Abstract
How the human pathogen Streptococcus pneumoniae coordinates cell-wall synthesis during growth and division to achieve its characteristic oval shape is poorly understood. The conserved eukaryotic-type Ser/Thr kinase of S. pneumoniae, StkP, previously was reported to phosphorylate the cell-division protein DivIVA. Consistent with a role in cell division, GFP-StkP and its cognate phosphatase, GFP-PhpP, both localize to the division site. StkP localization depends on its penicillin-binding protein and Ser/Thr-associated domains that likely sense uncross-linked peptidoglycan, because StkP and PhpP delocalize in the presence of antibiotics that target the latest stages of cell-wall biosynthesis and in cells that have stopped dividing. Time-lapse microscopy shows that StkP displays an intermediate timing of recruitment to midcell: StkP arrives shortly after FtsA but before DivIVA. Furthermore, StkP remains at midcell longer than FtsA, until division is complete. Cells mutated for stkP are perturbed in cell-wall synthesis and display elongated morphologies with multiple, often unconstricted, FtsA and DivIVA rings. The data show that StkP plays an important role in regulating cell-wall synthesis and controls correct septum progression and closure. Overall, our results indicate that StkP signals information about the cell-wall status to key cell-division proteins and in this way acts as a regulator of cell division.
The human pathogen Streptococcus pneumoniae is a Gram-positive coccus with a characteristic oval shape resembling an American football. This shape likely is achieved by the alternation of two separate biosynthetic events: peripheral cell-wall elongation and septal-wall synthesis (1). According to this model, dividing cells show an initial inward growth of the septal wall, but its progression is halted until the two newly synthesized internal hemispheres have reached the size of the external ones. At this point, septal-wall synthesis resumes rapidly, leading to cell division (1). Additional synthesis of cell-wall material is thought to be required to form mature cell poles (2). Although this model was established for Enterococcus faecalis (formerly Streptococcus faecalis), it is also consistent with observations in other oval streptococci such as S. pneumoniae (3). Although recently substantial progress has been made in understanding the molecular mechanisms that govern S. pneumoniae cell division (for a review, see ref. 4), the molecular mechanisms involved in the earlier events of the cell cycle, and what controls them, are largely unknown. In particular, what is not understood is how S. pneumoniae coordinates peripheral and septal cell-wall synthesis, by sensing the morphological changes that occur during growth and division, to achieve proper shape.
Prokaryotes often use phosphorylation/dephosphorylation cascades to monitor and to respond to environmental changes and cell-cycle signals. Two component systems, consisting of a histidine kinase with a cognate response regulator, are the most abundant signaling systems (5). Recent studies have shown that eukaryotic-type Ser/Thr protein kinases (STKs) also are present in a wide range of prokaryotic genomes and regulate complex and diverse cellular processes (6–13).
Gram-positive bacteria possess an ultraconserved subfamily of STKs specifically implicated in regulating growth and cell division (14–20). These STKs consist of a cytoplasmic kinase domain and an extracellular C-terminal region composed of several penicillin-binding protein and Ser/Thr kinase-associated (PASTA) domains. It was suggested that PASTA domains can bind peptidoglycan (PG) fragments that might act as a signaling molecule (21, 22). This hypothesis was supported by the finding that PASTA domains of protein kinase PrkC from Bacillus subtilis bind PG in vitro and activate spore germination in response to cell-wall–derived muropeptides (23). It was found that the minimal signal for PrkC is N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) linked to the pentapeptide (NAG/NAM-pp). PrkC responded only to a synthetic PG containing meso-diaminopimelate (m-DAP) at position three of the pentapeptide (24). This evidence suggests that this third residue, which is an l-lysine in pneumococci, likely represents the specificity-determining element for STKs.
Here we are interested in elucidating the biological function of the PASTA-containing STK of S. pneumoniae, StkP. Previous work has shown that StkP acts as a dimer (25) and forms a signaling couple with its cognate phosphatase PhpP, which is a predicted cytoplasmic protein (26, 27). PASTA domains of StkP were shown to bind synthetic and native PG subunits and β-lactam antibiotics (28). A strain in which stkP is deleted is still viable in vitro but grows more slowly, is less competent for genetic transformation, and is more susceptible to several environmental stresses (29–32). StkP also plays an essential role for in vivo survival, because stkP mutants were strongly attenuated in virulence in mouse models (29, 30). Phenotypic analysis, through both transmission electron microscopy and differential interference contrast microscopy, showed that stkP mutants often are elongated, suggesting a defect in cell division (30, 33). Several StkP substrates playing a role in cell-wall metabolism and cell division were identified, including phosphoglucosamine mutase GlmM and the cell-division proteins DivIVA and FtsZ (26, 33, 34). By using immunofluorescence it was shown that StkP localizes to cell-division sites (34), but the molecular mechanisms underlying the stkP-null phenotype remain unknown.
We previously developed a single-cell toolbox for S. pneumoniae that allowed in vivo protein-localization studies in live pneumococcal cells using a fast-folding variant of GFP (35).
With this tool we showed that S. pneumoniae DivIVA localizes to both the cell division sites and the cell poles (35). We now show that StkP also localizes to the midcell and that this localization pattern depends on its extracellular PASTA domains. Furthermore, we find that StkP and its phosphatase PhpP show a cell-cycle–dependent localization pattern and localize to cell-division sites at which active PG synthesis is occurring. We provide in vivo evidence that the signal for StkP to autophosphorylate is uncross-linked PG, likely NAG/NAM-pp, which is present mainly in growing cells. We developed automated fluorescent time-lapse microscopy of double-labeled strains to image live cells. Using this technique for S. pneumoniae, we show that StkP is recruited to the cell-division site shortly after the assembly of the early cell-division protein FtsA but before the recruitment of DivIVA. However, StkP remains at midcell longer than FtsA, until division is complete. In the absence of StkP, peripheral cell-wall synthesis is increased compared with septal-wall synthesis, resulting in the elongated phenotype characteristic of the stkP-null mutants. The data support a model wherein StkP coordinates streptococcal growth and division.
