characteristic of the tyrosine-dependent oxidoreductases SDR family of proteins. According to structure-based sequence alignment, the canonical catalytic triad T193, Y228, and K232 of RNRP is present at similar positions as found in other members of this family (Fig. S1C). One of the distinctive features in the N-terminal domain is the presence of a unique insertion of a helix-turn-helix motif (S90 to D119) embedded between β3 and β4 that has not been observed in other known structures from this family.
Fig. 3.
Analysis of RNRP crystal structure: Structure of reductase domain RNRP shows that it is composed of an N-terminal domain with the Rossmann fold and a unique C-terminal domain.
The C-terminal domain in SDR family of proteins is implicated in varied substrate recognition and binding. Sequence analysis indicated that the mycobacterial R domains possess a larger C-terminal domain (around 140 residues) than the other SDR family members (around 100 residues) (16). Structural comparisons show that C-terminal region (unlike N-terminal region) is very divergent with a DALI Z-score of 3.8 with 10% sequence identity and root-mean-square deviation (rmsd) of 3.4 Å over 87 Cα atoms aligned with guanosine diphosphate-fucose synthetase from E. coli (17). The DALI scores provide structural similarity between each pair of protein domains and structures that have significant similarities have a Z-score above 2. The low DALI Z-score along with the rmsd analysis indicates that the C-terminal domain is unique in its architecture. The C-terminal domain consists of four alpha helices (α9, 11 and 12, 14) and two beta strands (β9, 11). There are two unique insertions, a loop from F291 to A303 and a helix from D366 to L381 that is also conserved in other R domains from mycobacteria. The helical insertion (D366-L381) contains several hydrophobic residues (W370, L371, F374, L378) that in conjunction with other nonpolar residues F291, W253, and L354 form a large hydrophobic surface that could have a role in substrate binding. Interestingly, fatty acyl-CoAs could also be used as substrates by RNRP and RGPL, and both proteins could reduce lauroyl-CoA and palmitoyl-CoA into corresponding alcohol and aldehyde products (Fig. S3E).
Structural Analysis of NAD(P)H and Substrate-Binding Pocket.
Despite significant efforts no cofactor or substrate bound structures could be obtained (Fig. S4B). Based on the structural superimposition with other members, putative NAD(P)H binding pocket could be identified in the RNRP structure. The residues G59, G62, and G65 constitute the GxxGxxG Rossmann fold nucleotide-binding motif (Fig. S4C). In previous studies, the other conserved residues R87 and D127 were implicated in stacking interaction with the adenosine ring of the NAD(P)H, and the R97 could provide stabilizing interaction with the phosphate of NAD(P)H (18). Although the geometry and interacting residues of this cofactor-binding pocket are well conserved, this pocket is occupied by a loop (S153 to F166) in the RNRP structure (Fig. S4D). It is important to note that this loop between the β5 of the Rossmann fold and the following α-helix shows a distinctly different position in the other SDR structures that have been solved in the presence of NAD(P)H (19). We propose that this loop mobility is essential for generating a binding pocket for NAD(P)H. The average B-factor observed for the residues in this loop is 52.35 Å2 as compared with 44.13 Å2 for all atoms of RNRP crystal structure, which further provides support for the flexible nature of this loop.
In order to delineate the substrate-binding site, we superposed the RNRP structure on to the ternary complex structure of 2,4-dienoyl-CoA reductase (PDB ID 1W6U) with 2,4-hexadienoyl-CoA and β-NADP+ (Fig. S4 E and F). The conservation of NAD(P)H binding pocket and N terminus facilitated identification of the lipopeptidyl-CoA-binding pocket. The valeryl FTAA-ppant was docked into the binding site. Clearly, the valeryl-FTAA-ppant occupies the position adjacent to the conserved hydrophobic groove in the RNRP structure. The valeryl chain occupies only a small region of this hydrophobic groove, suggesting that a larger acyl chain could also be accommodated in this groove. The catalytic residues Y228 and K232 and the nicotinamide ring lie within catalytic distance from the thioester bond (orange, Fig. S5A).
