Abstract
Genome-wide studies have identified associations between polymorphisms in the IFN regulatory factor-5 (Irf5) gene and a variety of human autoimmune diseases. Its functional role in disease pathogenesis, however, remains unclear, as studies in Irf5−/− mice have reached disparate conclusions regarding the importance of this transcription factor in type I IFN production and antibody responses. We identified a spontaneous genomic duplication and frameshift mutation in the guanine exchange factor dedicator of cytokinesis 2 (Dock2) that has arisen in at least a subset of circulating Irf5−/− mice and inadvertently been bred to homozygosity. Retroviral expression of DOCK2, but not IRF-5, rescued defects in plasmacytoid dendritic cell and B-cell development, and Irf5−/− mice lacking the mutation in Dock2 exhibited normal plasmacytoid dendritic cell and B-cell development, largely intact type I IFN responses, and relatively normal antibody responses to viral infection. Thus, confirmation of the normal Dock2 genotype in circulating Irf5−/− mice is warranted, and our data may partly explain conflicting results in this field.
Keywords: immune cell development, autoimmunity, viruses
IFN regulatory factor-5 (IRF-5) is a transcription factor that has been reported to contribute to the induction of MyD88-dependent IFN responses after viral infection (1–3). The importance of IRF-5 in human disease is highlighted by numerous genome-wide association studies that implicate Irf5 alleles in a variety of autoimmune disorders including systemic lupus erythematosus, Sjögren syndrome, rheumatoid arthritis, inflammatory bowel diseases, and multiple sclerosis (4–8). Although no causal relationship between these Irf5 polymorphisms and autoimmune disease has been established definitively, earlier studies clearly showed that IFN-α therapy in cancer can trigger autoimmunity (9). Thus, it has been proposed that Irf5 variants may be linked to autoimmunity through their ability to induce elevated levels of type I IFNs.
Surprisingly, initial studies of Irf5−/− mice found no defects in type I IFN production by plasmacytoid dendritic cells (pDCs) (2), which are thought to be a major systemic source of IFN-α (10). Consistent with these results, virally infected Irf5−/− mice showed only modest reductions in serum type I IFN levels (11). Instead, Irf5−/− mice specifically failed to produce inflammatory cytokines such as TNF-α, IL-6, and IL-12 (2). However, subsequent studies reported a more dramatic reduction or complete abolishment of type I IFN production by Irf5−/− cells (12–14). Similarly, there are conflicting data regarding the requirement of IRF-5 in antibody production, as specific reductions in IgG2a or IgG1 and IgM levels after immunization with a T-cell–dependent antigen have been reported independently (15, 16). Thus, the importance of IRF-5 in type I IFN induction downstream of Toll-like receptor (TLR) engagement and antibody production remains unclear. As a consequence, the functional role of IRF-5 in modulating autoimmune diseases has been difficult to discern from the existing literature.
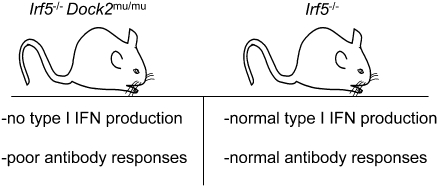
In studies aimed at identifying novel transcription factors important for DC development, we observed a number of unexpected phenotypes in Irf5−/− mice, including a reduction in peripheral pDC numbers, a complete inability of pDCs to produce type I IFN, a complete loss of marginal-zone B cells, and a reduction in antigen-specific IgG and IgM levels following West Nile virus (WNV) infection. However, further analysis demonstrated that these phenotypes were not dependent on the deficiency of IRF-5, but rather segregated with a spontaneous genomic duplication and frameshift mutation in dedicator of cytokinesis 2 (Dock2), a guanine nucleotide exchange factor that regulates Rac signaling, marginal-zone B-cell development, and pDC homing and function (17–19). Analysis of Irf5−/− mice in the absence of this secondary mutation in Dock2 revealed normal pDC and B-cell development, largely intact IFN-α production by pDCs after TLR stimulation, and relatively intact antibody responses to WNV infection. Our data suggest that at least some of the conflicts among published data regarding the functional properties of IRF-5 likely are the result of a mutation in Dock2 that arose spontaneously and is now widely distributed in Irf5−/− mouse strains.
Results
Irf5−/− Mice Have Impaired pDC and B-Cell Development.
As Flk2+ common myeloid progenitors (CMPs) precede common dendritic cell progenitors (CDPs) in hematopoietic cell development (20, 21), we sought to identify transcription factors that regulate dendritic cell (DC) lineage commitment and differentiation through comparisons of these two progenitor types. We focused our initial analysis on IRF family members, as several of these transcription factors have regulatory roles in DC development (22, 23). Among the differentially expressed genes between Flk2+ CMPs and CDPs was Irf5, which displayed a sevenfold (P < 0.05) increase in expression in CDPs relative to Flk2+ CMPs (Fig. 1A).
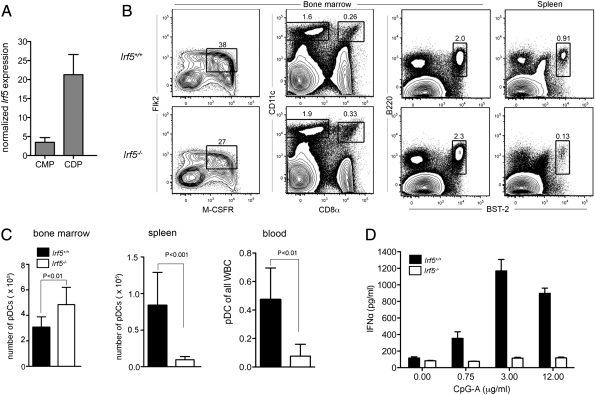
Fig. 1.