Results
StkP Localizes to Midcell.
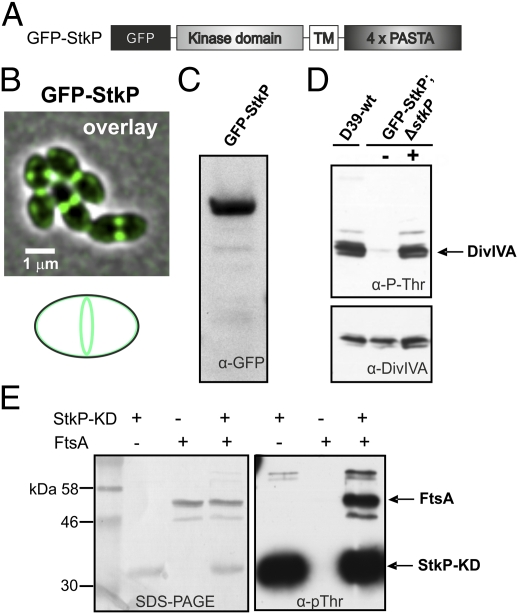
To determine the localization and dynamics of StkP in living cells, we constructed an N-terminal gfp-stkP fusion (Fig. 1A) by using vector pJWV25, which integrates by double-crossover in the chromosome at the nonessential bgaA locus and harbors the zinc-inducible PZn promoter (35). The resulting construct then was introduced into three closely related well-characterized and widely used S. pneumoniae strains: the encapsulated D39 and nonencapsulated R6 and Rx1 genetic backgrounds (36). Wild-type merodiploid strains carrying the gfp-stkP fusion were grown to midexponential phase, induced with 0.15 mM ZnSO4, and cells were collected for fluorescence microscopy 1 h later (OD600 ∼0.3). As shown in Fig. 1B, GFP-StkP localizes in the membrane with a clear enrichment at the cell-division sites. The same localization profile was observed when GFP-StkP was the only stkP copy present in the cell (Fig. S1) and was independent of the genetic background or growth medium used (Fig. 1B and Fig. S2). Western blot analysis of whole-cell extracts using either an anti-GFP antibody (Fig. 1C) or antibodies raised against StkP (Fig. S3) demonstrated the production of full-length GFP-StkP, absence of protein degradation, and only a slight (less than twofold) overproduction of StkP in the merodiploid strain.
Fig. 1.
StkP localizes to midcell and phosphorylates DivIVA. (A) Schematic outline of the GFP-StkP fusion. (B) Micrographs of strain KB02-20 (PZn-gfp-stkP) grown in C+Y medium at 37 °C. An overlay of the phase contrast (grey) and the GFP fluorescence (green) signal is shown. (Scale bar, 1 μm.) (C) Western blot analysis using anti-GFP antibodies (α-GFP). (D) Phosphoproteome analysis shows that GFP-StkP is functional and phosphorylates DivIVA efficiently. Total protein extracts were subjected to SDS/PAGE, and Western blotting was performed using anti-pThr antibodies (α–pThr) (Upper) or anti-DivIVA antibodies (α-DivIVA) (Lower). The plus sign indicates the addition of 0.15 mM of ZnCl2. (E) In vitro phosphorylation of FtsA by StkP. Recombinant His-FtsA was incubated with or without StkP-KD in kinase buffer. Proteins were separated by SDS/PAGE, electrotransferred to a PVDF membrane, and probed with anti-pThr antibody (α-pThr) to detect phosphorylation (Right). To detect the presence of proteins, the membrane was stained with amidoblack (Left). The arrows indicate the positions of FtsA and StkP-KD.
To test the catalytic activity of the GFP-StkP fusion, we examined the in vivo protein phosphorylation profile of a strain in which stkP was deleted but which harbored the zinc-inducible GFP-StkP fusion, using an antibody that reacts specifically to proteins that are phosphorylated on Thr residues (anti-pThr). Western blot analysis using antibodies raised against DivIVA served as a control of the whole-cell extracts, because DivIVA is the most abundant substrate of StkP (33). In the whole-cell extract of the wild-type strain, phosphorylated DivIVA (p-DivIVA) was detected readily, whereas in a strain that contained PZn-gfp-stkP as the only copy of stkP, p-DivIVA was detected only after GFP-StkP was induced (Fig. 1D and Fig. S4). Taken together, these data suggest that the GFP-StkP fusion protein is functional.
A recent global phosphoproteomic analysis showed that 84 S. pneumoniae proteins are phosphorylated on Ser, Thr, or Tyr residues, including the previously identified StkP substrate DivIVA (37). Which of the other proteins are true targets of StkP remains unclear. Interestingly, phosphorylated sites were identified in the essential cell-division protein FtsA at residues S113, T116, and T160 (37). However, phosphorylation profiling using anti-pThr did not detect migration of a protein that was the expected size of FtsA (Fig. S4). To test whether FtsA could be phosphorylated by StkP, we purified S. pneumoniae FtsA and performed in vitro phosphorylation assays using the purified StkP-kinase domain. As shown in Fig. 1E, FtsA was phosphorylated readily but only in the presence of the StkP-kinase domain. Nevertheless, singly (S113A, T116A, T160A) and doubly (T116A/T160A) mutated FtsA proteins still were phosphorylated efficiently by StkP in vitro (Fig. S5), raising a question as to whether FtsA is a bona fide target of StkP in vivo.
