Abstract
A key next step in synthetic biology is to combine simple circuits into higher-order systems. In this work, we expanded our synthetic riboregulation platform into a genetic switchboard that independently controls the expression of multiple genes in parallel. First, we designed and characterized riboregulator variants to complete the foundation of the genetic switchboard; then we constructed the switchboard sensor, a testing platform that reported on quorum-signaling molecules, DNA damage, iron starvation, and extracellular magnesium concentration in single cells. As a demonstration of the biotechnological potential of our synthetic device, we built a metabolism switchboard that regulated four metabolic genes, pgi, zwf, edd, and gnd, to control carbon flow through three Escherichia coli glucose-utilization pathways: the Embden–Meyerhof, Entner–Doudoroff, and pentose phosphate pathways. We provide direct evidence for switchboard-mediated shunting of metabolic flux by measuring mRNA levels of the riboregulated genes, shifts in the activities of the relevant enzymes and pathways, and targeted changes to the E. coli metabolome. The design, testing, and implementation of the genetic switchboard illustrate the successful construction of a higher-order system that can be used for a broad range of practical applications in synthetic biology and biotechnology.
As synthetic biology matures, the drive for higher-order systems and larger DNA assemblies is intensifying (1, 2). Recent successes include a sensing array for the detection of heavy metals and pathogens and a wide range of logic computations using simple circuits and chemical wires (3, 4). However, this push for complexity underscores the need for interoperable parts and expandable systems (5). Additional components that can be scaled up and operate orthogonally are needed for synthetic biology to continue to produce innovative systems and capitalize on its full potential in biotechnology (6). Previously, we introduced the synthetic riboregulator, an RNA-based gene-expression system, and noted its orthogonal expression capabilities (7, 8). Here, we present a genetic switchboard, a higher-order device that independently and tightly regulates multiple genes in parallel.
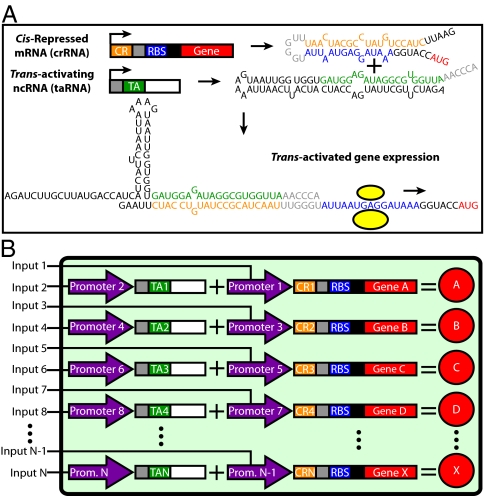
A switchboard is as an assembly of switches that is useful for controlling and linking electrical circuits. Here, we define a genetic switchboard as an assembly of orthogonal, genetic switches that is useful for controlling and linking biological circuits and pathways. The current iteration of our genetic switchboard combines four synthetic riboregulators serving as the orthogonal genetic switches for the platform. An individual riboregulator controls gene expression posttranscriptionally via two RNA species, a cis-repressed mRNA (crRNA) and a trans-activating RNA (taRNA) (Fig. 1A). Once transcribed, target-gene translation is blocked on the crRNA by the cis-repressive sequence forming a stem loop with the ribosome-binding site (RBS). Switching on target-gene expression requires the transcription of the taRNA, a small, noncoding RNA containing the trans-activating sequence, which destabilizes the crRNA stem loop and frees the RBS. Features of synthetic riboregulation that make it an attractive choice for the foundation of a switchboard include physiologically relevant protein production, component modularity, leakage minimization, tunability, fast response times, easy logic programmability, and negligible crosstalk between variants with different cis-repressive and trans-activating sequences (8).
Fig. 1.
Engineered riboregulation and the genetic switchboard. (A) Overview of engineered riboregulation. Mfold-predicted secondary structures of a crRNA and taRNA, along with the proposed structure of the crRNA–taRNA complex that promotes gene expression. Important features are color-coded: the cis-repressive sequence (orange), RBS (blue), target gene (red), trans-activating sequence (green), and crRNA–taRNA recognition bases (gray). The ribosome is presented as yellow circles. (B) Genetic switchboard schematic. Our higher-order synthetic device is composed of orthogonal riboregulators that have unique cis-repressive and trans-activating sequences. All components are modular, including the two promoters (purple) that regulate crRNA and taRNA for a single riboregulator. The ellipses indicate the potential for further expansion.
Originally, only two synthetic riboregulator variants were engineered with acceptable dynamic ranges, the RR10 and RR12 variants (7). In the present work, we first expanded the number of riboregulator variants; this expansion was needed to complete a genetic switchboard. We used two distinct rational design strategies that focused on different components of riboregulation. Using RR12 as the prototype, we replaced its RBS with an engineered RBS of similar strength and mutated the bases involved in the initial crRNA–taRNA recognition complex. After obtaining a set of four orthogonal variants, we assembled the riboregulators onto two plasmids in single cells to create the genetic switchboard and tested the performance of the switchboard in a biosensing setup with easily detectable outputs. The switchboard sensor simultaneously regulates four differentiable reporters, GFP, mCherry, β-galactosidase, and luciferase, with four environmentally sensitive promoters, pLuxI, PLlexO, PLfurO, and pMgrB, respectively. Measured reporter levels showcase the tight and powerful regulation, with minimal crosstalk, provided by the genetic switchboard.