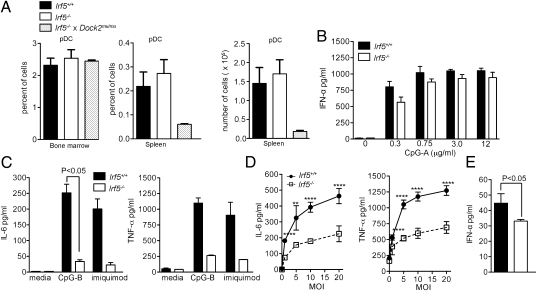
Irf5−/− mice have reduced peripheral pDCs and IFN-α production. (A) Irf5 expression in sorted Flk2+ CMPs and CDPs was determined by quantitative RT-PCR and normalized to β-actin expression. (B) Analysis of CDP (Left), CD11chighCD8α+ (second from left), CD8α− cDCs (second from left), and B220+ BST2+ pDC from the bone marrow (third from left) and spleen (Right) in Irf5−/− mice. (C) Numbers of pDC from bone marrow, and spleen, and frequency of pDC from blood in Irf5−/− mice. (D) IFN-α secretion by sorted pDCs stimulated with varying doses of CpG-A as measured by ELISA. Data shown are the mean and SD of one experiment performed in triplicate (A), three experiments with a total of five to eight mice (B and C), or representative of two experiments (D). P values were determined by using an unpaired, two-tailed t test.
To determine if IRF-5 had a role in DC development, we first assessed CDP and cDC frequencies in the bone marrow and spleen, respectively, of WT and Irf5−/− mice (1). Although no apparent defects were observed in CDPs or mature CD11chigh CD8α+ or CD8α− cDCs in Irf5−/− mice (Fig. 1 B and C), we did observe a slight increase in the frequencies and numbers of pDCs in the bone marrow of Irf5−/− mice, and this was accompanied by a sharp reduction of pDCs in the spleen and blood (8.7- and 6.2-fold; P < 0.01). Similarly, peripheral blood of Irf5−/− mice had a reduced percentage of pDCs relative to total leukocytes (Fig. 1C).
The role IRF-5 plays in type I IFN production by pDCs remains unclear, as different studies have conflicting results (2, 11–14), possibly because of differences in the source (in vitro- or in vivo-derived) of pDCs. To assess the ability of Irf5−/− pDCs obtained directly from mice to secrete type I IFN in response to TLR9 stimulation, WT or Irf5−/− pDCs were sorted from bone marrow and cultured for 24 h with increasing doses of CpG-A, a TLR9 agonist. Notably, IFN-α secretion by freshly sorted Irf5−/− pDCs was not detected at greater than background levels at any of the CpG-A doses tested (Fig. 1D).
Impaired B-Cell Development and Antibody Production in Irf5−/− Mice.
Given the postulated role of IRF-5 in B-cell development (15, 16), we analyzed the numbers and percentages of B-cell subsets by relative cell surface expression of B220, IgM, IgG, CD21, CD23, and CD93 (24). Fewer numbers of splenocytes were detected in 8-wk-old Irf5−/− compared with WT mice, which resulted in a significantly decreased number (3.1-fold; P < 0.001) and frequency (1.3-fold; P < 0.01) of B220+ cells (Fig. 2B). Within the B220+IgD+IgMlo cell gate, the Irf5−/− mice exhibited a small reduction in the frequency (89.5% vs. 84.5%; P < 0.001) and number (3.8-fold; P < 0.001) of follicular B cells (B220+IgD+IgMlowCD93−CD23+; Fig. 2 A and B). The most striking difference, however, was in the marginal zone (B220+IgD+IgMlowCD23−CD21+) B-cell compartment, which was virtually absent (11.0% vs. 1.2%; P < 0.0001) in Irf5−/− mice (Fig. 2 A and B). In the blood, we also observed an accumulation in the percentage (P < 0.001) of B220+ cells (Fig. S1 A and B). These cells in Irf5−/− mice, however, had an immature phenotype, as judged by an increased frequency of B220+CD93+ (4.0% vs. 7.7%; P < 0.0001) B cells and a decreased frequency of B220+CD23+CD93− B cells.
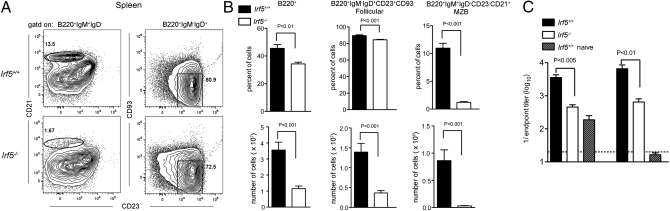
Fig. 2.