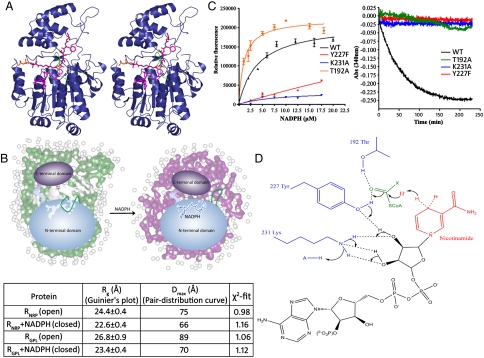
It is obvious that valeryl-FTAA thioester reduction to valeryl-FTA-Alaninol product requires two rounds of hydride transfer from two molecules of NADPH. Because in our cell-free assay there is no mechanism to regenerate spent NADPH in situ, it is mandatory to exchange the oxidized cofactor with the reducing equivalent to carry out a second reduction. Moreover, the SDR enzymes have been shown to follow a common sequential ordered bi-bi reaction mechanism, involving sequential cofactor binding, substrate binding, catalysis, product release, and finally release of coproduct (20). We thus asked the question whether cofactor recycling would involve the displacement of aldehyde intermediate from the active site. Based on our modeling studies for the RGPL domain, the cofactor is bound deep inside the active site pocket and the substrate is positioned such that it could possibly block the exit of the oxidized cofactor (Fig. 4A). Thus, it is also expected that the cofactor exchange will be associated with conformational changes. We assessed RNRP conformational rearrangements associated with NADPH binding by performing small angle X-ray scattering (SAXS) analysis. We observed a significant decrease in radius of gyration (Rg) and in maximum particle size (Dmax) upon addition of NADPH (Fig. 4B, Fig. S5B). Superposition of the SAXS envelope and the RNRP crystal structure revealed that our apo-RNRP structure actually represents the NADPH-bound or the closed conformation (Fig. S5C). An illustrative model for the possible loop movement along with compaction of N- and C-terminal domains resulting in decreased radius of gyration is shown in open and closed conformations of RNRP (Fig. 4B).
Fig. 4.
Dissecting the two-step 4e- reduction mechanism: (A) Stereoimage shows the cofactor NADPH (magenta) and substrate valeryl-FTAA-ppant (red) modeled in the R domain. A movement of the loop, S153-F166 (green) would provide a platform for binding of the NADPH followed by valeryl-FTAA-ppant. The extrusion of the aldehyde formed after first reduction might facilitate the exchange of oxidized cofactor NADP+ for NADPH. (B) Structural parameters determined for RGPL and RNRP along with NADPH using Guinier approximation and indirect Fourier transformation of SAXS data. Also shown is an illustrative model demonstrating loop movement along with compaction of N- and C-terminal domains in open and closed conformations. (C) NADPH binding to RGPL and Y227F, K231A, and T192A mutants as determined by intrinsic NADPH fluorescence (Left) and analysis of NADPH consumption in spectrophotometric assays of these mutants and WT RGPL using valeryl-FTA-Alaninal substrate (Right). (WT RGPL Kd for NADPH was calculated as 4.64 ± 0.94 μM.) (D) Proposed mechanism of R domain-catalyzed reduction of lipopeptidyl thioester substrate.
Dissecting the [2 + 2]e- Reduction Mechanism.
The tolerance of RGPL to utilize fatty acyl-CoAs provided a relatively easy assay system to initially investigate the details of catalytic mechanism. We first established that RGPL catalyzed lauroyl-CoA reduction proceeds through the formation of aldehyde product. Lauroyl alcohol appears later and eventually becomes the major product (Fig. S6A). Interestingly, we identified a positive correlation between protein concentration and alcohol product formation (Fig. S6B). Because the formation of alcohol product requires two rounds of reduction, the protein concentration dependence can have two possible explanations: these enzymes either reduce slowly or function in the nonprocessive mode (as proposed by the structural studies). Furthermore, if these R domains have to function as nonprocessive enzymes, then exogenously provided aldehyde should be a good substrate for these enzymes. We thus undertook chemical synthesis of RGPL aldehyde intermediate valeryl-FTA-Alaninal (Fig. S6 C and D). HPLC-MS analysis of RGPL and RNRP enzymatic assays with purified valeryl-FTA-Alaninal clearly showed formation of valeryl-FTAAlaninol (II) as the product in the presence of protein and NADPH (Fig. S6E). Kinetic analysis performed by monitoring NADPH utilization at 340 nm revealed that the turnover of aldehyde to alcohol was several times faster (kcat/kM[valeryl-FTAAlaninal] = 100 M-1 min-1, kcat[valeryl-FTAAlaninal] = 0.24 min-1) than thioester reduction (kcat/kM[valeryl-FTAA-CoA] = 7 M-1 min-1, kcat[valeryl-FTAA-CoA] = 0.003 min-1). The greater efficiency of the second step could be important, as several SDR proteins are shown to have a competing reversible reaction and would also ensure that there is no accumulation of aldehyde intermediate. Cell-free reactions with RGPL by using [1-14C] lauroylaldehyde also demonstrated formation of lauroyl alcohol in the presence of NADPH (Fig. S6F).