Localization of StkP Depends on Its Extracellular PASTA Domains.
StkP can be divided into three functional domains: a cytoplasmic kinase domain, a membrane-spanning (transmembrane, TM) domain, and the extracellular PASTA domains. To determine which part of StkP is the driving force in localizing protein to midcell, we constructed a set of N-terminal GFP fusions to the separate domains (Fig. 2A). To verify efficient production of the fusion proteins and the absence of protein degradation, wild-type strains carrying the PZn-gfp-kinase, PZn-gfp-kinase-TM, and PZn-gfp-TM-PASTA fusions were grown and induced with 0.15 mM ZnSO4, and cells were harvested at midexponential phase for Western blot analysis using anti-GFP polyclonal antibodies and for fluorescence microscopy. As shown in Fig. 2B, all fusion proteins were produced and were present mainly as full-length proteins. Fluorescence microscopy showed a cytoplasmic signal present in GFP fused to the kinase domain (Fig. 2C). The fusion of the kinase domain with the TM domain showed membrane localization with a slight enrichment at the septum, possibly caused by the presence of a double membrane (Fig. 2C). When GFP was fused to the PASTA domains without the kinase domain, a clear midcell localization was observed. Similar localization profiles also were observed in the absence of stkP (Fig. S6). A GFP fusion to a catalytically inactive mutant of StkP, StkP-K42R (26), also showed midcell localization (Fig. S7). Finally, localization of StkP did not depend on DivIVA (Fig. S8). Together, these results strongly suggest that the PASTA domains, and not the active kinase domain, are responsible for cellular targeting.
Fig. 2.
Localization of StkP depends on its PASTA domains. (A) Schematic outline of the GFP fusions used. (B) Exponentially growing cells in C+Y medium were harvested for Western blot analysis after 1 h of induction with 0.15 mM ZnSO4. (C) Cells were grown in GM17 medium and were analyzed by fluorescence microscopy. Note that some fusions expressed higher fluorescence levels than others, hence the differences in contrast of the micrographs. (Scale bar, 1 μm.) Phase contrast (PC), the GFP signal, and an overlay are shown. For clarity, schematic representations of the localization patterns are depicted next to the micrographs. (D) Micrographs of strains KB02-60 (PZn-gfp-stkP′prkC) (Upper) and KB02-61 (PZn-gfp-stkP′prkC, ΔstkP) (Lower) grown in C+Y medium. (Scale bar, 1 μm.)
PASTA domains of StkP bind muropeptides derived from S. pneumoniae, but they fail to bind PG preparations of Staphylococcus aureus (28). Furthermore, Shah et al. (23) showed that only NAG/NAM-pp with m-DAP at position three in the stem peptide was able to activate PrkC-dependent germination. Thus, if the PASTA domains of S. pneumoniae StkP (PASTAspn) indeed are responsible for targeting StkP to the cell-division site by specifically recognizing unlinked PG containing l-lysine at position three of the stem peptide, then an StkP mutant that contains the PASTA domains of B. subtilis PrkC should not localize properly. To test this hypothesis, we constructed a hybrid, StkP-PrkC, fused to GFP (Fig. 2A). When the stkP-prkC hybrid was the only stkP copy present in the genome, membrane localization and occasional polar enrichment were observed without the typical midcell localization (Fig. 2D). Interestingly, when wild-type StkP was present, the midcell localization pattern could be observed, but the ratio of midcell to membrane signal was reduced at higher levels of induction (Fig. S9). This finding suggests that the GFP-StkP-PrkC hybrid is able to interact with wild-type StkP, likely through the TM region, which is sufficient for dimerization of StkP in vivo (25).
PhpP Localizes to Midcell in a StkP-Dependent Manner.
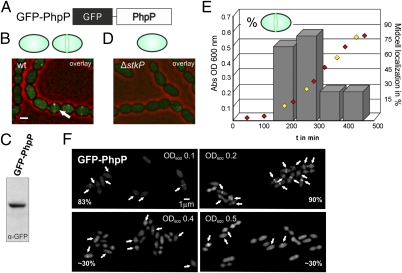
To examine the localization of StkP's cognate phosphatase, PhpP, we constructed a zinc-inducible N-terminal gfp-phpP fusion (Fig. 3A and Fig. S3) and inserted it in the D39, R6, and Rx1 genetic backgrounds. Wild-type strains carrying the gfp-phpP fusion were grown to midexponential phase, induced with 0.15 mM ZnSO4, and cells were analyzed by fluorescence microscopy. As shown in Fig. 3B, both cytoplasmic and midcell localization profiles were observed for GFP-PhpP, independent of the genetic background tested (Fig. S2). The phosphoprotein profile in a PZn-gfp-phpP, ΔphpP strain demonstrated that the GFP-PhpP fusion was at least partially functional, because induction resulted in dephosphorylation of DivIVA (Fig. S4). So far only StkP and RitR have been identified as targets of PhpP (26, 27, 38), but our analysis also identifies DivIVA as an in vivo target of PhpP.
Fig. 3.
PhpP is enriched at midcell in growing cells. (A) Schematic outline of the GFP-PhpP fusion. (B and D) Micrographs of strain KB01-14 (PZn-gfp-phpP) (B) and strain KB01-21 (PZn-gfp-phpP, ΔphpP-stkP) (D) taken at late exponential growth. Overlays between phase contrast (red) and the GFP signal (green) are shown. The arrow points to midcell-localized GFP-PhpP. (Scale bar, 1 μm.) For clarity, a schematic representation of the localization pattern is depicted above the micrographs. (C) Western blot analysis of strain KB01-14 using anti-GFP antibodies. (E) KB02-23 (PZn-gfp-phpP) cells were grown in C+Y medium at 37 °C (red and yellow diamonds) and were scored for the presence of midcell GFP-PhpP localization at different points in time (yellow diamonds). At least 500 cells per time point were analyzed. (F) Representative GFP micrographs of KB02-23 cells from the culture at different ODs. Arrows point to midcell-localized GFP-PhpP.