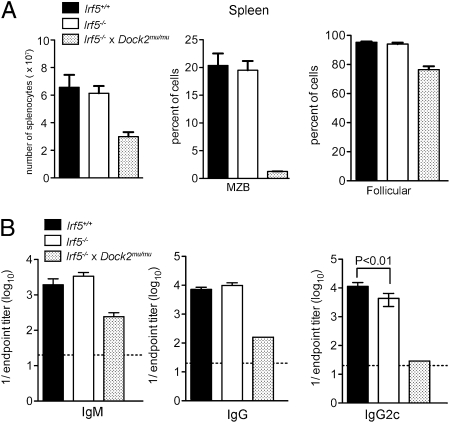
Irf5−/− mice have impaired peripheral B-cell development. (A) Flow cytometric analysis of marginal zone B cells (B220+IgM+IgD−CD23−CD21+) and follicular B cells (B220+IgM−IgD+CD23+CD93−) from the spleen. (B) Reduced frequency and number of B220+ splenocytes, marginal-zone B cells, and follicular B cells. (C) Titers of anti–WNV-envelope IgM (day 6, left) or IgG (day 9, right) in serum collected from infected or naive mice. Data shown are the mean and SD from three experiments with a total of nine mice per group. P values were determined by using an unpaired, two-tailed t test.
Because of the altered B-cell profiles, we assessed whether antibody responses were impaired in Irf5−/− mice by measuring serum antiviral antibody titers early after WNV infection by ELISA. Irf5−/− mice had decreased (sevenfold lower; P < 0.05) production of WNV-specific IgM at day 6 after infection compared with WT mice (Fig. 2C). At day 9 after infection, WNV-specific IgG titers also were reduced (43-fold lower; P < 0.01).
Reduction of pDCs and Marginal Zone B Cells Is Not Dependent on IRF-5.
Genetic background effects have been reported to affect peripheral pDC numbers (25). To more rigorously test the effects of an IRF-5 deficiency on B-cell and pDC development and function, we intercrossed Irf5+/− mice to generate littermate controls of both Irf5+/− and Irf5+/+ genotypes to compare with Irf5−/− mice. Offspring of each genotype were evaluated by flow cytometry for normal or abnormal marginal zone B-cell and pDC development (Figs. 1B and 2A). Of 39 F2 mice analyzed, 11 mice exhibited abnormal B-cell and pDC development. Surprisingly, within this group, five mice were Irf5+/−, three were Irf5−/−, and three were Irf5+/+, demonstrating independent segregation of Irf5 deficiency and the B-cell and pDC developmental phenotypes (Table S1). These results suggested that the B-cell and pDC developmental phenotypes were transmitted with a recessive Mendelian inheritance pattern, but neither defect segregated with a deficiency in IRF-5.
Abnormal B-Cell and pDC Phenotype Maps to Chromosome 11.
To identify the mutation responsible for impaired pDC and B-cell development, we used SNP mapping. The Irf5−/− mice used as described earlier had been backcrossed for eight generations onto the C57BL/6 background and maintained as homozygotes. We crossed these mice to WT BALB/c mice to generate Irf5+/− mice in a mixed C57BL/6-BALB/c background. These F1 mice subsequently were intercrossed and evaluated for abnormal marginal zone B-cell and pDC development, and genomic DNA was subjected to SNP analysis. All 10 animals in the first round of screening with defective B-cell and pDC development had conserved C57BL/6 DNA between 0 and 41.0 Mb on chromosome 11 (Tables S2 and S3). This result agreed with our heterozygous cross-analysis demonstrating that the B-cell and pDC developmental phenotype was not dependent on Irf5, which is located on chromosome 6. The presence of C57BL/6 DNA in unaffected mouse 24 further narrowed the region to 0 to 38.7 Mb. Analysis of mouse 63, which retained the B-cell and pDC developmental defect, identified the region between 30.0 and 38.7 Mb on chromosome 11 as responsible for transmitting the observed phenotype in the Irf5−/− mice.
Irf5−/− Mice Have a Genomic Duplication Within Dock2 Locus.
Interestingly, the gene encoding Dock2, which is required for activation of Rac2 and actin polymerization downstream of chemokine receptor signaling (17), is located within this region. Similar to the phenotype exhibited by the Irf5−/− mice, a genetic deficiency in Dock2 resulted in a loss of pDC and marginal zone B cells (17, 19). To test whether Irf5−/− mice had reduced levels of Dock2 expression, we performed quantitative RT-PCR on cells from the bone marrow and spleen. Dock2 expression was noticeably reduced in Irf5−/− bone marrow cells and splenocytes [9.5-fold (P < 0.01) and 5.6-fold (P < 0.001), respectively; Fig. 3A]. These data suggested that a reduction in the expression of Dock2, a gene located within the identified 8 Mb genomic region on chromosome 11, may be responsible for the impaired development of B cells and pDCs in Irf5−/− (now referred to as Irf5−/− × Dock2mu/mu) mice.
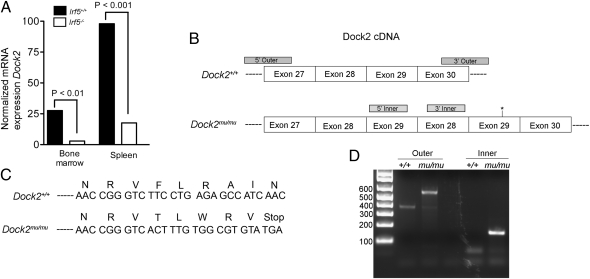
Fig. 3.