The functional importance of active site residues Y227, K231, and T192 in NAD(P)H binding was determined by assaying the intrinsic fluorescence of NAD(P)H in the presence and absence of WT RGPL, Y227F, K231A, and T192A RGPL mutant proteins. We observed increase in NAD(P)H fluorescence emission in the presence of WT and T192A RGPL but not with Y227F and K231A mutants (Fig. 4C). Similar spectral changes were observed for FabG protein, where Tyr151 was shown to be necessary for high-affinity productive cofactor binding (21). All three mutant proteins are catalytically inactive in cell-free assays set up with either valeryl-FTAA-CoA or valeryl-FTA-Alaninal substrates (Fig. 4C; Fig. S6G and H). Based on these evidences, we propose the NADPH and Thr-Tyr-Lys dependent mechanism for RGPL catalyzed reduction of its cognate lipopeptide thioester and aldehyde substrates (Fig. 4D). To our surprise, all three mutant proteins could convert lauroyl aldehyde to lauroyl alcohol. Although the exact mechanism for this conversion is not clear, this result reflects the difference between valeryl-FTA-Alaninal and smaller substrate lauroyl aldehyde. An analogous catalytic Tyr-independent mechanism of reduction has been previously reported for an SDR family protein pteridine reductase, PTR1 (22), and an aldo-ketoreductase (AKR), AKR1C9 (23).
Modelling of Nrp Termination Module.
We further attempted to place the R domain in the context of the entire module based on termination module structure of srfA-C from Bacillus subtilis (B. subtilis) (PDB code 2VSQ) (24). Fig. S7A shows the superposition of RNRP with the thioesterase domain of the termination module from B. subtilis. Interestingly, careful structural analysis of the hydrolase fold of the TE domain and the Rossmann fold showed remarkable structural superposition for the central β-sheet architecture (Fig. S7A, Inset). This conserved core along with few surrounding helices permitted placement of the RNRP active site in the vicinity of TE active site residues with respect to other domains of 2VSQ, as shown in Fig. S7B. The entire termination module of Nrp protein could be modeled using 2VSQ as a template. A reasonably high conservation of the overall architecture and position of active site residues were noted (Fig. S8). Our studies thus suggest a similar overall organization of catalytic centers for the two termination modules.
Discussion
An important feature of the assembly-line multifunctional NRPS and PKS biosynthetic enzymes is the presence of chain-releasing domain (25). Apart from performing the crucial off-loading step to maintain enzymatic turnover, both TE and R domains mediate different outcomes of the final product. While TE domains belong to α/β hydrolase fold and involve mandatory formation of oxyester intermediate, R domains have been proposed to reductively release acyl chains directly from the ppant arm of the upstream T domain (5). In this study, we have characterized R domains from two mycobacterial NRPS proteins that perform 4e- reductions to release lipidic metabolites as alcohols. Our studies suggest a nonprocessive mechanism of catalysis for 4e- reductases, wherein at the end of each reductive cycle the product extrudes out from the active site pocket to facilitate exchange of cofactor. Such a mechanism has been recently proposed for Dot1 enzyme that is involved in mono-, di-, or trimethylation of lysine residues in histone proteins (26).