Interestingly, the midcell-localization profile was not evident in late, exponentially grown cells. Western blot analysis of whole-cell extracts using an anti-GFP antibody demonstrated the production of full-length GFP-PhpP, indicating that the cytoplasmic signal was not caused by protein degradation (Fig. 3C). Because PhpP is the cognate phosphatase of StkP, it seemed reasonable that the enriched localization of PhpP at midcell was caused by a direct interaction with StkP. To test this possibility, we examined localization of PhpP in the absence of StkP. As expected, in the stkP-null mutant background, GFP-PhpP was localized exclusively in the cytoplasm (Fig. 3D and Fig. S10). Notably, enrichment of GFP-PhpP at midcell also was lost completely in the catalytically inactive stkP-K42R mutant background (Fig. S10).
Because GFP-PhpP was not enriched at midcell in all cells, we wondered whether this enrichment was dependent on the growth phase. Therefore, we performed a time-series experiment taking cells for fluorescence microscopy analysis at different stages of the growth curve (Fig. 3E). As shown in Fig. 3F, most exponentially growing cells (83–90%) displayed the midcell-enriched localization pattern. However, when cells entered the stationary growth phase, this number decreased significantly (to 31%). These results indicate that midcell localization of GFP-PhpP is a good proxy for StkP activity.
Localization and Activation of StkP Requires Ligand Availability.
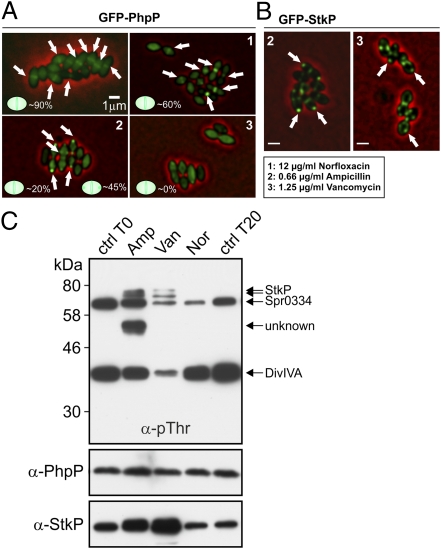
To substantiate that newly synthesized, uncross-linked PG subunits (NAG/NAM-pp) are the ligand of StkP, we made use of the fact that part of the cellular pool of PhpP is enriched at cell division sites in the presence of active StkP. If binding or recognition of uncross-linked PG is required for localization and activation of StkP, the addition of antibiotics that perturb the formation or availability of uncross-linked PG, such as ampicillin and vancomycin, should perturb localization of PhpP. Vancomycin binds to the terminal d-Ala-d-Ala moieties of NAG/NAM-pp, thus blocking the subsequent enzymatic reactions of the penicillin-binding proteins (PBPs) (39). Ampicillin binds to the transpeptidase and the carboxypeptidase domains of PBPs, thus blocking PG cross-linking and PG maturation (40). As shown in Fig. 4A, in the absence of antibiotics, GFP-PhpP shows an enriched localization to midcell in about 90% of exponentially growing cells. When incubated for 20 min with an IC50 concentration of 12 μg/mL of norfloxacin (41), an inhibitor of topoisomerases (42), most (∼60%) of the cells still displayed a normal midcell GFP-PhpP localization. However, when incubated with vancomycin (1.25 μg/mL; IC50 value), midcell localization of GFP-PhpP was no longer observed (Fig. 4A). Addition of ampicillin (0.66 μg/mL; IC50 value) also caused a delocalization of GFP-PhpP. Strikingly, however, GFP-PhpP did not delocalize to the cytoplasm completely; in a large portion (∼45%) of cells, GFP-PhpP localized to the cell poles, indicating improper production of PG in the presence of ampicillin (Fig. 4A). Similar results were observed for GFP-StkP: In the presence of vancomycin (1.25 μg/mL), GFP-StkP delocalized from the septum and displayed a spotty pattern, and in the presence of ampicillin (0.66 μg/mL) many cells showed polarly localized GFP-StkP (Fig. 4B).
Fig. 4.
Localization of GFP-PhpP and GFP-StkP is perturbed by antibiotics that inhibit cell-wall synthesis. (A and B) KB02-23 (PZn-gfp-phpP) cells (A) or KB02-20 (PZn-gfp-stkP) (B) cells were grown in C+Y medium at 37 °C to an OD600 0.2 and were incubated for 20 min with the indicated amount of antibiotics: (1) 12 μg/mL norfloxacin (Nor); (2) 0.66 μg/mL ampicillin (Amp); (3) 1.25 μg/mL vancomycin (Van). The percentage of cells with midcell-localized GFP-PhpP in each micrograph is indicated in A. Arrows indicate regions of enriched localization. (Scale bar, 1 μm.) (C) Protein phosphorylation in wild-type S. pneumoniae grown in the presence of antibiotics. At OD600 0.2, antibiotics were added, and cells were harvested for analysis after 20 min of incubation. Proteins were separated by SDS/PAGE and immunoblotted using anti-pThr antibody. Protein phosphorylation of untreated cultures before (lane 1) and after (lane 5) 20-min incubation and of cultures treated with 0.66 μg/mL ampicillin (lane 2), 1.25 μg/mL vancomycin (lane 3), or 12 μg/mL norfloxacin (lane 4). Native StkP and PhpP were detected in the same samples using specific anti-PhpP and anti-StkP antibodies, respectively. Arrows indicate protein bands corresponding to the previously identified proteins StkP, DivIVA, and Spr0334 and an unknown substrate.