Biological circuitry that regulates many genes in parallel lends itself to a variety of biotechnological applications and particularly to metabolic engineering. Synthetic biology has a history of providing components for metabolic engineering, such as biosynthetic pathways and enzyme scaffolds (9, 10). Adding to this toolbox, the genetic switchboard is a well-defined, biological module that possesses the flexibility to aid different metabolic engineering strategies. As proof of concept, we constructed a metabolism switchboard that controls carbon flux through three Escherichia coli glucose-utilization pathways, the Embden–Meyerhof (EMP), Entner–Doudoroff (EDP), and pentose phosphate (PPP) pathways, via the regulation of four different genes, pgi, zwf, edd, and gnd. The performance of the metabolism switchboard over multiple biological scales, namely, at the RNA, protein, and metabolome levels, showcases the real-world potential of our higher-order control system.
Results and Discussion
Rational Design of Riboregulator Variants.
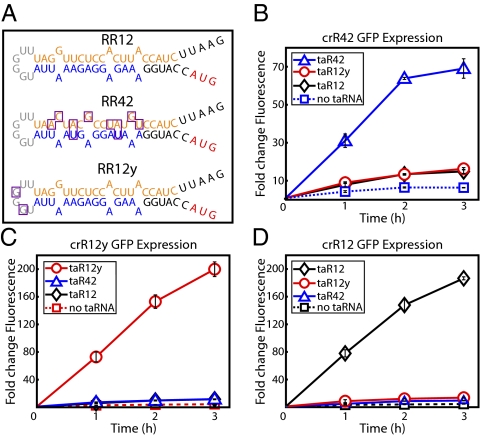
In the initial synthetic riboregulator study, the RR10 variant was constructed successfully, and, in an attempt to improve the dynamic range of the system, the RR12 variant was built as a rationally designed refinement (7). Here, RR42 and RR12y, the two riboregulator variants required to create the genetic switchboard, also were rationally designed, but via two unique strategies. RR42 was the result of RBS manipulation, and RR12y was the result of mutating the crRNA–taRNA recognition sequence. In both cases, RR12 was the parent riboregulator variant, and we attempted to minimize the changes to the successful RR12 blueprint while introducing enough mutations to generate orthogonal activity. Therefore, critical specifications of RR12, such as the Mfold-predicted secondary structures and thermodynamic values, were preserved in the designs of RR42 and RR12y (11).
When constructing the RR42 riboregulator, we targeted the RBS of the RR12 variant. Using the RBS Calculator (12), we identified a synthetic RBS candidate that had the same length and similar predicted strength as the RBS in the RR12 riboregulator. We subcloned this synthetic RBS into the RR12 variant to generate the riboregulator RR42. The cis-repressive sequence is highly complementary to the RBS, and replacing the RBS in the RR12 variant mandated that mutations be made in the cis-repressive sequence to maintain this strong complementation. The RR42 crRNA secondary structure is presented in Fig. 2A; the mutations in the RBS and corresponding mutations in the cis-repressive sequence are outlined in purple.
Fig. 2.
Characterization of riboregulators RR42 and RR12y. (A) Mfold-predicted secondary structures of crR12, crR42, and crR12y. Mutations in variants crR42 and crR12y, relative to the parent crR12 variant, are outlined in purple. Important features are color-coded: cis-repressive sequence (orange), RBS (blue), target gene start codon (red), and taRNA recognition bases (gray). (B–D) Fold changes in GFP expression from fully induced crRNA cotranscribed with cognate and noncognate taRNA. (B) crR42. (C) crR12y. (D) crR12. All values were normalized by OFF state (no crRNA or taRNA induction). Graphs depict the triplicate mean ± SEM.
When constructing the RR12y riboregulator, we targeted the recognition sequence of the RR12 variant. The first step in trans-activation of cis-repressed gene expression is the interaction between the linear region of the taRNA and the loop region of the crRNA (13). Without efficient recognition, we speculate that the rate of crRNA–taRNA duplex formation will drop dramatically, regardless of complementation between the cis-repressive and trans-activating sequences. Keeping the RBS and cis-repressive sequence of the RR12 variant intact, we mutated only two bases in the recognition loop region of the crRNA, along with the corresponding taRNA bases, to generate the riboregulator RR12y. The RR12y crRNA secondary structure is presented in Fig. 2A; the two mutated bases are outlined in purple.