Reduced expression of Dock2 mRNA and mutation in Dock2 cDNA in Irf5−/− mice. (A) Expression of Dock2 mRNA in total bone marrow or spleen cells normalized to Gapdh expression. Data shown are from two independent experiments with a total of four mice per group. (B) Representation of Dock2 cDNA from exon 27 to exon 30 in WT and Irf5−/− × Dock2mu/mu mice as determined by sequencing. Asterisks represent location of premature stop codon in mutated cDNA. Gray blocks represent primer-binding sites for genotyping PCR as demonstrated in D. (C) DNA sequence of either Dock2+/+ or Irf5−/− × Dock2mu/mu demonstrating introduction of stop codon in Irf5−/− × Dock2mu/mu mice. (D) PCR results from genotyping of WT or Irf5−/− × Dock2mu/mu mice with outer primers (amplifying the region flanking the duplication) or inner primers (amplifying exon 29 to exon 28).
To identify a potential mutation, splenic cDNA was sequenced across the Dock2 coding region. We observed a repeated sequence corresponding to exons 28 and 29, likely indicating a genomic duplication of this region (Fig. 3A). This exon duplication resulted in a frameshift mutation and premature stop codon after exon 29 (Fig. 3C). As Dock2 possesses 52 exons, the introduction of a stop codon after exon 29 likely results in nonsense-mediated decay of the transcript (26), perhaps explaining the low levels of mRNA expression that we observed. To further confirm this mutation, we designed primers flanking the region of gene duplication and performed RT-PCR on splenocyte RNA from the Irf5−/− × Dock2mu/mu mice. We observed a larger PCR product in the Irf5−/− × Dock2mu/mu compared with WT control mice (Fig. 3D). Furthermore, PCR primers that amplify a region spanning exons 29 and 28 generated a product from Irf5−/− × Dock2mu/mu cDNA but not from the WT template. These data confirm that the Irf5−/− mice carried an exon duplication within the Dock2 locus, likely resulting in a nonfunctional allele.
To assess the prevalence of the mutation outside of our facility, Irf5−/− mice maintained by three separate laboratories were screened by RT-PCR for the aberrant Dock2 transcript. Although the mutant Dock2 gene was not identified in Irf5−/− mice maintained in Japan, the Dock2 mutation was detected in Irf5−/− mice from two other laboratories in the United States (Fig. S2). Therefore, the mutation is present in multiple colonies within the United States.
Dock2 Expression Rescues B-Cell and pDC Development in Irf5−/− × DOCK2mu/mu Mice.
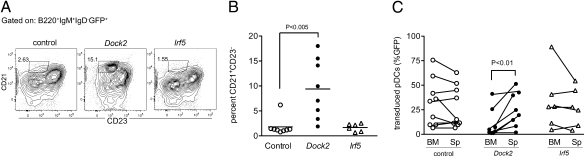
To test whether ectopic expression of DOCK2 could restore marginal zone B-cell and pDC development in Irf5−/− × Dock2mu/mu mice, we transduced hematopoietic stem cells (HSCs) from CD45.2 Irf5−/− × Dock2mu/mu mice with a GFP-marked retrovirus encoding Dock2 or a control MSCV and reconstituted sublethally irradiated CD45.1 mice. Retroviral expression of DOCK2 resulted in a fivefold increase (P < 0.001) in the frequency of marginal zone B cells compared with transduction with the control retrovirus (Fig. 4 A and B). Similarly, the percentage of GFP+ Dock2-transduced donor splenic pDCs was consistently higher than that of bone marrow pDCs in the same mice, demonstrating a rescue of the pDC homing defects (Fig. 4C). No such increase was observed in mice reconstituted with the control retrovirus (Fig. 4C). As predicted, mice reconstituted with IRF-5–transduced Irf5−/− × Dock2mu/mu HSCs still exhibited marginal zone B-cell and pDC defects and were indistinguishable from recipients receiving HSCs transduced with control retrovirus (Fig. 4 B and C). Together, these data conclusively demonstrate that the developmental defect in the Irf5−/− × Dock2mu/mu mice was not dependent on Irf5 deficiency, but rather was the consequence of a recessive Dock2 mutation.
Fig. 4.
Ectopic expression of Dock2 rescues marginal zone B-cell and pDC development in Irf5−/− mice. (A) Flow cytometry plots of marginal zone B cells (CD21+CD23−) within the B220+IgM+IgD− gate from Irf5−/− × Dock2mu/mu mice reconstituted with stem cells transduced with control (Left), Dock2 (Center), or Irf5 (Right) expressing retrovirus. (B) Percentage of marginal zone B cells from Irf5−/− × Dock2mu/mu mice reconstituted with stem cells as described in A. (C) Percentage of GFP+ control or Dock2 transduced pDCs (B220+BST2+) in the spleen or bone marrow. P values were determined by using an unpaired, two-tailed t test (B) or a paired, two-tailed t test (C) using data from three independent experiments.
Normal pDC and B-Cell Development in Irf5−/− Mice of 13th Generation.
To conclusively determine the role of IRF-5 in immune cell development, Irf5−/− mice lacking the Dock2 mutation were generated by backcrossing to C57BL/6 mice for an additional five generations, and analyzed for bone marrow and splenic pDCs by flow cytometry. In contrast to Irf5−/− × Dock2mu/mu mice, Irf5−/− mice had normal frequencies of pDCs in the spleen (Fig. 5A). Furthermore, we measured IFN-α production after CpG stimulation in vitro from sorted bone marrow pDCs from Irf5−/− or WT mice. Whereas IFN-α production from Irf5−/− × Dock2mu/mu pDCs was nearly undetectable (Fig. 1D), Irf5−/− and WT pDCs produced essentially equivalent levels of IFN-α, with a nonsignificant (P > 0.2) trend toward a small reduction only at the lowest dose of 0.3 μg/mL CpG-A (Fig. 5B). These data differ from those from previous studies, which reported minimal type I IFN production by in vitro-derived Irf5−/− pDCs (12). It is unclear if these discrepancies are related to the Dock2 mutation or differences in assay conditions.
