Abstract
Diffuse large B-cell lymphomas in humans are associated with chromosomal rearrangements (∼40%) and/or mutations disrupting autoregulation (∼16%) involving the BCL6 gene. Studies of lymphoma development in humans and mouse models have indicated that lymphomagenesis evolves through the accumulation of multiple genetic alterations. Based on our prior studies, which indicated that carcinogen-induced DNA mutations enhance the incidence of lymphomas in our mouse model expressing a human BCL6 transgene, we hypothesized that mutated genes are likely to play an important cooperative role in BCL6-associated lymphoma development. We used retroviral insertional mutagenesis in an effort to identify which genes cooperate with BCL6 in lymphomagenesis in our BCL6 transgenic mice. We identified PIM1 as the most frequently recurring cooperating gene in our murine BCL6-associated lymphomas (T- and B-cell types), and we observed elevated levels of PIM1 mRNA and protein expression in these neoplasms. Further, immunohistochemical staining, which was performed in 20 randomly selected BCL6-positive human B- and T-cell lymphomas, revealed concurrent expression of BCL6 and PIM1 in these neoplasms. As PIM1 encodes a serine/threonine kinase, PIM1 kinase inhibition may be a promising therapy for BCL6/PIM1-positive human lymphomas.
Diffuse large B-cell lymphomas in humans are associated with a high mortality rate, and the majority of these contain mutations or chromosomal rearrangements that involve the BCL6 gene (1). Prior studies of deregulated BCL6 expression (2, 3) in mouse models have suggested that lymphoma development in these animals is hastened through the accumulation of multiple oncogenic hits. In an effort to define more targeted therapies for BCL6-associated lymphomas, we sought to identify which mutated genes cooperate with BCL6 in lymphomagenesis. Here, through the use of retroviral insertional mutagenesis in our mouse model of the human BCL6 transgene (2), we detected multiple retroviral insertion sites in the lymphomas (T- and B-cell types) in our animals, a phenomenon that resembles the mutational load noted in human B-cell lymphomas (4). We identified the gene called proviral integration site for Moloney murine leukemia virus 1 (PIM1) as the most frequently recurring cooperating gene in our murine BCL6-associated lymphomas. Immunohistochemical staining, which was performed in 20 randomly selected BCL6-positive human B- and T-cell lymphomas, revealed concurrent expression of BCL6 and PIM1. Because PIM1 encodes a serine/threonine kinase, our findings suggest that PIM1 specific kinase inhibitors may be promising therapies for BCL6/PIM1-associated lymphomas.
Results
MOL4070LTR Infection Accelerates Lymphomagenesis in BCL6 Transgenic Mice.
Our BCL6 transgenic mice are doubly transgenic animals, containing two transgenes: (i) human BCL6 cDNA under control of the tetracycline-responsive minimal promoter (tet-o-BCL6) and (ii) the tetracycline-transactivating protein (tTA) under control of the Ig heavy-chain enhancer and the SRα promoter (EμSR-tTa) (2, 5). As previously described (2), we generated two transgenic mouse lines, one containing four copies of the human BCL6 transgene, the other 20 copies. We used the retrovirus MOL4070LTR (6) for retroviral insertional mutagenesis in our BCL6 transgenic and control animals (nontransgenic mice of the same background). All animals were followed until natural death or necessity for euthanasia. It was possible to analyze the tissues from 92 of 99 transgenic mice injected with retrovirus and from 32 of 33 nontransgenic injected controls. Two animals in the transgenic group died on weaning, and tissues from all other excluded animals were too autolyzed to be analyzed.
Of 53 animals in the four-copy transgenic line, 24 (45.3%) developed lymphomas (18 T-cell, six B-cell; representative histology and flow data shown in Fig. 1). Of 39 mice in the 20-copy line, 10 (25.6%) developed lymphomas (three T-cell, seven B-cell). Lymphomas (BCL6-negative) also were anticipated in the control group, as the MOL4070LTR retrovirus is known to cause lymphomas and leukemias in mice (23% of FVB mice develop T-cell lymphomas by 270 d and approximately 50% of mice develop myeloid leukemias) (6). In our study, of 32 injected control (i.e., nontransgenic) animals, 11 (34.4%) developed lymphomas (five T-cell, six B-cell), with an unexpectedly high incidence of B-cell tumors compared with what is reported in the literature (6). Of 124 evaluable animals, 20 did not develop hematopoietic neoplasms (lymphoid or other, e.g., myeloid leukemias). Of these, three were in the control (i.e., nontransgenic) group and lived an average of 155 d (range 53–246 d), five were in the four-copy line and survived an average of 312 d (range, 254–392 d), and 12 were in the 20-copy line and lived an average of 249 d (range, 180–338 d).