If uncross-linked PG (NAG/NAM-pp) is the real in vivo target of StkP, the addition of ampicillin should increase StkP activity, and the addition of vancomycin, which binds to NAG/NAM-pp, should reduce StkP activity. To test this notion, we examined the in vivo Thr protein phosphorylation profile of cells incubated with vancomycin or ampicillin. A phosphoprotein pattern was observed that included the StkP substrates DivIVA and Spr0334, as previously identified (33). In line with the localization results, protein phosphorylation was reduced significantly by adding vancomycin (Fig. 4C). When norfloxacin was added, a slightly reduced protein phosphorylation pattern was observed, perhaps caused by an arrested growth in the presence of this antibiotic, consistent with previous reports (33). Strikingly, we observed hyperphosphorylation in the presence of ampicillin, and a previously faintly phosphorylated band (26), representing an as yet unknown protein, became robustly phosphorylated (Fig. 4C). Although cell morphologies remained normal 20 min after the addition of these antibiotics, we cannot exclude the possibility that partial autolysis already was induced, liberating PG fragments that might act as ligands for StkP. We also cannot exclude the possibility that vancomycin blocks the binding of another protein that produces a ligand. Nevertheless, these data suggest that cell-wall synthesis and uncrossed-linked PG not only are the signal for StkP to localize to cell division sites but also stimulate its autophosphorylation activity.
Temporal Order of Assembly of FtsA, StkP, and DivIVA.
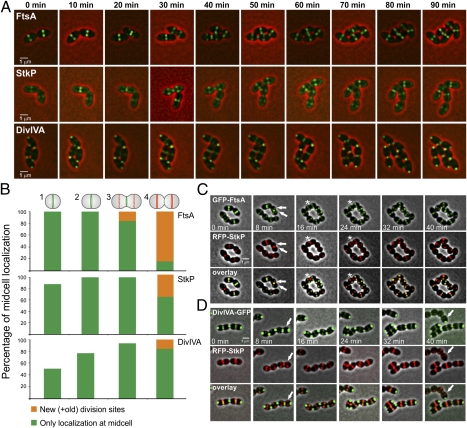
FtsA, together with FtsZ, is one of the earliest proteins to localize at the division sites, whereas DivIVA arrives somewhat later (43, 44). Interestingly, we now have shown that StkP phosphorylates DivIVA and FtsA in vitro (Fig. 1 D and E). How are we to reconcile these biochemical data with the previously observed temporal localization patterns? To establish when StkP arrives at the cell division sites relative to FtsA and DivIVA, we performed time-lapse and time-series microscopy using GFP fusions to FtsA, StkP, and DivIVA. For time-lapse microscopy, cells first were grown to midexponential phase and then were transferred to a microscope slide containing medium with 1.5% agarose and inducer (see Materials and Methods for more details). Images were acquired every 10 min. As shown in Fig. 5A and Movies S1 and S2, GFP-FtsA localizes to midcell early in the pneumococcal cell cycle, whereas GFP-StkP arrives somewhat later. The DivIVA-GFP fusion clearly has the latest arrival time and remains polarly localized (Fig. 5A and Movie S3).
Fig. 5.
Temporal hierarchy in the assembly of the pneumococcal divisome. (A) Fluorescence time-lapse microscopy of strains KB02-62 (PZn-gfp-ftsA), KB02-20 (PZn-gfp-stkP), and KB02-63 (PZn-divIVA-gfp). Overlays between phase contrast (red) and GFP (green) are shown. Stills are from Movies S1, S2, and S3. An arbitrary time point within the time-lapse experiment was chosen and set to T = 0. (Scale bar, 1 μm.) (B) Time-series analysis of the abovementioned strains. Cells were grown in C+Y medium and sorted into four distinct cell types that can be distinguished easily based on membrane staining. At least 500 cells were scored, and the proportion of cells displaying each pattern was plotted. (C and D) Fluorescence time-lapse microscopy of strains HK95 (PZn-gfp-ftsA, PZn-rfp-stkP) and HK96 (PZn-divIVA-gfp, PZn-rfp-stkP). Overlays between phase contrast (gray), GFP (green), and RFP (red) are shown. Stills are from Movies S4 and S5. (Scale bars, 1 μm.) In C, arrows point to cells with FtsA rings at new cell division sites that do not yet contain StkP; asterisks show cells with no FtsA ring at the old septum but in which StkP is still present. In D, arrows point to cells in which StkP is present at new cell division sites but DivIVA is not yet present.
To quantify these arrival and departure times in more detail, we performed time-series microscopy and stained the membranes of the cells with the lipophilic dye Nile red. This analysis shows that within an exponentially growing culture of S. pneumoniae, roughly four categories of cells were distinguished: (i) newly divided short cells of about 1 μm; (ii) slightly elongated cells of about 1.2 μm; (iii) elongated cells of about 1.5 μm; and (iv) diplococci of 2 μm that are just closing the septa. Scoring and assigning midcell localization of GFP-FtsA, GFP-StkP, and DivIVA-GFP to these categories clearly showed different distributions for the three cell-division proteins (Fig. 5B). GFP-FtsA is the first of the three proteins to localize to midcell, and its localization at future division sites can be observed in category 3 cells, but localization at this time is never observed for GFP-StkP and DivIVA-GFP (Fig. 5B). GFP-StkP arrives at midcell somewhat later; among category 4 cells, about 40% show GFP-StkP midcell localization, but only about 10% show DivIVA-GFP midcell localization. These results indicate a temporal order of division-protein assembly. Remarkably, although StkP localizes to midcell relatively early, it remains there for a significantly longer time than FtsA; this observation might explain how StkP can interact with both earlier and later cell-division proteins.