We characterized the RR42 and RR12y variants by riboregulating GFP (Fig. 2 B and C). In addition, we investigated each combination of crRNA–taRNA to assess orthogonality (Fig. 2 B–D). For example, we measured GFP expression from three plasmids, each containing crR42 but with three different taRNA variants, taR42, taR12y, and taR12 (Fig. 2B). The RR42 and RR12y riboregulators showed activation of GFP expression only when the cognate taRNA was present and induced (Fig. 2 B and C). At 3 h after induction, the RR42 variant (crR42-taR42) displayed an ∼70-fold change in fluorescence, significantly higher than fold changes from the crR42–taR12 and crR42–taR12y control plasmids (Fig. 2B). More impressively, the RR12y variant (crR12y–taR12y) exhibited a 200-fold increase in GFP expression at 3 h postinduction, outperforming the parent variant RR12 (Fig. 2C). Again, noncognate GFP expression was reduced substantially in the crR12y–taR42 and crR12y–taR12 control plasmids. We address the two- to three-times lower GFP expression observed for the RR42 riboregulator in detail in SI Results and Discussion. Importantly, the RR42 and RR12y variants will not disrupt the performance of the RR12 riboregulator, because the parent variant also showed minimal amounts of GFP induction from the control plasmids crR12–taR42 and crR12–taR12y (Fig. 2D).
In the genetic switchboard setting, the amount of crosstalk undoubtedly would be lower than the GFP expression levels measured from noncognate crRNA–taRNA binding in the control plasmids (Fig. 2 B–D). If multiple riboregulators are present in the same cell, there should be a degree of preferential binding between cognate crRNA and taRNA molecules. With these control plasmids, the preferred cognate crRNA–taRNA interaction was not available; the noncognate duplex was the only possible interaction. Furthermore, part of the measured crosstalk was caused solely by transcription of more crRNA and, thus, by more opportunities for leakage. The contribution of this leakage factor was highlighted by GFP expression when the taRNA remained uninduced (Fig. 2 B–D; dotted line with squares). When integrating simple modules into a higher-order system, it is imperative that the building blocks perform well individually. These characterization assays demonstrated the generous range of expression, low leakage, and orthogonality of the individual riboregulator variants, thus highlighting their suitability for use in the genetic switchboard.
Switchboard Sensor.
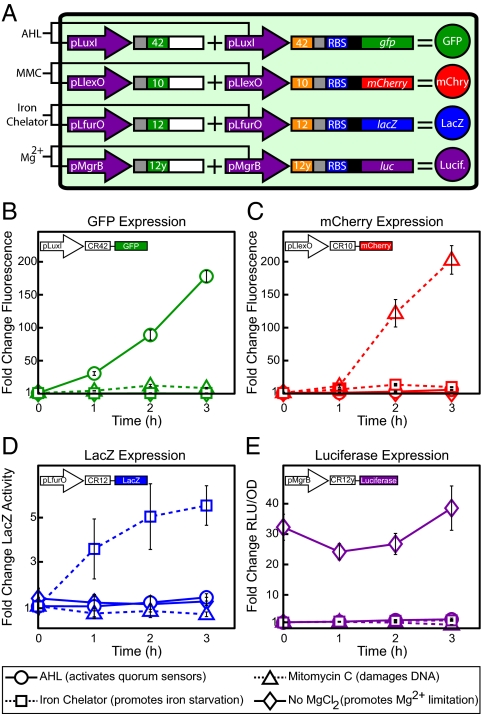
We introduced the four riboregulator variants into single cells and used these variants to regulate four reporters—GFP, mCherry, β-galactosidase (LacZ), and firefly luciferase—in parallel (Fig. 3A) to test the effectiveness of the genetic switchboard directly. GFP and mCherry are fluorescent proteins with independent excitation and emission spectra, and LacZ and luciferase are identified in colorimetric and luminescent assays, respectively. This combination of reporters allowed straightforward, fast, and differentiable detection of the four switchboard-regulated genes.
Fig. 3.
Switchboard sensor. (A) Switchboard sensor schematic. (B) Fold changes in levels of the four reporters for each induction condition. All values were normalized by OFF state (expression of all reporters repressed). Graphs depict the triplicate mean ± SEM.
We chose four promoters that were sensitive to environmental conditions, thereby creating a switchboard sensor. The promoters pLuxI, PLlexO, PLfurO, and pMgrB regulated GFP, mCherry, LacZ, and luciferase, respectively (Fig. 3A). Derived from a quorum-sensing pathway in Vibrio fischeri, pLuxI responds to the addition of the quorum-signaling molecule acyl-homoserine lactone (AHL) (14). PLlexO and PLfurO were based on the design of the synthetic PLlacO-1 promoter, with LexA and Fur binding sites replacing the LacR operators (15, 16). The LexA repressor undergoes RecA-mediated autocleavage upon recognition of DNA damage, and the transcription factor Fur de-represses iron uptake and utilization genes when the concentration of extracellular iron drops (17, 18). Thus, PLlexO functioned as a DNA-damage sensor, and PLfurO functioned as an iron sensor. We used the DNA-damaging drug mitomycin C (MMC) and the iron chelator 1,10-phenanthroline to drive expression from PLlexO and PLfurO, respectively. Finally, pMgrB, a natural E. coli promoter modulated by the magnesium-sensitive regulator PhoQ, functioned as a magnesium sensor (19). Mg2+ concentration is inversely related to pMgrB activity, and we added MgCl2 to repress pMgrB expression. All switchboard sensor promoters regulated both the crRNA and taRNA for a given riboregulator (Fig. 3A).