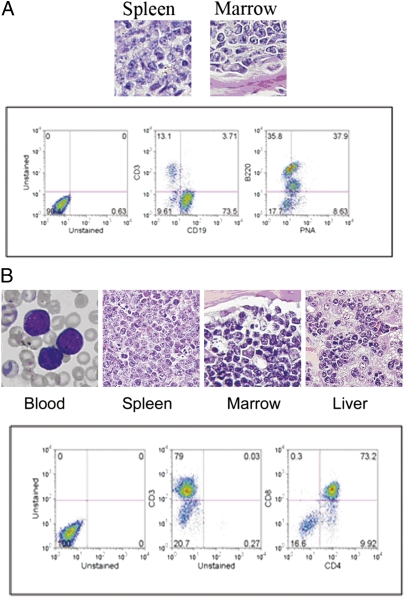
Fig. 1.
Representative lymphomas in BCL6 transgenic mice injected with MOL4070LTR retrovirus. (A) Mature B-cell aggressive lymphoma. Upper: Representative histologic sections. The spleen and bone marrow are infiltrated with large malignant lymphoid cells. Lower: The tumor is a B-cell lymphoma that is CD19+ (Center) and B220+ (Right); the peanut agglutinin positivity (Right) indicates a possible germinal center origin, which is a feature often associated with BCL6-positive B-cell tumors in humans. (B) Precursor T-cell lymphoblastic leukemia/lymphoma. Upper: Representative histologic sections. The peripheral blood smear shows many lymphoblasts. The architecture of the spleen is effaced by lymphoma, and the marrow is packed with lymphoma cells. The liver sinusoids are engorged with large numbers of these cells. Lower: Representative flow cytometry of the splenic lymphoma cells shows that most are CD3 positive (Center) and immature (CD4+/CD8+; Right).
We hypothesized that genes cooperating with BCL6 in lymphomagenesis would be likely to promote faster lymphoma development in transgenic animals compared with control animals. Thus, for statistical purposes, we primarily focused on the first 180 d. Our four-copy BCL6 transgenic line comprised 58% (53 of 92) of the analyzable mice in the study group, and the majority of the early lymphomas developed in this line (14 T-cell, two B-cell). Fig. 2 shows that the study (i.e., transgenic) group in this four-copy line had worse survival (hazard ratio, 3.8; 95% confidence interval, 1.1–13.0; P = 0.03, Cox regression) and, specifically, had a significantly worse 180-d lymphoma mortality rate (30.2%) than the control (i.e., nontransgenic) group (9.4%; P = 0.03, Fisher exact test).
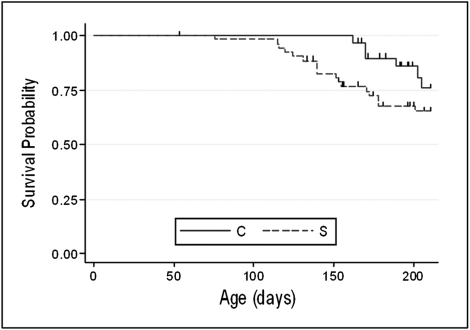
Fig. 2.
Lymphoma-related survival, BCL6 transgenic (i.e., four-copy) line. Cox regression and Fisher exact test show a significantly shorter latency to develop lymphoma for study (S; i.e., transgenic) mice (dashed line) compared with controls (C; i.e., nontransgenic) mice (solid line) at no more than 180 d; the 180-d lymphoma mortality rate was 9.4% in the control group and 30.2% in the study group (P = 0.03).
The lymphomas usually were aggressive neoplasms. Most of the B-cell lymphomas were mature B-cell (7), CD19+, B220+, with associated splenomegaly (Fig. 1A) and often lymphadenopathy. The majority of T-cell tumors (Fig. 1B) could be classified as precursor T-cell lymphoblastic lymphoma/leukemia (7), often manifesting with very large thymic masses as well as infiltration in multiple organs.