To confirm this quantitative analysis and to visualize StkP with FtsA and DivIVA simultaneously in real time, we constructed double-labeled strains using a red fluorescent protein (RFP) fused to StkP (PZn-gfp-ftsA, PZn-rfp-stkP, and PZn-divIVA-gfp, PZn-rfp-stkP). Fluorescence time-lapse microscopy of these strains again showed that FtsA is the first division protein to arrive at midcell, followed by StkP and subsequently by DivIVA (Fig. 5 C and D and Movies S4 and S5).
Absence of StkP Results in Defects in Growth and Division.
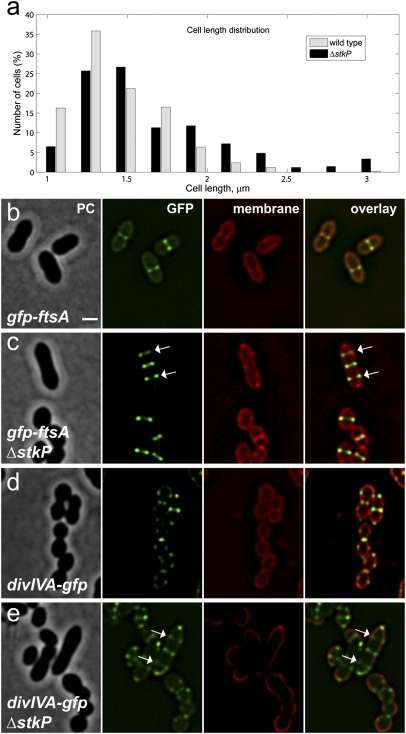
S. pneumoniae cells in which stkP is deleted frequently display elongated morphologies (30, 33). A quantitative cell-length analysis of phase-contrast images of exponentially growing cells indeed shows that ΔstkP mutants are significantly longer than wild-type cells [1.43 ± 0.29 μm for wild type (n = 426 measured cells) vs. 1.66 ± 0.48 μm for ΔstkP (n = 417 measured cells); P < 0.001, Mann–Whitney rank sum test) (Fig. 6A; ± indicates SD). Importantly, the increased cell length is a direct consequence of the lack of phosphorylation of StkP's targets, because overexpression of the PhpP phosphatase in an otherwise wild-type genetic background results in a phenotype comparable to ΔstkP cells [1.59 ± 0.42 μm (n = 183 measured cells), P = 0.172, Mann–Whitney rank sum test). Similar observations were made for S. pneumoniae cells expressing catalytically inactive StkP-K42R (33) (Fig. S10). To investigate the effects of StkP depletion or overexpression on cell morphology, we constructed a complementation strain in which a stkP-null mutant carries a copy of wild-type stkP at the ectopic bgaA locus under the control of the PZn promoter (ΔstkP, PZn-stkP). Wild-type shape was recovered upon induction of stkP expression with 0.25 mM ZnCl2 (Fig. S11 A–D). S. pneumoniae cells overexpressing StkP, in the presence of 0.45 mM ZnCl2, were significantly smaller and rounder (P = 0.0384, Mann–Whitney rank sum test) than wild-type cells grown under the same conditions (Fig. S11 E and F). Cells also were found to be shorter and rounder when GFP-StkP was induced in the absence of PhpP (Fig. S11 G and H). Interestingly, we observed a strong selective pressure against the characteristic elongated phenotype of ΔstkP cells, and putative suppressors rapidly arise in R6 and Rx1 genetic backgrounds, resulting in a pleiotropic array of cell morphologies including chains of cells and round cells (Fig. S12). These morphologies were observed to a lesser extent in the D39 strain, perhaps because the capsule can change and/or mask certain division phenotypes (45).
Fig. 6.
Perturbed assembly of FtsA and DivIVA in the absence of StkP. (A) Increased cell length in ΔstkP cells. The lengths of at least 400 cells of strains R6 (wild type) and KB02-29 (ΔstkP), based on phase-contrast images, were measured automatically using Microbetracker. (B–E) Strains KB02-62 (PZn-gfp-ftsA) (B), KB02-65 (PZn-gfp-ftsA, ΔstkP) (C), KB02-63 (PZn-divIVA-gfp) (D), and KB02-64 (PZn-divIVA-gfp, ΔstkP) (E) were grown in C+Y medium and induced with 0.1 mM ZnSO4. Membranes were stained with Nile red and analyzed by fluorescence microscopy. Arrows indicate elongated cells with aberrant GFP-FtsA or DivIVA-GFP localization. (Scale bar, 1 μm.)
Because StkP phosphorylates DivIVA and, at least in vitro, also FtsZ (34) and FtsA (Fig. 1E), it is tempting to speculate that these cell-division proteins are perturbed in their localization in the absence of StkP. To test this hypothesis, we performed time-lapse fluorescence microscopy of GFP-FtsA and DivIVA-GFP in a ΔstkP mutant background. As shown in Fig. 6 B–E and Movies S6 and S7, elongated cells of the stkP-null mutant frequently display multiple and often unconstricted FtsA and DivIVA rings that never are observed in the wild type. To confirm the effects of the stkP deletion on DivIVA localization in otherwise wild-type cells (i.e., not using GFP reporters), we performed immunofluorescence using specific antibodies raised against S. pneumoniae DivIVA. As shown in Fig. S13, multiple unconstricted bands of DivIVA could be observed in elongated cells.