We induced the reporters individually and normalized all values by the switchboard sensor OFF state, in which the expression of each reporter was repressed. Switchboard sensor induction data are presented in Fig. 3 B–E. Upon addition of the quorum signal AHL, expression of GFP increased dramatically (Fig. 3B). At 2 h after induction, GFP levels were nearly 100 times higher than in the OFF state, and by 3 h GFP levels were well over 150 times greater. MMC powerfully increased mCherry expression, with a rise of more than 200-fold at 3 h posttreatment (Fig. 3C). Moreover, MMC-treated cells showed decreased activity relative to the OFF state in the LacZ and luciferase assays (Fig. 3 D and E), most likely because of cells shifting resources from reporter production to surviving the DNA stress.
Iron chelator boosted β-galactosidase activity by more than five times relative to the OFF state, a significant jump as compared with the other treatments (Fig. 3D). Notably, previous studies have shown DNA damage can affect Fur-mediated iron regulation (16). In this case, MMC-treated cells did not show a bump in PLfurO de-repression, thus providing further evidence of riboregulator orthogonality. In the luminescence dataset, growing switchboard sensor cells without exogenous MgCl2, and thus in a state of Mg2+ limitation, resulted in approximately constant luciferase expression that was 25- to 40-fold higher than in the OFF state (Fig. 3E). Because we established Mg2+ limitation at inoculation, we detected luciferase induction at time 0 (OD600 ∼0.3). We added the inducers for GFP, mCherry, and LacZ at time 0; hence, we did not detect expression of the fluorescent or colorimetric reporters at that time point. Also, the low luciferase levels from the iron chelator treatment demonstrated the specificity of 1,10-phenanthroline chelation.
The switchboard sensor exhibited tight and powerful regulation of four reporters in parallel. We measured very large expression ranges, between 100- and 200-fold inductions for the fluorescent reporters. In addition, leakage and crosstalk between riboregulators were minimal in each treatment; no significant unintended gene expression was captured. These results confirmed the orthogonality of the RR42 and RR12y variants first observed in the characterization experiments. Together, the biosensing tests established the high functionality of our genetic switchboard.
Metabolism Switchboard.
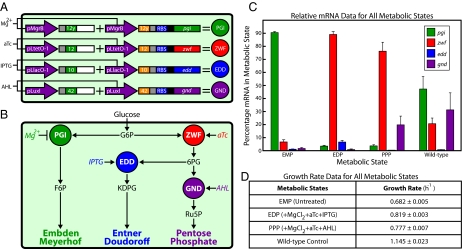
We chose to showcase the biotechnological potential of the genetic switchboard in a metabolic engineering proof-of-concept application. Our metabolism switchboard riboregulated four E. coli genes, pgi, zwf, edd, and gnd, which control carbon flux through three glucose-utilization pathways: EMP (familiarly, glycolysis), EDP, and PPP (Fig. 4 A and B). Manipulating these metabolic enzymes and pathways provided an ideal situation to determine the effectiveness of the metabolism switchboard. Pgi, Zwf, Edd, and Gnd comprise major branch points at the beginning of their respective metabolic pathways and have no redundant counterparts (Fig. 4B) (20). Therefore, the riboregulated enzymes are responsible for the catabolism of all glucose imported into the cell, and shifts among the pathways are clearly defined and measurable. Furthermore, these pathways have been identified as potential metabolic engineering targets for increased production of industrial chemicals (21).
Fig. 4.
Metabolism switchboard. (A) Metabolism switchboard schematic. (B) Overview of glucose-utilization pathways controlled by our metabolism switchboard (27). Glucose imported into the cell is converted quickly to glucose-6-phosphate (G6P). Phosphoglucose isomerase (Pgi; green), regulated here by Mg2+-sensitive pMgrB, converts G6P to fructose-6-phosphate (F6P), which is the first step in the EMP. Glucose-6-phosphate dehydrogenase (Zwf; red), regulated here by aTc-sensitive PLtetO-1, converts G6P to 6-phosphogluconolactone, which then is converted to 6-phosphogluconate (6PG) by 6-phosphogluconolactonase (Pgl; not shown). Phosphogluconate dehydratase (Edd; blue), regulated here by IPTG-sensitive PLlacO-1, converts 6PG to 2-keto-3-deoxy-6-phosphogluconate (KDPG), which is the first of only two steps in the EDP. 6-Phosphogluconate dehydrogenase (Gnd; purple), regulated here by AHL-sensitive pLuxI, converts 6PG to ribulose-5-phosphate (Ru5P), which is the distinguishing step in the PPP. (C) Metabolism switchboard performance at the RNA scale. Relative mRNA concentrations for the target metabolic genes, presented as the percentage of the total of all switchboard-regulated mRNA for each metabolic state. (D) Exponential growth rates per hour. The graph in C and table in D depict the triplicate mean ± SEM.