The 34 lymphomas from transgenic mice (24 from the four-copy line and 10 from the 20-copy line) contained three to 29 insertion sites (average of 11); the 11 neoplasms from the controls contained between eight and 20 insertion sites (average of 12.7). Sequencing of 377 insertion sites identified in the transgenic animals and 140 in the controls led to the identification of 15 insertion sites that were present in at least two transgenic animals but not in any controls. A number of these had been previously reported in the Retrovirus Tagged Cancer Gene Database (July 2007). We elected to focus attention on lymphomas containing retroviral integration sites in or near the PIM1 gene, which represented the most commonly recurring integration site in our transgenic animals. Specifically, the MOL4070LTR retrovirus had inserted in or near the PIM1 gene in seven of 34 lymphomas (21%) in transgenic mice, but not in any of the 11 lymphomas that developed in the 32 control (i.e., nontransgenic) animals. Assuming that the retrovirus has an equal likelihood of inserting anywhere within the mouse genome (∼25,000 genes), the finding that the lymphomas from seven transgenic mice (but no control animals) contained insertion sites within or near the PIM1 gene is highly statistically significant (P < 0.0001, binomial probability test).
The lymphomas containing insertion sites in or near the PIM1 gene were identified in four female and three male mice, representing both of our BCL6 transgenic lines, ranging in age from 4 to 7.6 mo (average, 5.5 mo) at death. The viral integration site was 5′ of the PIM1 gene in two lymphomas, 3′ in three lymphomas, and within the fifth intron of the PIM1 gene in two additional cases (Table 1). One was a B-cell lymphoma (CD19+, B220+) with spread to multiple organs, and the others were T-cell tumors (precursor T-cell lymphoblastic lymphoma/leukemia per Bethesda classification system for murine lymphoid neoplasms) (7). They were aggressive malignancies, usually presenting as large thymic masses with spread in multiple organs.
Table 1.
Relative PIM1 RNA in lymphomas from transgenic mice
| Type | Location of insert | Expression level vs. control |
| T | Intron 5 | 63.3-fold |
| T | Intron 5 | 46.8 |
| T | 92.4 kb 5′ to PIM1 | 3.8 |
| T | 92.4 kb 5′ to PIM1 | 3 |
| T | 4.4 kb 3′ to PIM1 | 5.8 |
| B | 5.1 kb 3′ to PIM1 | 2.2 |
| T | 5.2 kb 3′ to PIM1 | 1.3 |
Enhanced PIM1 RNA Expression in Lymphomas Containing Retroviral Insertions in or Near PIM1.
Total RNA extracted from mouse lymphomas was reverse-transcribed and subjected to real-time quantitative RT-PCR. Each PCR yielded a single product, without nonspecific amplification. Overall, the six T-cell tumors in transgenic mice contained 20.7-fold more PIM1 RNA (mean ± SEM, 0.083 ± 0.044) than the T-cell tumors from three randomly selected controls (mean ± SEM, 0.004 ± 0.0007; P = 0.04; Fig. 3). The two T-cell lymphomas containing retroviral inserts within the PIM1 gene had the highest PIM1 RNA levels (63.3- and 46.8-fold more, respectively, than the mean level found in the T-cell tumors from the nontransgenic retrovirus-injected mice; Table 1). These two insertion sites were within the fifth intron of the PIM1 gene (mouse chromosome 17; nucleotide positions 29,631,139 and 29,631,068, respectively). Table 1 shows that three lymphomas (one B-cell, two T-cell) contained inserts 3′ of PIM1, a region previously reported to be frequently affected by proviral integrations in murine T-cell lymphomas and associated with enhanced levels of PIM1 RNA (8). The only lymphoma that had a less than twofold elevation in PIM1 RNA level compared with the controls was a T-cell tumor with a 3′ viral insertion site.
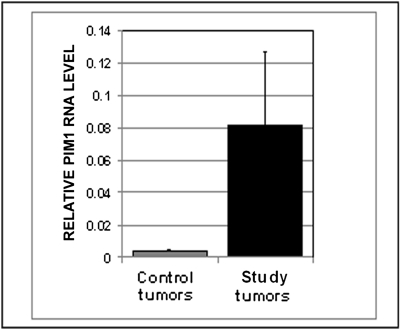
Fig. 3.
PIM1 RNA levels in the six T-cell lymphomas from virus-injected BCL6 transgenic mice (study tumors, black bar) are significantly higher (20.7-fold; P = 0.04) than in T-cell lymphomas from virus- injected nontransgenic controls (control tumors, gray bar).