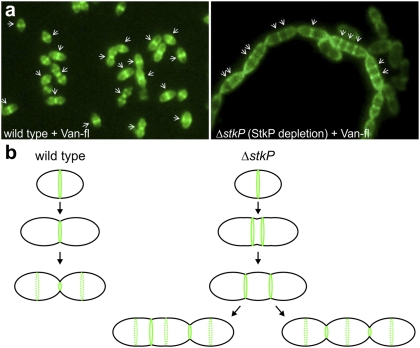
To determine if the cell-division defects observed in elongated ΔstkP cells were the results of unbalanced cell-wall assembly, we took advantage of fluorescently labeled vancomycin (Van-FL) as a probe for nascent sites of PG synthesis (46). Wild-type cells mainly show a midcell staining pattern (i.e., showing current and future division sites) whereas cells in which StkP is depleted (ΔstkP, PZn-stkP grown without zinc) clearly showed an elongated phenotype, with cell-wall synthesis occurring along the peripheral side between the septal zones (Fig. 7A and Fig. S14). Taken together, these results suggest that, in the absence of StkP, cells can start the division process but do not know where and when to divide: Elongation prevails over division, or cell division is less active, thus providing an explanation for the ΔstkP phenotype (Fig. 7B).
Fig. 7.
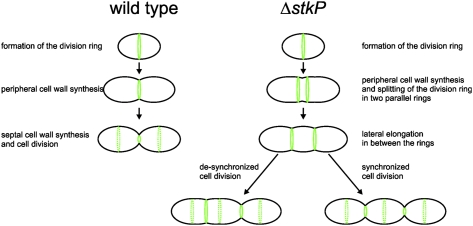
Unbalanced cell growth and division in the absence of StkP. (A) Van-FL staining of the Rx1 wild- type strain (Left) and in cells depleted for StkP (strain Sp38; ΔstkP, PZn-stkP) (Right). Sites of PG insertion are indicated by arrows. Note multiple sites of PG insertion in the ΔstkP mutant, in contrast to the wild-type Rx1 strain, in which PG is inserted only at midcell. (B) Model for StkP function. Cells lacking StkP still are able to assemble a cell-division ring at midcell. However, instead of progressing to septal cell-wall synthesis and constriction, the ring is split into two or more rings, and lateral elongation occurs between the rings. The cells eventually divide, generating three or more daughter cells instead of two.
Discussion
StkP Is Part of the S. pneumoniae Divisome.
Here we show that the Ser/Thr kinase StkP has a crucial role in controlling cell division in S. pneumoniae. Consistent with this role, StkP is recruited to the midcell relatively early in the cell cycle and remains there until division is complete (Fig. 5). Recently, Giefing et al. (34) reported a similar pattern of localization and proposed a model wherein StkP is targeted to cell-division sites via the interaction of the StkP-kinase domain with FtsZ. Although an interaction between StkP and FtsZ is likely, our results indicate that the PASTA domains, rather than the StkP-kinase domains, are responsible for targeting StkP to midcell (Fig. 2). Targeting likely occurs by direct binding to uncross-linked PG (28). Notably, the site of active cell-wall synthesis not only seems to be the marker for StkP's PASTA domains but also is the most probable trigger for StkP autophosphorylation and consequently phosphorylation of well-established and putative cell-division substrates (Figs. 1–4).
Previously it was shown that the kinase domain of StkP, together with the TM domain, was able to dimerize (25). However, the truncated enzyme does not phosphorylate substrate proteins in vivo (33), indicating that dimerization itself is not sufficient for kinase activation and supporting a model of ligand-dependent activation of StkP. As we show in this study, the truncated protein is localized homogenously in the membrane. Thus, it is tempting to speculate that the locally high concentration of StkP at midcell stimulates StkP-dimer formation in a ligand-dependent manner, in turn triggering its autophosphorylation activity. This specific localization of StkP brings it into close contact with its substrates: Once active and present at the division site, StkP will phosphorylate DivIVA and putatively also FtsZ and FtsA (Fig. 1D and refs. 26, 33, 34, 37). The activity of StkP is kept in check by its cognate phosphatase, PhpP, which is present at midcell only when StkP is active (Fig. 3).
StkP Arrives at Midcell After FtsA but Before DivIVA.
As in the model organisms E. coli and B. subtilis, FtsZ and FtsA are the first proteins to arrive at midcell in S. pneumoniae and form the Z-ring before cell division initiates (47, 48). Once formed, the Z-ring recruits the later cell-division proteins such as the membrane connectors, DivIVA, and the enzymes required for septal cell-wall synthesis [PBPs, likely PBP2x and PBP1a (4, 43)]. However, before septation can proceed in oval cells such as S. pneumoniae, peripheral cell-wall synthesis must occur (1). We have shown here that StkP arrives at midcell after FtsA and before DivIVA (Fig. 5), consistent with the observation that new PG insertion at midcell provides the signal recognized by StkP's PASTA domains. This timing of localization resembles the assembly of cell-division proteins by discrete steps that has been shown to occur in the model organisms (44, 49).
StkP Controls Correct Septum Progression and Closure.
In the absence of StkP, cells still are able to divide, although the cell size at which they divide is highly heterogeneous, with cells often elongating before division. These elongated cells contain multiple, unconstricted cell-division rings (Movies S6 and S7 and Fig. S13). Image analysis reveals that ΔstkP cells show a different mode of cell division: After the division ring forms at midcell, instead of constricting at the center, the ring splits, generating two or more rings. New cell-wall material then is inserted in between these rings, resulting in the elongated phenotype. Consistently, vancomycin staining revealed the presence of multiple sites of new cell-wall material along ΔstkP elongated cells, in sharp contrast to wild-type diplococci in which the only site for new cell-wall insertion is at midcell (Fig. 7A and Fig. S14). However, the multiple septa eventually close, generating three or more daughter cells (Fig. 7B and Movies S6 and S7). Together, our data support a model whereby StkP, septally localized through its PASTA domains, is necessary in S. pneumoniae to control septum progression and closure properly.