To transfer control of glucose metabolism to the genetic switchboard, pgi, zwf, edd, and gnd were removed from the MG1655Pro (F-, λ-, SpR, lacR, tetR) genome, and cells were grown in modified M9 minimal medium with 0.5% glucose as the sole carbon source (15). Notably, the experimental design included an overnight incubation in which switchboard cells were grown in the EMP, the default metabolic state of our system. The EMP was chosen as the default pathway because it is a major component of E. coli central carbon metabolism. Nearly 80% of glucose is metabolized via the EMP in wild-type cells, and shifting flux to less-used pathways is a significant departure from the typical metabolic state of the cell (20, 21). In each experiment, the EMP remained the target pathway after the overnight incubation, or inducers were added to shift carbon flux to the EDP or PPP. These three states of the metabolism switchboard were compared with untreated MG1655Pro cells without plasmids (wild-type control), representing normal glucose metabolism.
As with the switchboard sensor, one promoter regulated both the crRNA and taRNA for each riboregulator variant in our metabolism switchboard. Specifically, pMgrB, anhydrotetracycline (aTc)-sensitive PLtetO-1, isopropyl-β-d-thio-galactoside (IPTG)-sensitive PLlacO-1, and pLuxI regulated pgi, zwf, edd, and gnd, respectively (Fig. 4A). The combination of inducers that determined the target pathway of the metabolism switchboard is illustrated in Fig. 4B. Briefly, without any inducers, the EMP (Pgi ON) was the target metabolic state. When we boosted the Mg2+ concentration with exogenous MgCl2, the EMP was shut down via pMgrB repression. We then shifted carbon flux to the EDP (Zwf and Edd ON) by inducing with aTc and IPTG, or we shunted flux to the PPP (Zwf and Gnd ON) by inducing with aTc and AHL.
Cells in each experiment were harvested for analysis at late exponential growth phase (OD600 ∼0.8). At this OD600, the growth rate was approximately constant, and cells were expected to reach a metabolic quasi-steady state in which carbon fluxes and metabolite concentrations were approximately constant (22). Hence, we avoided measuring metabolic transition states when switching metabolic pathways. Importantly, there was not a significant difference in the growth rates of switchboard cells when targeting the EMP or shifting to the EDP or PPP (Fig. 3D). Similar growth rates enabled a clear comparison of metabolic states.
We measured the performance of the metabolism switchboard across the RNA, protein, and metabolome biological scales. Because the switchboard relied on the production of multiple RNA species, we used quantitative PCR (qPCR) to measure relative mRNA concentrations; qPCR reported on the performance of the metabolism switchboard at its most basic level. Relative mRNA concentrations, the qPCR output, described the abundance of crRNA molecules for each target gene in each metabolic state. This measurement provided a check that the switchboard was built correctly and that the promoters used were performing as expected and were independently activated.
qPCR data are presented in Fig. 4C. For each metabolic state, the relative mRNA concentrations for pgi, zwf, edd, and gnd were totaled, and the percentage of each gene in this total was calculated. For example, in the wild-type control, pgi mRNA accounted for nearly 50% of the total mRNA measured between all four metabolic genes. The distribution of relative mRNA concentrations in the wild-type sample agreed with carbon flux data in the literature. In normal glucose metabolism, the distribution of flux through the EMP, PPP, and EDP is ∼75%, 25%, and <1%, respectively (20, 21). Here, pgi mRNA was present in the highest percentage in the wild-type sample, with decreasing percentages of gnd > zwf > edd mRNA.
In the default EMP state of the metabolism switchboard, pgi mRNA represented 90% of the switchboard-regulated mRNA, a positive result because pgi was the only induction target in this state. When the target pathway was shifted to the EDP and PPP, zwf mRNA dominated, with percentages >75% in each case. In addition, both edd and gnd mRNA were present at significantly higher percentages in the metabolic states in which they were induced. edd mRNA constituted almost 10% of the total in the EDP, compared with <1.5% in the EMP or PPP, and gnd mRNA constituted 20% of the total in the PPP, compared with <2% in the EMP or EDP.
The qPCR data illustrated that the metabolism switchboard was functional at the RNA level and that the desired amount of crRNA was present for each gene when targeting the EMP and shifting to the EDP and PPP. Beyond the RNA level, we used enzyme activity assays to determine whether translation was occurring as desired and whether the target enzymes were active. Shunting of carbon through a target pathway requires shifting the activities of the relevant enzymes (23). Although activity also can depend on covalent modifications and effector interactions, the metabolism switchboard was designed to change activity through alterations of enzyme concentrations (23).