Fig. 5.
Irf5−/− mice have normal pDC development. (A) Frequency of pDCs (B220+BST2+) from bone marrow and spleen and number of pDCs from spleen from Irf5+/+, Irf5−/−, or Irf5−/− × Dock2mu/mu mice. (B) IFN-α production by sorted pDCs from Irf5−/− stimulated with CpG as measured by ELISA. (C) Primary bone marrow pDCs from Irf5+/+ and Irf5−/− mice were stimulated with CpG-B or imiquimod, and levels of IL-6 (Left) and TNF-α (Right) secretion in the supernatant were measured by flow cytometry with beads containing capture antibodies. P values were determined by a paired, two-tailed t test from four mice from two independent experiments. (D) Levels of IL-6 (Left) and TNF-α (Right) production from sorted CD11b+ cells infected with HSV-1 at varying multiplicities of infection (MOIs). P values (****P < 0.0001, **P < 0.01) were determined by using an unpaired, two-tailed t test. (E) IFN-α production from HSV-1–infected (multiplicity of infection of 20) splenic CD11b+ cells measured by ELISA. P values were determined by using an unpaired, two-tailed t test. Data shown are from a total of six mice from two experiments (A), a representative of two experiments with two mice each (B and C), or a total of four mice from two experiments (D and E).
Previous studies reported a defect in the production of inflammatory cytokines by Irf5−/− cells in response to TLR agonists or viral infection (2, 11). To verify these findings, in our cells that have a WT Dock2 gene, we stimulated primary bone marrow pDCs with CpG-B or imiquimod and measured IL-6 and TNF-α secretion. Both TLR agonists stimulated less IL-6 production in Irf5−/− pDCs relative to WT cells, although this difference did not reach statistical significance in imiquimod-treated cultures. Similarly, although the magnitude of reduction was variable, TNF-α production by Irf5−/− pDCs was diminished relative to WT cells under both conditions. Analogously, HSV-1–infected splenic Irf5−/− CD11b+ cells showed a statistically significant reduction in IL-6 (2.0–2.4 fold; P < 0.001), TNF-α (5.6–7.1-fold; P < 0.0001), and IFN-α (1.3-fold; P < 0.05) secretion relative to WT counterparts. Thus, these data confirm an IRF-5–dependent pathway for induction of inflammatory cytokines in pDCs and CD11b+ cells. Moreover, the data demonstrate that, under certain conditions, IRF-5 can modestly regulate type I IFN production.
The number and frequency of B220+ cells and B-cell subsets, including follicular B cells (B220+IgD+IgMlowCD93−CD23+) and marginal zone B cells (B220+IgD+IgMlowCD23−CD21+), was also normal in Irf5−/− mice (Fig. 6 A and B). To test the antibody response, Irf5−/− or WT mice were infected with WNV, and IgM or IgG titers were measured at days 6 and 9, respectively. The titer of anti–WNV-E–specific IgM antibody at day 6 and total anti-WNV IgG at day 9 were normal However, we did observe a small yet statistically significant (2.6-fold; P < 0.01) reduction in the anti-WNV IgG2c isotype response at day 9 in Irf5−/− mice (Fig. 6C), consistent with one previous study (15). Together, these data demonstrate that pDC numbers, type I IFN secretion, and B-cell development and function are largely normal in Irf5−/− mice.
Fig. 6.
Normal B-cell development in Irf5−/− mice. (A) Number of total splenocytes and frequency of marginal zone and follicular B cells from Irf5+/+, Irf5−/− mice, or Irf5−/− × Dock2mu/mu mice. (B) Titers of anti–WNV-envelope IgM (day 6) or IgG (day 9) or IgG2c (day 9) in serum collected from infected mice. Data shown are the mean and SD of two experiments with a total of five or six mice Irf5+/+ or Irf5−/− mice. P values were determined by using an unpaired, two-tailed t test.
Discussion
Our initial analysis of Irf5−/− mice yielded unexpected results, including a lack of peripheral pDCs and marginal zone B cells, dramatically reduced IFN-α secretion by pDCs, and marked reductions in antigen-specific IgG and IgM following WNV infection. However, through additional crosses with C57BL/6 mice, we demonstrated an independent segregation of Irf5-deficiency and these observed defects. A genomic duplication within Dock2 resulted in a frameshift mutation and premature stop codon, and ectopic expression of DOCK2 by retroviral transduction restored pDC and marginal-zone B-cell development. Finally, Irf5−/− mice lacking the Dock2 mutation had largely normal IFN-α production by pDCs and normal IgM and IgG titers with only a subtle decrease in antiviral IgG2c production.
Although the Dock2 mutation was introduced spontaneously and inadvertently bred to homozygosity, it remains unclear how and when the Irf5−/− mouse line acquired the mutation. The Dock2 mutation was observed in two separate Irf5−/− colonies initiated from mice obtained independently from the original colony (Fig. S2). Thus, it is likely that at least a subset of Irf5−/− mice from the original colony possessed the Dock2 mutation. The identification of a mutation that was maintained in a broadly distributed strain through intercrossing highlights the importance of periodic backcrossing and verification of findings through genetic complementation studies. Given the implications of Irf5 in association with autoimmune disease, many studies have analyzed the functional contribution of IRF-5 to type I IFN production and to antibody responses, often with different conclusions. Some of these disparities may be explained by the distinct assays or cells used in each study. Indeed, DOCK2 is not required for monocyte development or migration (17), and we have no reason to question previous findings regarding Irf5−/− macrophages (11, 27). Nevertheless, our findings suggest that a spontaneous Dock2 mutation in Irf5−/− mice may have affected some previous conclusions, and provide an additional explanation for overt discrepancies among published results. Clearly, repetition of viral pathogenesis and autoimmune studies are warranted with Irf5−/− mice on a background lacking the Dock2 mutation to conclusively define the function of IRF-5 in priming innate and adaptive immune responses.