PIM1 Protein Levels in Lymphomas from BCL6 Transgenic Mice and Human BCL6-Positive Lymphomas.
PIM1 protein levels in murine B- and T-cell tumors detected by immunohistochemical staining correlated generally with PIM1 RNA levels from murine lymphomas quantified by real-time PCR (Fig. 4). As B5 fixation interfered with anti-PIM1 staining, only formalin-fixed tissues were used. PIM1 protein was detectable in all of 20 randomly selected BCL6-positive human tumors (10 B-cell, 10 T-cell). PIM1 expression was variably cytoplasmic and/or nuclear. Nuclear expression, which is believed to correlate with PIM1 biologic effects (9), was detected at least focally in all the murine and human BCL6-positive lymphomas.
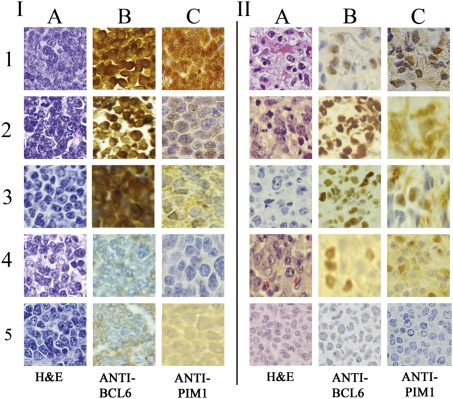
Fig. 4.
Concurrent expression of PIM1 and BCL6 in lymphomas, mouse and human, B- and T-cell, by immunohistochemistry. I, Representative murine lymphomas. In each case, histologic analysis shows malignant tumor cells (H&E stain in column A). 1–3, BCL6-positive lymphomas (brown nuclei, B) from BCL6 transgenic mice containing viral insertions within or near the PIM1 gene (first two rows are T-cell tumors as demonstrated by flow cytometry; Fig. 1B), and row 3 is a B-cell tumor (Fig. 1A). PIM1 levels are positive in all these lymphomas (brown nuclei, C), and the level of staining correlates roughly with the intensity of BCL6 expression. Rows 4 and 5 are lymphomas from control mice (row 4 is T-cell, row 5 is B-cell); BCL6 (B) and PIM1 staining (C) are negative in both controls. II, Representative randomly selected human lymphomas. Histology analysis indicates malignant neoplasm in each case (A, H&E stain). 1–3, Randomly chosen BCL6-positive B-cell tumors previously studied for diagnostic purposes. 4, Randomly selected BCL6-positive lymphoma previously classified diagnostically as T-cell. BCL6 and PIM1 staining are positive (brown nuclei) in each case. 5, Previously studied T-cell lymphoma that does not stain for BCL6 or PIM1.
Discussion
Through the use of retroviral insertional mutagenesis in our unique human BCL6 transgenic mouse model, we identified PIM1 as a gene that cooperates with BCL6 to promote the development of lymphomas, and we showed the concurrent expression of the proteins encoded by these two genes in 20 randomly selected BCL6-positive human B- and T-cell lymphomas. It has long been known that BCL6 and PIM1 are both protooncogenes that play roles in the pathogenesis of lymphoma, but, in each case, supplemental factors (i.e., further oncogenic “hits”) are thought to be necessary for malignant transformation to take place (2, 3, 10). BCL6 expression is believed to become deregulated as a result of mutations that affect binding sites in the first (i.e., noncoding) exon (1) or chromosomal rearrangements in which normal BCL6 regulatory sequences are replaced by heterologous promoters (11). PIM1 has been reported as one of the multiple translocation partners that have been identified in rearrangements with BCL6 (12). PIM1 and BCL6 are associated with the somatic hypermutation events that occur in Ig genes (13, 14), a process that, when aberrant, is thought to contribute to malignant transformation (14); and both are expressed predominantly in lymphocytes (9, 15). In humans, overexpression of PIM1 has been noted in B-cell lymphomas (16) and has been associated with genomic instability (17) in prostate cells. However, in PIM1 transgenic mice, retroviral infection induces T-cell tumors (8). Even in PIM1 transgenic mice containing a leukemia virus long-terminal repeat supplemented with an upstream Ig enhancer and expressing high PIM1 RNA levels in both B and T cells, development of T-cell lymphomas was noted. Interestingly, these T-cell neoplasms were found to contain activated MYC (18), a gene usually associated with B-cell lymphomas in humans. Similarly, in MYC transgenic mice (5), and also in BCL6 (2) transgenic mice, predominantly T-cell lymphomas develop spontaneously (in the case of MYC) (5) or, in the case of BCL6, after induction with a retrovirus (as described here) or mutagen (2), even though these genes are associated predominantly with B-cell neoplasms in humans. Further, it has long been known that BCL6 is expressed not only in human B-cell neoplasms, but also in certain T-cell lymphomas (19), which are more recalcitrant to current treatment modalities than are B-cell tumors. Recently the role of BCL6 in T-cell development and function has received increasing attention (20–26). It is now believed that, even though BCL6 is not required for B or T-cell development, it is absolutely essential for germinal center B-cell (27, 28) and T follicular helper cell differentiation (20–22) and is important in other stages/subsets as well.