Processive enzymes perform consecutive rounds of reduction without substrate dissociation, whereas nonprocessive or hit-and-run enzymes dissociate and reassociate to allow exchange of cofactor and reorientation of substrate. Our structural studies showed that aldehyde intermediate generated during the first reductive step could physically obstruct the exchange of the oxidized cofactor. This persuasive evidence is also supported by biochemical studies: (i) the relative ratio of formation of 4e- reduction product to 2e- reduction product increases with increasing amount of enzyme and (ii) both RGPL and RNRP overwhelmingly catalyze reduction of lipopetidyl aldehyde over the cognate lipopetidylthioester substrate. It is important to note that lipopeptidyl-CoA is not the native substrate and so kinetic parameters need to be compared with caution. On the other hand, precise kinetic studies for T-R didomain by loading substrates through ppant transferase only permit single-turnover assays. Crystallographic studies in conjunction with SAXS and fluorescence data suggest an important role of protein conformational changes in response to cofactor NAD(P)H. In the apo-structure of RNRP, the NAD(P)H-binding site is occupied by a loop S153 to F166. We propose that this loop will have to move out to accommodate NAD(P)H, as observed for other homologous structures crystallized with this cofactor (19). The correlation of this loop movement to the [2 + 2]e- reduction cycle could in future provide further insights into this nonprocessive mode of catalysis.
Catalysis by R domains could be described by minimal 10 steps: (1) NADPH binding, (2) open to close conformation, (3) thioester binding, (4) thioester reduction to aldehyde, (5) close to open conformation, (6) aldehyde followed by NADP dissociation, (7) second NADPH binding, (8) open to close conformation, (9) aldehyde reduction to alcohol, and (10) alcohol followed by NADP dissociation. The difference in catalysis observed between valeryl-FTA-Alaninal and lauroylaldehyde could be due to less steric hindrance from lauroylaldehyde and therefore less conformational restraint for Steps 7 and 8. Tyr and Lys may be essential for conformational change (Steps 7 and 8) with the natural substrate mimic (val-FTAA-alaninal) and not so essential with the smaller substrate (lauroylaldehyde), which may also possess a relatively lower activation energy barrier. The contrast probably also reflects in the observed large rate difference between thioester reduction (Steps 1–9) and aldehyde reduction (Steps 7–9). An analogous catalytic Tyr-independent mechanism of reduction is reported from an SDR family protein pteridine reductase, PTR1, and an aldo-ketoreductase (AKR), AKR1C9. Other factors like propinquity effect proposed in earlier study cannot be ruled out (23).
The functional analysis of RGPL clarifies the crucial step in GPL biosynthesis, as the formation of this terminal alcohol is mandatory for the subsequent glycosylation. Although the implications of the Nrp protein in Mtb biology are not clear, our studies predict that this cluster would produce a lipophilic peptide containing a terminal alcoholic group. Such R domains are indeed widely distributed in several NRPS biosynthetic clusters (5, 27) and are also found in fungal peptaibol synthetases (28) and more recently in Dictyostelium PKSs (29). Analogous to other off-loading TE domains, the broad substrate tolerance of the R domains may be crucial to the assembly-line logic for maintaining the enzymatic flux. In conclusion, our structural and biochemical studies reveal an unconventional two-step catalytic mechanism of reduction, which provides interesting insights into the evolution of enzymatic functions.
Methods
The Bacterial Artificial Chromosome library of M. tuberculosis H37Rv was obtained from Pasteur Research Institute (Paris). Genomic DNA was isolated from M. smegmatis Mc2. Radiolabeled compounds were obtained from American Radiolabeled Chemicals. All chemicals of analytical grade were purchased from Sigma, New England Biolabs, and Merck.
Cloning, Expression, and Purification of Proteins.
Primers used for PCR amplification of respective T-R and R domains and genes are listed in Table S2, and purification was performed using affinity chromatography followed by size exclusion chromatography. Details are provided in SI Text.
Enzymatic Assays.
Valeryl-D-Phe-D-Thr-D-Ala-L-Ala-OH and valeryl-L-Phe-L-Thr-L-Ala-L-Ala-OH were synthesized in solution phase. Purified RGPL/RNRP 5–120 μM, valeryl-FTAA-CoA 200 μM, and NADPH 2 mM were used for enzymatic assays, as described in SI Text.
Crystallization, Data Processing, and Refinement of RNRP.
Crystals of RNRP were obtained by sitting-drop method at 20 °C using 1 μL of a premix of protein, NADPH, and Lauroyl-CoA in 1∶10∶10 ratio. Structure was solved by single wavelength anomalous dispersion method (SI Text) with data collected at X-12 beamline at European Molecular Biology Laboratory, Hamburg, Germany.
Structural Modeling and SAXS Studies.