Materials and Methods
Mice.
C57BL/6 WT mice and B6.SJL mice were commercially obtained from The Jackson Laboratory Irf5−/− mice were originally generated by T. Mak (University of Toronto, Toronto, ON, Canada) (2), maintained by T. Taniguchi (University of Tokyo, Tokyo, Japan), and obtained in the United States from I. Rifkin (Boston University School of Medicine, Boston, MA) after eight generations of backcrossing on the C57BL/6 background. Genotyping primers spanning the Dock2 mutation from cDNA are as follows: Dock2 27–30, 5′-GGATGCGGCCTTCACTTA-3′; and Dock2 27–30, 5′-TCCACAGCTGGAACTCAAAG-3′. Genotyping primers that detect the specific Dock2 mutation from cDNA are as follows: Dock2 29–28, 5′-CAAGGACCTCATTGGGAAGAA-3′; and Dock2 29–28, 5′-CTGAGCTGGTCTGGAAGGTCT-3′
Viruses.
The WNV strain (3000.0259) was isolated in New York in 2000 and passaged once in C6/36 Aedes albopictus cells. Mice were inoculated s.c. in the footpad with 102 pfu of WNV diluted in HBSS and 1% heat-inactivated FBS. The KOS strain of HSV-1 was provided by D. Leib (Dartmouth College, Hanover, NH).
Flow Cytometry.
Single cell suspensions were depleted of erythrocytes by using pure Histopaque-1119 or Histopaque-1119 mixed with Histopaque-1077 (Sigma-Aldrich) and differential centrifugation. Cells were stained at 4 °C in PBS solution containing 2% FBS. Data were collected on a FACSCalibur or LSRII device (BD Biosciences) and analyzed with FlowJo software (Tree Star).
Antibodies.
Monoclonal antibodies were purified from the following hybridomas by Bio-X-Cell: TER119 (anti-Ter119), 1D3 (anti-CD19), and A20.1.7 (anti-CD45.2). Antibodies were conjugated to Pacific blue or Alexa Fluor 647 (Invitrogen) according to the manufacturer's instructions. The following were purchased from eBiosciences: anti–c-kit (2B8) and anti-B220 (6B2) conjugated to PE-Cy7; anti-CD135 (A2F10) and anti-CD23 (B3B4) conjugated to PE; anti-CD11c conjugated to PerCp-Cy5.5; anti-Sca-1 (D7) conjugated to Alexa Fluor 700; anti-CD45.1 (A20) conjugated to APC-Cy7; anti-CD21 (4E3) conjugated to Pacific blue; and anti-IgD (clone 11–26) conjugated to biotin; and anti-BST2 (129-c) conjugated to APC. The following antibodies were purchased from BD Biosciences: anti-IgM (11/41) conjugated to PE-Cy5.5 and anti-CD11b (M1/70) conjugated to Alexa Fluor 647. Anti-Ly6D (114.8.1) conjugated to FITC was purchased from Sigma.
WNV-Specific Antibody ELISA.
Serum antibody titers were calculated as previously described (27). Briefly, 96-well plates (Nalge Nunc) were coated with 5 μg/mL recombinant WNV envelope protein overnight at 4 °C. After blocking for 1 h with PBS solution supplemented with 2% (wt/vol) BSA and 0.05% (vol/vol) Tween 20 at 37 °C, diluted serum samples were added to wells and incubated for 1 h at room temperature. Plates were washed with PBS solution with 0.05% (vol/vol) Tween 20, and incubated with 1 μg/mL of biotinylated anti-IgG or IgM (Sigma Aldrich) for 1 h. After washing, streptavidin conjugated horseradish peroxidase (2 μg/mL; Zymed) was added to each well and plates were incubated for 1 h. Plates were developed by using tetramethylbenzine substrate (Dako) and quenched with H2SO4. The optical density was read at 450 nm, and endpoint dilutions were calculated as three SDs above the background.
DNA Constructs and Retroviral Production.
The coding region of Dock2 cDNA was obtained by RT-PCR from mouse bone marrow RNA as a NotI-SalI fragment, and cloned into the same sites of murine stem cell virus (MSCV)–internal ribosomal entry site (IRES)–GFP (28, 29) with sequence verification. Transfection and retroviral infection of 293FT cells (Invitrogen) was used to confirm expression of DOCK2 by Western blot with commercially available antibodies (Chemicon). The 293FT cells were cultured to approximately 60% density on 10-cm2 tissue culture plates and transfected by using calcium phosphate coprecipitation with 3.5 μg pMD2.G (a VSV-G-encoding expression vector), 6.5 μg of an expression plasmid encoding MMLV gag-pol (30), and 10 μg of MSCV-IRES-GFP, MSCV-Irf5-IRES-GFP, or MSCV-Dock2-IRES-GFP. Medium was changed 4 to 6 h after transfection, and supernatant was harvested at 48 and 72 h later and filtered through 0.45-μM syringe filters. Twelve milliliters of viral supernatant was mixed with 3 mL of 1× PBS solution/25% polyethylene glycol 8000 (Sigma) and incubated overnight at 4 °C. The mixture was centrifuged at 2,000 × g for 10 min, supernatant was removed, and the pellet was resuspended in 100 μL PBS solution. Aliquots were frozen and stored at −80 °C until use.