Human PIM1 encodes a 313-aa serine-threonine kinase that has antiapoptotic activity and is involved in cell survival, proliferation, differentiation, transformation, tumor progression, as well as angiogenesis (29–31). Its protein is expressed in the thymus, liver, and spleen, and in hematopoietic progenitor cells in the bone marrow during development (32), as well as in the central nervous system (33). Although its cellular localization may be nuclear and/or cytoplasmic, nuclear localization is believed to be necessary for its biologic effects (9). In humans, PIM1 and c-MYC appear to have a cooperative role in prostate cancer (34), and increased expression of PIM1 has been noted in gastrointestinal cancers (35), pancreatic adenocarcinoma (36), and oral squamous-cell carcinoma (37). PIM1 also has been noted to be overexpressed in myeloid and lymphoid leukemias (32) and has been associated with B-cell lymphomas (4, 9). Because loss of PIM1 does not adversely affect the fertility or survival of KO mice (38), there has been intense recent interest in small molecules that inhibit PIM1 expression. SGI-1776, a small-molecule inhibitor of PIM1, PIM2, and PIM3 kinases, induced apoptosis in a human mantle cell lymphoma line and acute myeloid leukemia cell lines when studied as mouse xenografts and in acute myeloid leukemia cell lines in cell culture with little toxicity to normal human lymphocytes (39). This drug, which was in a phase I study in patients with refractory prostate cancer and lymphoma, was associated with cardiac toxicity, and further use in human trials was halted (http://clinicaltrials.gov/ct2/show/NCT00848601).
In conclusion, we have identified PIM1 as a cooperating gene with BCL6 in lymphomas that develop in retrovirus-injected mice expressing the human BCL6 transgene, and we have shown in our studies the concurrent expression of PIM1 and BCL6 in a limited number of randomly selected BCL6-positive human lymphomas. Although PIM1 has long been known to be a protooncogene, its association with BCL6 has not been described to our knowledge. It would be of interest to analyze further the cooperation between PIM1 and BCL6 by breeding Eμ-pim-1 transgenic mice (18) with our mice transgenic for human BCL6 (2) and with the engineered mice of Cattoretti et al. (3). We suggest that study of human lymphomas for PIM1 expression (which is not currently a part of the standard diagnostic lymphoma assessment panel) be added to the routine testing that is currently performed on human B- and T-cell tumors. Our results imply that inhibition of PIM1 kinase in human BCL6-positive and/or PIM1-positive lymphomas may be a useful therapeutic modality in this disease.
Materials and Methods
Retroviral Insertional Mutagenesis.
Under approved institutional protocols, we injected 99 BCL6 transgenic mice (2) and 33 nontransgenic controls [animals of the same background, WT or positive for either (but not both) the Eμ-tTA or BCL6 transgenes] (2) with MOL4070LTR recombinant murine retrovirus (6), 105 infectious units intraperitoneally 1 d after birth, and followed them until euthanasia or death, at which time all animals underwent necropsy.
Histology and Flow Cytometry.
Tissues were fixed in 10% (vol/vol) buffered formalin, paraffin-embedded, sectioned, stained with H&E, and studied with an experienced hematopathologist who was blinded whether the mouse was transgenic or control. Hematologic malignancies were analyzed by FACS whenever possible. Single-cell suspensions were incubated for 15 min with anti-2.4G2 to inhibit nonspecific binding, then labeled with fluorochrome-conjugated antibodies (BD Biosciences/Pharmingen or Sigma-Aldrich) according to the manufacturer's protocol. Analysis was performed on an LSRII flow cytometer (BD Biosciences). Markers included antibodies directed against murine B and T cells, macrophages, myeloid and erythroid cells, immature cells (e.g., Sca-1, CD117), immunoglobulins, κ-, λ-, and germinal center cells (e.g., peanut agglutinin). Lymphoid neoplasms were classified according to the Bethesda proposals (7).