Models were generated using MODELLER. SAXS measurements were recorded using in-house SAXS instrument, S3 Micro (Hecus X-ray systems GMBH, Graz), as described in the SI Text.
Coordinates.
The coordinates of the Reductase domain (RNRP) have been deposited in Protein Data Bank (accession code 4DQV).
Supplementary Material
Acknowledgments.
We thank Dr. Souvik Maiti and Dr. Kausik Chakraborty for their valuable discussion. A.C., A.S.H., and A.G. are Senior Research Fellows of the Council of Scientific and Industrial Research, India. R.S. is supported by a Swarnajayanti Fellowship from the Department of Science and Technology, India. R.S.G. is a Howard Hughes Medical Institute International Fellow and is also supported by a Swarnajayanti Fellowship from the Department of Science and Technology, India. This work is also supported by grants from the Council of Scientific and Industrial Research, India, three Council of Scientific and Industrial Research laboratories and grants from the Department of Biotechnology, India, and the National Institute of Immunology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4DQV).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118680109/-/DCSupplemental.
References
- 1.Gokhale RS, Saxena P, Chopra T, Mohanty D. Versatile polyketide enzymatic machinery for the biosynthesis of complex mycobacterial lipids. Nat Prod Rep. 2007;24:267–277. doi: 10.1039/b616817p. [DOI] [PubMed] [Google Scholar]
- 2.Gokhale RS, Sankaranarayanan R, Mohanty D. Versatility of polyketide synthases in generating metabolic diversity. Curr Opin Struct Biol. 2007;17:736–743. doi: 10.1016/j.sbi.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 3.Walsh CT, Gehring AM, Weinreb PH, Quadri LE, Flugel RS. Post-translational modification of polyketide and nonribosomal peptide synthases. Curr Opin Chem Biol. 1997;1:309–315. doi: 10.1016/s1367-5931(97)80067-1. [DOI] [PubMed] [Google Scholar]
- 4.Gokhale RS, Hunziker D, Cane DE, Khosla C. Mechanism and specificity of the terminal thioesterase domain from the erythromycin polyketide synthase. Chem Biol. 1999;6:117–125. doi: 10.1016/S1074-5521(99)80008-8. [DOI] [PubMed] [Google Scholar]
- 5.Du L, Lou L. PKS and NRPS release mechanisms. Nat Prod Rep. 2010;27:255–278. doi: 10.1039/b912037h. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Walsh CT. Cyclopiazonic acid biosynthesis in Aspergillus sp: Characterization of a reductase-like R* domain in cyclopiazonate synthetase that forms and releases cyclo-acetoacetyl-L-tryptophan. Biochemistry. 2009;48:8746–8757. doi: 10.1021/bi901123r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kavanagh KL, Jornvall H, Persson B, Oppermann U. Medium- and short-chain dehydrogenase/reductase gene and protein families : the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci. 2008;65:3895–3906. doi: 10.1007/s00018-008-8588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Billman-Jacobe H, McConville MJ, Haites RE, Kovacevic S, Coppel RL. Identification of a peptide synthetase involved in the biosynthesis of glycopeptidolipids of Mycobacterium smegmatis. Mol Microbiol. 1999;33:1244–1253. doi: 10.1046/j.1365-2958.1999.01572.x. [DOI] [PubMed] [Google Scholar]
- 9.Schorey JS, Sweet L. The mycobacterial glycopeptidolipids: Structure, function, and their role in pathogenesis. Glycobiology. 2008;18:832–841. doi: 10.1093/glycob/cwn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 11.Kopp F, Mahlert C, Grunewald J, Marahiel MA. Peptide macrocyclization: the reductase of the nostocyclopeptide synthetase triggers the self-assembly of a macrocyclic imine. J Am Chem Soc. 2006;128:16478–16479. doi: 10.1021/ja0667458. [DOI] [PubMed] [Google Scholar]
- 12.Yadav G, Gokhale RS, Mohanty D. SEARCHPKS: a program for detection and analysis of polyketide synthase domains. Nucleic Acids Res. 2003;31:3654–3658. doi: 10.1093/nar/gkg607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gokhale RS, Tsuji SY, Cane DE, Khosla C. Dissecting and exploiting intermodular communication in polyketide synthases. Science. 1999;284:482–485. doi: 10.1126/science.284.5413.482. [DOI] [PubMed] [Google Scholar]