Retroviral Infections and HSC Transplantation.
Bone marrow was harvested from donor mice by crushing bones and removing debris on a density gradient by using Histopaque 1119 (Sigma). Bone marrow was then enriched for c-kit+ cells by using CD117+ microbeads (Miltenyi Biotec). Cells were stained with antibodies (described earlier), and HSCs were isolated by FACS based on previously defined reactivity for particular cell surface markers (Lineage− c-kit+ Sca-1+ Flk2−) on a FACSAria device (BD Biosciences). Cells (n = 5,000–10,000) were cultured overnight in 200 μL DMEM F12 media with Hepes containing 10% FBS (HyClone), nonessential amino acids, sodium pyruvate, GlutaMAX, 50 μM β-mercaptoethanol (Gibco), 50 ng/mL stem cell factor (Peprotech), and 50 ng/mL thrombopoietin (Peprotech) per well of 96-well round-bottom plates (BD Biosciences). Five microliters of polyethylene glycol-concentrated virus was added to the cultures the following day. One day after infection, cells were washed extensively and transplanted by retroorbital injection into mice irradiated with 600 cGy.
Quantitative Real-Time RT-PCR.
First-strand cDNA was synthesized from RNA by using SuperScript III (Invitrogen) and random hexamers according to the manufacturer's instructions. Amplifications were performed by using SYBR Green JumpStart TaqReadyMix (Sigma) on an ABI 7000 Sequence Detection System (Applied Biosystems) using at least 1,000 cell equivalents per reaction. Primer sequences are as follows: β-actin, 5′-GTCTGAGGCCTCCCTTTTT-3′ and 5′-GGAGACCAAAGCCTTCATA-3′; Irf5, 5′-GGAAGAAATGAAGCCAGCAG-3′ and 5′-ACCCTGGGGTAATTGGACTC-3′; Dock2, 5-CTTCTTCCAAGTCTCAGATGG-3′ and 5′-TTCCCACAGTGCTCG GCTCA -3′; and GAPDH, 5-GGCAAATTCAACGGCACAGT-3′ and 5-GATGGTGATGGGCTTCCC-3′.
Cell Culture and Cytokine Detection.
pDCs from bone marrow were isolated by surface markers (BST2+B220+) by using FACS (Aria-II; BD) and cultured in 96-well plates with 50,000 pDCs per well in 200 μL RPMI medium supplemented with 10% FBS, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 50 μM β-mercaptoethanol, and varying doses of CpG-A 2216 (Operon) or ODN-1585 (InvivoGen). After 18 h in culture, supernatant was harvested and levels of IFN-α were measured by ELISA per the manufacturer's instructions (PBL IFN Source). Similarly, 25,000 flow cytometry-sorted bone marrow pDCs were cultured with 6 μg/mL CpG-B (Operon) or 6 μM imiquimod (InvivoGen). CD11b+ cells were sorted from the spleen and infected with HSV-1 in 96-well plates with 100,000 cells per well in 200 μL RPMI medium supplemented with 10% FBS, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, and 50 μM β-mercaptoethanol. After 24 h in culture, supernatant was harvested and IL-6 and TNF-α levels were measured by flow cytometry with the Mouse Inflammation CBA kit (BD Biosciences) for pDC and CD11b+ cultures.
SNP Analysis.
Irf5−/− × DOCK2mu/mu mice were crossed with WT BALB/c mice. The resulting F1 mice were intercrossed, and F2 mice were analyzed for normal or abnormal marginal zone B-cell development. DNA from 10 normal and 10 abnormal mice was used for SNP analysis conducted by Jackson Laboratory.
Supplementary Material
Acknowledgments
We thank I. Rifkin, P. Pitha, and T. Taniguchi for genotyping results of Irf5−/− mice; H. Suleiman, A. Shaw, and M. Cella for assistance with pDC assays; and M. Noll for animal colony maintenance. This work was supported by National Institutes of Health Grant K01DK078318 (to D.B.) and Pacific Northwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research Grants U54 AI081680 and U19 AI083019 (to M.S.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 5565 (volume 109, number 15).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118155109/-/DCSupplemental.