Inverse PCR and Database Searches.
DNA from all analyzable lymphomas was purified and subjected to inverse PCR as described (40), except that 29 cycles of PCR amplification were performed in a 20-μL final volume, with annealing at 61 °C for the 5′ LTR primers and 56 °C for the 3′ LTR primers, and the final extension step was 15 min. Pools of PCR products were cloned and transformed with a TOPO TA Cloning Kit for sequencing (Invitrogen). Purified DNA from ampicillin-resistant colonies was sequenced with a Big Dye Terminator cycle sequencing kit (v3.1; Applied Biosystems) on a 3730xl DNA Analyzer (Applied Biosystems) by using primers from the pCR4-TOPO vector (Invitrogen) in which the inserts had been cloned. Sequences were matched to murine databases of the University of California Santa Cruz (genomic build of July 2007) genome browser and the National Institutes of Health. As it is known that gene expression can be affected over hundreds of kilobases by retroviral integrations (41) and chromosomal translocations (42), we studied potentially affected genes within at least a 100-kb window of an insertion site. Candidate genes identified in this way were considered as possibly relevant if they were noted in at least two transgenic mice but not in any controls (43, 44).
Real-Time Quantitative PCR.
Total RNA was extracted from mouse lymphomas, DNase-1-digested, reverse transcribed, and subjected to real-time RT-PCR (along with “no-RT” controls) with the use of SYBR Green Master Mix (Takara Bio) according to the manufacturer's instructions and a LightCycler 480 (Roche Applied Science). Primers included β-actin (Promega) that amplifies 285 bp (2) of cDNA (vs. 396 bp for genomic DNA) and primers that amplify 176 bp of cDNA of the murine PIM1 gene (5′ primer in exon 5, 3′ primer in exon 6; Real Time Primers). Controls included water template and RNA extracted from randomly selected lymphomas (three T-cell and three B-cell lymphomas) of the comparable organs of retrovirus-injected nontransgenic mice. Results from triplicate wells (for each study animal) or duplicate wells (multiple control mice) were averaged and compared.
Immunohistochemistry.
Human lymphoma paraffin-embedded blocks were obtained from the surgical pathology archives under an institutional review board-approved protocol. Immunohistochemical staining on human tissues was performed with mouse monoclonal anti-human BCL6 (clone LN22; Novocastra) at a dilution of 1:20 on the automated Bond system (Leica Biosytems) with Bond Polymer Refine Detection. Antigen retrieval was performed in Epitope Retrieval Solution 2 for 20 min (Leica Microsystems). A modified protocol included antibody incubation for 25 min, post-primary reagent 15 min, polymer 25 min, peroxide block 8 min, 3,3′-diaminobenzidine 10 min, and hematoxylin 8 min. BCL6 staining on mouse tissues was performed as described previously (2). Additionally, paraffin-embedded formalin-fixed human and murine lymphomas were stained with anti-PIM1 (purified rabbit polyclonal antibody, AP7932d at dilution 1:50; Abgent). For anti-PIM1 staining, antigen retrieval was performed in Bond Epitope Retrieval Solution 2 for 20 min; the peroxide block, post-primary rabbit anti-mouse IgG, and hematoxylin applications were omitted, and the antibody was applied twice (25 min each). Images were taken with a BX41 microscope (Olympus), a DP72 digital camera, and cellSens Standard imaging software (Olympus).
Statistical Analysis.
Lymphoma-related survival in the study vs. control mice was compared with the use of Cox regression and Fisher exact test (to look further at lymphoma-related survival at specific time points). Kaplan–Meier curves also were generated for each group. The data from real-time PCR in the study and control groups were analyzed by the comparative CT method (45) and compared by the Wilcoxon rank-sum test. Statistical analyses were performed by using Stata software (version 12; StataCorp).