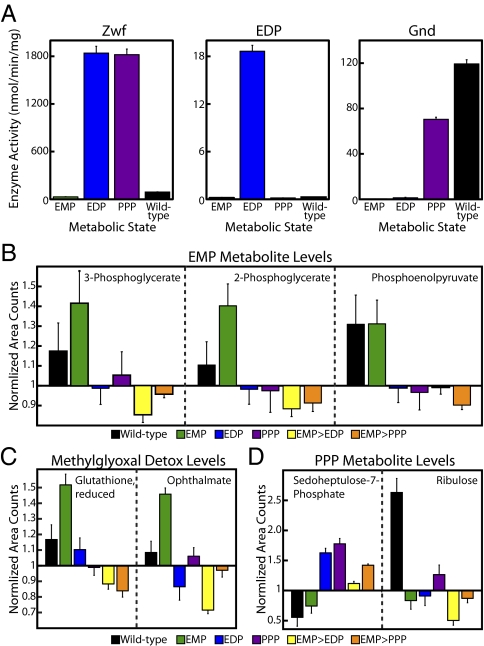
Enzyme activity data, expressed in nanomolars per minute per milligram, for Zwf, the EDP, and Gnd are presented in Fig. 5A. Zwf is the gateway enzyme to the EDP and PPP (Fig. 4B). When we shifted carbon flux to the EDP or PPP, Zwf activity increased by a factor of more than 60. Shifting metabolic flux away from Pgi and the EMP is a severe change for the cell, and increasing Zwf activity to high levels is the key step in this shift. Because Pgi and Zwf form a branch point for glucose, this impressive boost in Zwf activity ensured that a significant amount of carbon would be shunted to the EDP and PPP, even if there were no change in Pgi activity. Notably, the induced Zwf activity was 20-fold higher than the wild-type activity, and the uninduced Zwf activity was threefold lower than wild-type activity; these results underscored the tightness and range of switchboard protein production.
Fig. 5.
Metabolism switchboard performance. (A) Protein scale. Activities of Zwf, the EDP, and Gnd in nanomolars per minute per milligram. Graphs depict the triplicate mean ± SEM. (B–D) Metabolome scale. Using mass spectrometry, raw area counts for identified biochemicals were normalized by the total protein concentration and then rescaled by setting the median value equal to 1.0. Missing values were imputed with the minimum value. Graphs depict the sextuplicate mean ± SEM. (B) Levels of EMP intermediates. (C) Levels of methylglyoxal detoxification intermediates. (D) Levels of PPP intermediates.
We measured the activity of the entire EDP, instead of just determining the Edd enzyme activity. The EDP consists of only two enzymes, Edd and Eda, both of which are required to produce the output molecule of the activity assay, pyruvate (24). When we shunted carbon from the EMP to the EDP, pathway activity increased by two orders of magnitude (Fig. 5A). Furthermore, the very low wild-type EDP activity highlighted how minor the pathway is in normal glucose metabolism and thus how dramatically the switchboard altered wild-type carbon flow.
When we shifted carbon flow to the PPP, Gnd activity rose by more than 50 times (Fig. 5A). Again, the metabolism switchboard significantly up-regulated the target enzyme activity while tightly repressing unintended activity in the EMP and EDP states. Interestingly, Gnd activity was slightly higher in the wild-type control than in the PPP switchboard state. This result underlined the link between the RNA and protein scales, because wild-type Gnd mRNA was more abundant than switchboard-regulated Gnd mRNA (Fig. 4C).
Unfortunately, Pgi activity could not be measured reliably. The Pgi assay required the presence of Zwf to produce the assay output NADPH (23). Because Zwf activity increased nearly 100 times when carbon flux was shifted from the EMP to the EDP or PPP, consistent Pgi activity values across metabolic states were not obtained, even when exogenous Zwf was added to all samples. However, the measured enzyme activities showed a clear shift away from the EMP via the activation of the EDP and PPP. These two pathways remained tightly off, with activity below that of wild-type levels, until the powerful, switchboard-mediated induction of the relevant enzymes.
Finally, we examined the performance of the metabolism switchboard on the metabolome scale. Metabolic-profiling was performed to identify changes in the levels of a large number of metabolites, intermediates, and biochemicals. A major goal of the metabolomics study was to provide the direct evidence of switchboard-mediated EMP activation and deactivation that was not found in the RNA- and protein-scale data.
The metabolic-profiling output consisted of normalized area values for each metabolite and biochemical detected via GC/MS, liquid chromatography (LC)/MS, and LC/MS/MS. In addition to the usual EMP, EDP, PPP, and wild-type samples, we investigated two additional metabolism switchboard samples, EMP > EDP and EMP > PPP. These samples were switched from the EMP state to the EDP or PPP state during exponential growth and metabolic steady state, as opposed to shifting from the EMP state after the overnight incubation. Thus, the EMP > EDP and EMP > PPP samples provided different and more rigorous pathway-switching conditions.