- 14.Trivedi OA, et al. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature. 2004;428:441–445. doi: 10.1038/nature02384. [DOI] [PubMed] [Google Scholar]
- 15.Arora P, et al. Mechanistic and functional insights into fatty acid activation in Mycobacterium tuberculosis. Nat Chem Biol. 2009;5:166–173. doi: 10.1038/nchembio.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oppermann U, et al. Short-chain dehydrogenases/reductases (SDR): the 2002 update. Chem Biol Interact. 2003;143–144:247–253. doi: 10.1016/s0009-2797(02)00164-3. [DOI] [PubMed] [Google Scholar]
- 17.Somers WS, Stahl ML, Sullivan FX. GDP-fucose synthetase from Escherichia coli: structure of a unique member of the short-chain dehydrogenase/reductase family that catalyzes two distinct reactions at the same active site. Structure. 1998;6:1601–1612. doi: 10.1016/s0969-2126(98)00157-9. [DOI] [PubMed] [Google Scholar]
- 18.Alphey MS, Yu W, Byres E, Li D, Hunter WN. Structure and reactivity of human mitochondrial 2,4-dienoyl-CoA reductase: enzyme-ligand interactions in a distinctive short-chain reductase active site. J Biol Chem. 2005;280:3068–3077. doi: 10.1074/jbc.M411069200. [DOI] [PubMed] [Google Scholar]
- 19.Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Molecular basis for severe epimerase deficiency galactosemia. X-ray structure of the human V94m-substituted UDP-galactose 4-epimerase. J Biol Chem. 2001;276:20617–20623. doi: 10.1074/jbc.M101304200. [DOI] [PubMed] [Google Scholar]
- 20.Jornvall H, et al. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 21.Price AC, Zhang YM, Rock CO, White SW. Cofactor-induced conformational rearrangements establish a catalytically competent active site and a proton relay conduit in FabG. Structure. 2004;12:417–428. doi: 10.1016/j.str.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Gourley DG, et al. Pteridine reductase mechanism correlates pterin metabolism with drug resistance in trypanosomatid parasites. Nat Struct Biol. 2001;8:521–525. doi: 10.1038/88584. [DOI] [PubMed] [Google Scholar]
- 23.Schlegel BP, Ratnam K, Penning TM. Retention of NADPH-linked quinone reductase activity in an aldo-keto reductase following mutation of the catalytic tyrosine. Biochemistry. 1998;37:11003–11011. doi: 10.1021/bi980475r. [DOI] [PubMed] [Google Scholar]
- 24.Tanovic A, Samel SA, Essen LO, Marahiel MA. Crystal structure of the termination module of a nonribosomal peptide synthetase. Science. 2008;321:659–663. doi: 10.1126/science.1159850. [DOI] [PubMed] [Google Scholar]
- 25.Ehmann DE, Gehring AM, Walsh CT. Lysine biosynthesis in Saccharomyces cerevisiae: Mechanism of alpha-aminoadipate reductase (Lys2) involves posttranslational phosphopantetheinylation by Lys5. Biochemistry. 1999;38:6171–6177. doi: 10.1021/bi9829940. [DOI] [PubMed] [Google Scholar]
- 26.Frederiks F, et al. Nonprocessive methylation by Dot1 leads to functional redundancy of histone H3K79 methylation states. Nat Struct Mol Biol. 2008;15:550–557. doi: 10.1038/nsmb.1432. [DOI] [PubMed] [Google Scholar]
- 27.Schuttelkopf AW, Hardy LW, Beverley SM, Hunter WN. Structures of Leishmania major pteridine reductase complexes reveal the active site features important for ligand binding and to guide inhibitor design. J Mol Biol. 2005;352:105–116. doi: 10.1016/j.jmb.2005.06.076. [DOI] [PubMed] [Google Scholar]
- 28.Wiest A, et al. Identification of peptaibols from Trichoderma virens and cloning of a peptaibol synthetase. J Biol Chem. 2002;277:20862–20868. doi: 10.1074/jbc.M201654200. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh R, et al. Dissecting the functional role of polyketide synthases in Dictyostelium discoideum: Biosynthesis of the differentiation regulating factor 4-methyl-5-pentylbenzene-1,3-diol. J Biol Chem. 2008;283:11348–11354. doi: 10.1074/jbc.M709588200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.