Acknowledgments
We thank Dr. B. Gladstone for technical assistance, Dr. I. Aifantis for antibodies for flow cytometry, D. Kane for expertise in immunohistochemistry, R. Duggan for assistance with FACS, and G. Musa for assistance with illustrations. This work was supported by the Department of Pathology at the University of Chicago (B.W.B.), University of Chicago Cancer Center Support Grant P30 CA014599 (to B.W.B.), and Hematology Research Funds at the University of Chicago donated by S. Samsky and E. Lanzl (to J.M.B.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.Pasqualucci L, et al. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood. 2003;101:2914–2923. doi: 10.1182/blood-2002-11-3387. [DOI] [PubMed] [Google Scholar]
- 2.Baron BW, et al. The human BCL6 transgene promotes the development of lymphomas in the mouse. Proc Natl Acad Sci USA. 2004;101:14198–14203. doi: 10.1073/pnas.0406138101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cattoretti G, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 4.Pasqualucci L, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 6.Wolff L, Koller R, Hu X, Anver MR. A Moloney murine leukemia virus-based retrovirus with 4070A long terminal repeat sequences induces a high incidence of myeloid as well as lymphoid neoplasms. J Virol. 2003;77:4965–4971. doi: 10.1128/JVI.77.8.4965-4971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morse HC, 3rd, et al. Hematopathology subcommittee of the Mouse Models of Human Cancers Consortium Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. 2002;100:246–258. doi: 10.1182/blood.v100.1.246. [DOI] [PubMed] [Google Scholar]
- 8.Selten G, Cuypers HT, Berns A. Proviral activation of the putative oncogene Pim-1 in MuLV induced T-cell lymphomas. EMBO J. 1985;4:1793–1798. doi: 10.1002/j.1460-2075.1985.tb03852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ionov Y, et al. Pim-1 protein kinase is nuclear in Burkitt's lymphoma: Nuclear localization is necessary for its biologic effects. Anticancer Res. 2003;23(1A):167–178. [PubMed] [Google Scholar]
- 10.Saris CJM, Domen J, Berns A. The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG. EMBO J. 1991;10:655–664. doi: 10.1002/j.1460-2075.1991.tb07994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye BH, et al. Chromosomal translocations cause deregulated BCL6 expression by promoter substitution in B cell lymphoma. EMBO J. 1995;14:6209–6217. doi: 10.1002/j.1460-2075.1995.tb00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akasaka T, et al. Nonimmunoglobulin (non-Ig)/BCL6 gene fusion in diffuse large B-cell lymphoma results in worse prognosis than Ig/BCL6. Blood. 2000;96:2907–2909. [PubMed] [Google Scholar]
- 13.Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 14.Pasqualucci L, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 15.Allman D, et al. BCL-6 expression during B-cell activation. Blood. 1996;87:5257–5268. [PubMed] [Google Scholar]
- 16.Mahadevan D, et al. Transcript profiling in peripheral T-cell lymphoma, not otherwise specified, and diffuse large B-cell lymphoma identifies distinct tumor profile signatures. Mol Cancer Ther. 2005;4:1867–1879. doi: 10.1158/1535-7163.MCT-05-0146. [DOI] [PubMed] [Google Scholar]
- 17.Roh M, et al. Overexpression of the oncogenic kinase Pim-1 leads to genomic instability. Cancer Res. 2003;63:8079–8084. [PubMed] [Google Scholar]
- 18.van Lohuizen M, et al. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell. 1989;56:673–682. doi: 10.1016/0092-8674(89)90589-8. [DOI] [PubMed] [Google Scholar]
- 19.Hyjek E, Chadburn A, Liu YF, Cesarman E, Knowles DM. BCL-6 protein is expressed in precursor T-cell lymphoblastic lymphoma and in prenatal and postnatal thymus. Blood. 2001;97:270–276. doi: 10.1182/blood.v97.1.270. [DOI] [PubMed] [Google Scholar]
- 20.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu D, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Chang CC, et al. BCL6 is required for differentiation of Ig-like transcript 3-Fc-induced CD8+ T suppressor cells. J Immunol. 2010;185:5714–5722. doi: 10.4049/jimmunol.1001732. [DOI] [PubMed] [Google Scholar]