EMP activation and deactivation were demonstrated by both direct and indirect means in the metabolic-profiling data. First, when the EMP was the target metabolic pathway, the metabolism switchboard enriched three downstream intermediates in the EMP: 3-phosphoglycerate, 2-phosphoglycerate, and phosphoenolpyruvate (Fig. 5B). The magnitude of enrichment was above wild-type levels, a sign of an overactive EMP, and resembled previous studies measuring carbon flux in PPP and EDP knockouts (20, 21). Moreover, EMP intermediates were less abundant in the EDP, PPP, EMP > EDP, and EMP > PPP metabolic states than in the wild-type control. This decrease in metabolite levels was particularly striking for the EMP > EDP and EMP > PPP samples, because here the costly shift from the EMP was instigated during the metabolically active exponential growth phase. Similarly, the major detoxification pathway for methylglyoxal, a toxic byproduct formed primarily by the EMP, was enriched only in the EMP samples (Fig. 5C) (25). The switchboard not only altered the direct metabolic intermediates of a target pathway; it also maintained the secondary changes associated with using a specific pathway.
Notably, the metabolic-profiling data also supported PPP activation. In both samples in which carbon flow was shunted to the PPP state, our metabolism switchboard enriched the level of sedoheptulose-7-phosphate, a downstream PPP intermediate and one of the very few metabolites that was detected in the PPP or EDP (Fig. 5D). Also, ribulose, a sugar that can be formed from the important PPP intermediate ribulose-5-phosphate, was enriched in the relevant metabolic states (Fig. 5D). Thus, metabolic-profiling provided direct evidence of switchboard-mediated EMP activation and also offered insights on the shift of carbon flux to the PPP.
Conclusions
Here we describe the process of constructing and testing a genetic switchboard, culminating in the demonstration of controlling carbon flux through E. coli glucose-utilization pathways. Our metabolism switchboard riboregulated the enzymes Pgi, Zwf, Edd, and Gnd and determined the flow of glucose-derived carbon through the EMP, EDP, and PPP. Despite the strong preference of the cell for using the EMP, we shunted carbon to much less commonly used pathways, as evidenced by data over three biological scales. On the RNA scale, the metabolism switchboard successfully produced the desired amounts of mRNA in each metabolic state, highlighting tight and independent regulation at the simplest level. On the protein scale, our system dramatically shifted metabolic enzyme and pathway activities, showcasing a strikingly large range of protein production. On the metabolome scale, the switchboard altered, in a targeted manner, the levels of relevant metabolites and biomolecules, displaying control of even small subpathways.
There are a number of practical future uses for the genetic switchboard in biotechnology and metabolic engineering. For example, when shunting carbon through an industrially important pathway leads to the accumulation of toxic byproducts, the switchboard could switch pathways temporarily to alleviate stress on the cell. Also, our device could sample a variety of metabolic states at once in an effort to maximize the efficiency of an industrially attractive pathway. In almost every potential application, linking gene expression to bioreactor conditions would be a valuable innovation. Bioreactor feedback would allow the switchboard to sense the environment and respond appropriately to optimize carbon flux.
Materials and Methods
Plasmids.
Basic molecular biology techniques were implemented as previously described (26). All riboregulator systems were based on the published design (7). Genetic switchboards consist of two plasmids containing two riboregulators each, two different origins of replication, and two different resistance markers. See SI Materials and Methods for details on plasmid construction.
Flow Cytometry.
GFP and mCherry fluorescence were measured using a Becton Dickinson FACSAria II flow cytometer. For each sample we recorded 100,000 events gated by a forward scatter and side scatter threshold. See SI Materials and Methods for more details.
Colorimetric and Luminescent Reporter Assays.
β-Galactosidase activity was measured using the Yeast β-Galactosidase Assay Kit (Thermo Scientific), and luciferase activity was measured using the Luciferase Assay System (Promega). See SI Materials and Methods for more details.
qPCR.
qPCR was performed using the Roche LightCycler 480. The mRNA concentrations for pgi, zwf, edd, and gnd were estimated using relative quantification, with lplT, a lysophospholipid transporter, and rrsH, 16S ribosomal RNA, as reference genes. A no-cDNA template control was analyzed also. See SI Materials and Methods for more details.
Activity Assays.
Cells were lysed with a microtip on the Branson Digital Sonifier, and the Coomassie Plus Assay Kit (Pierce) was used to measure total protein concentration. Protocols for measuring activity from cell lysate were based on Peng and Shimizu (23). The output of the Zwf and Gnd assays was NADPH, and the output of the EDP assay was pyruvate. NAPDH and pyruvate, corresponding to absorbance at 340 nm and 450 nm, respectively, were measured using a SpectraMax M5. See SI Materials and Methods for more details.
Growth Analysis.
To calculate exponential growth rate for the metabolism switchboard and wild-type control samples, OD600 was measured every 30 min, from 0–7 h, using the SPECTRAFluor Plus (Tecan).
Metabolic-Profiling.
To prepare cells for metabolic profiling, 30-mL cell cultures were washed and resuspended in 3 mL 1× PBS, pelleted by centrifugation at 3,000 × g and 4 °C for 3 min, and flash frozen in an ethanol-dry ice bath. Samples were shipped overnight to Metabolon on dry ice. See SI Materials and Methods for details on the metabolomics analysis.