References
- 1.Barnes BJ, Moore PA, Pitha PM. Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon alpha genes. J Biol Chem. 2001;276:23382–23390. doi: 10.1074/jbc.M101216200. [DOI] [PubMed] [Google Scholar]
- 2.Takaoka A, et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 3.Savitsky D, Tamura T, Yanai H, Taniguchi T. Regulation of immunity and oncogenesis by the IRF transcription factor family. Cancer Immunol Immunother. 2010;59:489–510. doi: 10.1007/s00262-009-0804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miceli-Richard C, et al. Association of an IRF5 gene functional polymorphism with Sjögren's syndrome. Arthritis Rheum. 2007;56:3989–3994. doi: 10.1002/art.23142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham RR, et al. Argentine and Spanish Collaborative Groups A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 6.Kristjansdottir G, et al. Interferon regulatory factor 5 (IRF5) gene variants are associated with multiple sclerosis in three distinct populations. J Med Genet. 2008;45:362–369. doi: 10.1136/jmg.2007.055012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sigurdsson S, et al. Association of a haplotype in the promoter region of the interferon regulatory factor 5 gene with rheumatoid arthritis. Arthritis Rheum. 2007;56:2202–2210. doi: 10.1002/art.22704. [DOI] [PubMed] [Google Scholar]
- 8.Dideberg V, et al. An insertion-deletion polymorphism in the interferon regulatory Factor 5 (IRF5) gene confers risk of inflammatory bowel diseases. Hum Mol Genet. 2007;16:3008–3016. doi: 10.1093/hmg/ddm259. [DOI] [PubMed] [Google Scholar]
- 9.Burman P, Karlsson FA, Oberg K, Alm G. Autoimmune thyroid disease in interferon-treated patients. Lancet. 1985;2:100–101. [PubMed] [Google Scholar]
- 10.Cella M, et al. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat Med. 1999;5:919–923. doi: 10.1038/11360. [DOI] [PubMed] [Google Scholar]
- 11.Yanai H, et al. Role of IFN regulatory factor 5 transcription factor in antiviral immunity and tumor suppression. Proc Natl Acad Sci USA. 2007;104:3402–3407. doi: 10.1073/pnas.0611559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yasuda K, et al. Murine dendritic cell type I IFN production induced by human IgG-RNA immune complexes is IFN regulatory factor (IRF)5 and IRF7 dependent and is required for IL-6 production. J Immunol. 2007;178:6876–6885. doi: 10.4049/jimmunol.178.11.6876. [DOI] [PubMed] [Google Scholar]
- 13.Gratz N, et al. Type I interferon production induced by Streptococcus pyogenes-derived nucleic acids is required for host protection. PLoS Pathog. 2011;7:e1001345. doi: 10.1371/journal.ppat.1001345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai P, et al. Myxoma virus induces type I IFN production in murine plasmacytoid dendritic cells via a TLR9/MyD88, IRF5/IRF7, and IFNAR-dependent pathway. J Virol. 2011;85:10814–10825. doi: 10.1128/JVI.00104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savitsky DA, Yanai H, Tamura T, Taniguchi T, Honda K. Contribution of IRF5 in B cells to the development of murine SLE-like disease through its transcriptional control of the IgG2a locus. Proc Natl Acad Sci USA. 2010;107:10154–10159. doi: 10.1073/pnas.1005599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lien C, et al. Critical role of IRF-5 in regulation of B-cell differentiation. Proc Natl Acad Sci USA. 2010;107:4664–4668. doi: 10.1073/pnas.0911193107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukui Y, et al. Haematopoietic cell-specific CDM family protein DOCK2 is essential for lymphocyte migration. Nature. 2001;412:826–831. doi: 10.1038/35090591. [DOI] [PubMed] [Google Scholar]
- 18.Gotoh K, et al. Selective control of type I IFN induction by the Rac activator DOCK2 during TLR-mediated plasmacytoid dendritic cell activation. J Exp Med. 2010;207:721–730. doi: 10.1084/jem.20091776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gotoh K, et al. Differential requirement for DOCK2 in migration of plasmacytoid dendritic cells versus myeloid dendritic cells. Blood. 2008;111:2973–2976. doi: 10.1182/blood-2007-09-112169. [DOI] [PubMed] [Google Scholar]
- 20.Liu K, et al. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324:392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onai N, et al. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat Immunol. 2007;8:1207–1216. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki S, et al. Critical roles of interferon regulatory factor 4 in CD11bhighCD8alpha- dendritic cell development. Proc Natl Acad Sci USA. 2004;101:8981–8986. doi: 10.1073/pnas.0402139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schiavoni G, et al. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8alpha(+) dendritic cells. J Exp Med. 2002;196:1415–1425. doi: 10.1084/jem.20021263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allman D, et al. Resolution of three nonproliferative immature splenic B cell subsets reveals multiple selection points during peripheral B cell maturation. J Immunol. 2001;167:6834–6840. doi: 10.4049/jimmunol.167.12.6834. [DOI] [PubMed] [Google Scholar]
- 25.Asselin-Paturel C, Brizard G, Pin J-J, Brière F, Trinchieri G. Mouse strain differences in plasmacytoid dendritic cell frequency and function revealed by a novel monoclonal antibody. J Immunol. 2003;171:6466–6477. doi: 10.4049/jimmunol.171.12.6466. [DOI] [PubMed] [Google Scholar]
- 26.Chang Y-F, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 27.Oliphant T, et al. Induction of epitope-specific neutralizing antibodies against West Nile virus. J Virol. 2007;81:11828–11839. doi: 10.1128/JVI.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharya D, Logue EC, Bakkour S, DeGregori J, Sha WC. Identification of gene function by cyclical packaging rescue of retroviral cDNA libraries. Proc Natl Acad Sci USA. 2002;99:8838–8843. doi: 10.1073/pnas.132274799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ranganath S, et al. GATA-3-dependent enhancer activity in IL-4 gene regulation. J Immunol. 1998;161:3822–3826. [PubMed] [Google Scholar]
- 30.Bhattacharya D, Lee DU, Sha WC. Regulation of Ig class switch recombination by NF-kappaB: Retroviral expression of RelB in activated B cells inhibits switching to IgG1, but not to IgE. Int Immunol. 2002;14:983–991. doi: 10.1093/intimm/dxf066. [DOI] [PubMed] [Google Scholar]