- 24.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol. 2010;11:114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mondal A, Sawant D, Dent AL. Transcriptional repressor BCL6 controls Th17 responses by controlling gene expression in both T cells and macrophages. J Immunol. 2010;184:4123–4132. doi: 10.4049/jimmunol.0901242. [DOI] [PubMed] [Google Scholar]
- 26.Baumjohann D, Okada T, Ansel KM. Cutting edge: Distinct waves of BCL6 expression during T follicular helper cell development. J Immunol. 2011;187:2089–2092. doi: 10.4049/jimmunol.1101393. [DOI] [PubMed] [Google Scholar]
- 27.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 28.Ye BH, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, et al. Pim-1 negatively regulates the activity of PTP-U2S phosphatase and influences terminal differentiation and apoptosis of monoblastoid leukemia cells. Arch Biochem Biophys. 2001;390:9–18. doi: 10.1006/abbi.2001.2370. [DOI] [PubMed] [Google Scholar]
- 30.Hu XF, et al. PIM-1-specific mAb suppresses human and mouse tumor growth by decreasing PIM-1 levels, reducing Akt phosphorylation, and activating apoptosis. J Clin Invest. 2009;119:362–375. doi: 10.1172/JCI33216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zippo A, De Robertis A, Bardelli M, Galvagni F, Oliviero S. Identification of Flk-1 target genes in vasculogenesis: Pim-1 is required for endothelial and mural cell differentiation in vitro. Blood. 2004;103:4536–4544. doi: 10.1182/blood-2003-11-3827. [DOI] [PubMed] [Google Scholar]
- 32.Amson R, et al. The human protooncogene product p33pim is expressed during fetal hematopoiesis and in diverse leukemias. Proc Natl Acad Sci USA. 1989;86:8857–8861. doi: 10.1073/pnas.86.22.8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eichmann A, Yuan L, Bréant C, Alitalo K, Koskinen PJ. Developmental expression of pim kinases suggests functions also outside of the hematopoietic system. Oncogene. 2000;19:1215–1224. doi: 10.1038/sj.onc.1203355. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, et al. Pim1 kinase synergizes with c-MYC to induce advanced prostate carcinoma. Oncogene. 2010;29:2477–2487. doi: 10.1038/onc.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen CN, et al. Gene expression profile predicts patient survival of gastric cancer after surgical resection. J Clin Oncol. 2005;23:7286–7295. doi: 10.1200/JCO.2004.00.2253. [DOI] [PubMed] [Google Scholar]
- 36.Reiser-Erkan C, et al. Hypoxia-inducible proto-oncogene Pim-1 is a prognostic marker in pancreatic ductal adenocarcinoma. Cancer Biol Ther. 2008;7:1352–1359. doi: 10.4161/cbt.7.9.6418. [DOI] [PubMed] [Google Scholar]
- 37.Chiang WF, et al. Up-regulation of a serine-threonine kinase proto-oncogene Pim-1 in oral squamous cell carcinoma. Int J Oral Maxillofac Surg. 2006;35:740–745. doi: 10.1016/j.ijom.2006.01.027. [DOI] [PubMed] [Google Scholar]
- 38.Laird PW, et al. In vivo analysis of Pim-1 deficiency. Nucleic Acids Res. 1993;21:4750–4755. doi: 10.1093/nar/21.20.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen LS, Redkar S, Taverna P, Cortes JE, Gandhi V. Mechanisms of cytotoxicity to Pim kinase inhibitor, SGI-1776, in acute myeloid leukemia. Blood. 2011;118:693–702. doi: 10.1182/blood-2010-12-323022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slape C, et al. Retroviral insertional mutagenesis identifies genes that collaborate with NUP98-HOXD13 during leukemic transformation. Cancer Res. 2007;67:5148–5155. doi: 10.1158/0008-5472.CAN-07-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lazo PA, Lee JS, Tsichlis PN. Long-distance activation of the Myc protooncogene by provirus insertion in Mlvi-1 or Mlvi-4 in rat T-cell lymphomas. Proc Natl Acad Sci USA. 1990;87:170–173. doi: 10.1073/pnas.87.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeidler R, et al. Breakpoints of Burkitt's lymphoma t(8;22) translocations map within a distance of 300 kb downstream of MYC. Genes Chromosomes Cancer. 1994;9:282–287. doi: 10.1002/gcc.2870090408. [DOI] [PubMed] [Google Scholar]
- 43.Mikkers H, et al. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet. 2002;32:153–159. doi: 10.1038/ng950. [DOI] [PubMed] [Google Scholar]
- 44.Caudell D, et al. Retroviral insertional mutagenesis identifies Zeb2 activation as a novel leukemogenic collaborating event in CALM-AF10 transgenic mice. Blood. 2010;115:1194–1203. doi: 10.1182/blood-2009-04-216184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]