Supplementary Material
Acknowledgments
We thank Dan Dwyer, Ahmad Khalil, Raffi Afeyan, Kevin Litcofsky, Henry Lee, and Jonathan Winkler for helpful discussions and Metabolon for performing the metabolic-profiling analysis. This work was supported by National Institutes of Health Director's Pioneer Award Program Grant DP1 OD003644 and by the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203808109/-/DCSupplemental.
References
- 1.Lu TK, Khalil AS, Collins JJ. Next-generation synthetic gene networks. Nat Biotechnol. 2009;27:1139–1150. doi: 10.1038/nbt.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellis T, Adie T, Baldwin GS. DNA assembly for synthetic biology: From parts to pathways and beyond. Integr Biol (Camb) 2011;3:109–118. doi: 10.1039/c0ib00070a. [DOI] [PubMed] [Google Scholar]
- 3.Prindle A, et al. A sensing array of radically coupled genetic 'biopixels'. Nature. 2012;481:39–44. doi: 10.1038/nature10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamsir A, Tabor JJ, Voigt CA. Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’. Nature. 2011;469:212–215. doi: 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao CV. Expanding the synthetic biology toolbox: Engineering orthogonal regulators of gene expression. Curr Opin Biotechnol. 2012 doi: 10.1016/j.copbio.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 6.Khalil AS, Collins JJ. Synthetic biology: Applications come of age. Nat Rev Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isaacs FJ, et al. Engineered riboregulators enable post-transcriptional control of gene expression. Nat Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- 8.Callura JM, Dwyer DJ, Isaacs FJ, Cantor CR, Collins JJ. Tracking, tuning, and terminating microbial physiology using synthetic riboregulators. Proc Natl Acad Sci USA. 2010;107:15898–15903. doi: 10.1073/pnas.1009747107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyle PM, Silver PA. Parts plus pipes: Synthetic biology approaches to metabolic engineering. Metab Eng. 2011 doi: 10.1016/j.ymben.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynch SA, Gill RT. Synthetic biology: New strategies for directing design. Metab Eng. 2011 doi: 10.1016/j.ymben.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franch T, Petersen M, Wagner EG, Jacobsen JP, Gerdes K. Antisense RNA regulation in prokaryotes: Rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J Mol Biol. 1999;294:1115–1125. doi: 10.1006/jmbi.1999.3306. [DOI] [PubMed] [Google Scholar]
- 14.Sitnikov DM, Schineller JB, Baldwin TO. Transcriptional regulation of bioluminescence genes from Vibrio fischeri. Mol Microbiol. 1995;17:801–812. doi: 10.1111/j.1365-2958.1995.mmi_17050801.x. [DOI] [PubMed] [Google Scholar]
- 15.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Little JW. Mechanism of specific LexA cleavage: Autodigestion and the role of RecA coprotease. Biochimie. 1991;73:411–421. doi: 10.1016/0300-9084(91)90108-d. [DOI] [PubMed] [Google Scholar]
- 18.Escolar L, Pérez-Martín J, de Lorenzo V. Opening the iron box: Transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minagawa S, et al. Identification and molecular characterization of the Mg2+ stimulon of Escherichia coli. J Bacteriol. 2003;185:3696–3702. doi: 10.1128/JB.185.13.3696-3702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hua Q, Yang C, Baba T, Mori H, Shimizu K. Responses of the central metabolism in Escherichia coli to phosphoglucose isomerase and glucose-6-phosphate dehydrogenase knockouts. J Bacteriol. 2003;185:7053–7067. doi: 10.1128/JB.185.24.7053-7067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J, Baba T, Mori H, Shimizu K. Global metabolic response of Escherichia coli to gnd or zwf gene-knockout, based on 13C-labeling experiments and the measurement of enzyme activities. Appl Microbiol Biotechnol. 2004;64:91–98. doi: 10.1007/s00253-003-1458-5. [DOI] [PubMed] [Google Scholar]
- 22.Zamboni N, Fendt SM, Rühl M, Sauer U. (13)C-based metabolic flux analysis. Nat Protoc. 2009;4:878–892. doi: 10.1038/nprot.2009.58. [DOI] [PubMed] [Google Scholar]
- 23.Peng L, Shimizu K. Global metabolic regulation analysis for Escherichia coli K12 based on protein expression by 2-dimensional electrophoresis and enzyme activity measurement. Appl Microbiol Biotechnol. 2003;61:163–178. doi: 10.1007/s00253-002-1202-6. [DOI] [PubMed] [Google Scholar]
- 24.Canonaco F, et al. Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble transhydrogenase UdhA. FEMS Microbiol Lett. 2001;204:247–252. doi: 10.1111/j.1574-6968.2001.tb10892.x. [DOI] [PubMed] [Google Scholar]
- 25.Ferguson GP, Tötemeyer S, MacLean MJ, Booth IR. Methylglyoxal production in bacteria: Suicide or survival? Arch Microbiol. 1998;170:209–218. doi: 10.1007/s002030050635. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Fraenkel D, Vinopal RT. Carbohydrate metabolism in bacteria. Annu Rev Microbiol. 1973;27:69–100. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.