Although DivIVA has been shown to be the main substrate for StkP phosphorylation in S. pneumoniae (this work and ref. 33), the cell-division phenotype of mutants lacking functional StkP cannot be ascribed solely to the lack of phosphorylation of DivIVA, because a divIVA-null mutant shows a distinguishably different phenotype with rounder and unseparated cells instead of elongated cells (50). Preliminary studies based on the in vivo effects of DivIVA mutant derivatives in key amino acid residues that abolish phosphorylation and/or mimic constitutive phosphorylation agree with this interpretation. Thus, it is more likely that the cell-division defect of ΔstkP is the cumulative effect of the lack of well-timed phosphorylation of a number of proteins involved in growth and division: the GlmM enzyme, which catalyzes one of the first steps of cell-wall biosynthesis, DivIVA, possibly also FtsZ and FtsA, and others with still unknown function (26). Identifying bona fide targets of StkP to unravel the molecular mechanism by which StkP controls cell division remains a major challenge, because in vitro phosphorylation assays do not necessarily correspond to in vivo results. For instance, although StkP efficiently phosphorylates FtsZ in vitro, negligible (if any) differences in phosphorylation patterns were observed using anti-pThr antibodies after immunoprecipitation in stkP mutants (34). However, FtsZ also was shown to be phosphorylated in Corynebacterium glutamicum, Streptomyces coelicolor, and Streptococcus agalactiae (20, 51, 52), suggesting that STK-dependent phosphorylation of FtsZ is widespread in Gram-positive bacteria.
How Is the Oval Shape Maintained During Growth and Division: An Evolutionary Solution Without MreB?
The morphology of the bacterial cell is ultimately the result of a complex interplay of many proteins that make up the cell-wall biosynthetic apparatus. In most rod-shaped bacteria, actin-like MreB proteins control peripheral growth synthesis by linking MreCD with the PBPs specifically involved in lateral elongation (53, 54), whereas the tubulin-like FtsZ protein controls septal cell-wall synthesis (55). However, how peripheral vs. septal growth is controlled in bacteria such as oval cocci, which must elongate before dividing but lack MreB, is still unknown. Perhaps oval cocci, such as S. pneumoniae, that have lost MreB but have retained the PBPs devoted to lateral elongation, RodA and MreC/D (4, 56), take advantage of the cytoskeletal protein FtsZ to fulfill this function. Consistent with this notion, FtsZ has been found to interact directly with PBP2b (i.e., the S. pneumoniae PBP most likely involved in lateral cell-wall elongation) in E. coli two-hybrid assays (57, 58). Interestingly, S. pneumoniae MreC/D recently were shown to direct peripheral PG synthesis (59). Also, Lactococcus lactis, a phylogenetically close relative of S. pneumoniae, has been shown recently to undergo a coccus-to-rod transition as a result of unbalanced activity of the cell-synthesis machineries (60).
We now have shown that, in the absence of StkP, the control between peripheral and septal cell-wall synthesis is lost, giving stkP mutants their elongated shape (Fig. 6) (30, 33). Correct septal progression and closure, driven by rapid disassembly of the Z-ring, is crucial for cell division, and it is this activity that seems most perturbed in the absence of functional StkP. Whether this impairment of septal progression and closure is a consequence of an excessive peripheral cell-wall extension or a block in cell division remains unclear. However, cells overexpressing StkP display a shorter and rounder phenotype (Fig. S11), indicating that in this situation septal cell-wall synthesis outruns peripheral cell-wall synthesis. In this sense, StkP may be seen as a “molecular switch” that, through phosphorylation of key division substrates, signals the shift from peripheral to septal cell-wall synthesis.
Rod-shaped bacteria that must elongate in the absence of MreB, such as Mycobacterium tuberculosis and C. glutamicum, need to do exactly the opposite: They need to make the switch from septal growth to polar growth. Indeed, a phenotypic study of Mycobacterium smegmatis showed that depletion of the essential protein kinase PknB resulted in narrowing and marked elongation of the mycobacterial cells. On the other hand, cells overexpressing PknB were broad and of irregular shape (15). Moreover, a recent study showed that PknB localizes to midcell and to the cell poles and that the PASTA domains are required for its localization (61). These data suggest that mycobacterial PknB has a function similar to that of pneumococcal StkP in coordinating cell morphogenesis during growth and division. The presence of a similar mechanism to control cell division by a eukaryotic-type STK in two unrelated species demonstrates the universality and importance of such signaling systems in bacteria.
Materials and Methods
Strains, Plasmids and Growth Conditions.
S. pneumoniae strains R6, Rx1, and D39 were grown at 30 °C or 37 °C in C+Y or in GM17 medium. Blood agar plates were made from Columbia agar containing 3% defibrinated sheep blood. For induction of PczcD (here noted as PZn), ZnSO4 or ZnCl2 was added to liquid medium and blood agar plates. Competent S. pneumoniae R6, Rx1, and D39 cells were prepared by the addition of competence-inducing peptide CSP-1. Media composition, strains, and plasmids construction are detailed in SI Materials and Methods. Strains and plasmids are listed in Table S1.
Recombinant DNA Techniques, Oligonucleotides, and Western Blotting.
Procedures such as DNA isolation, restriction, ligation, gel electrophoresis, Western blotting, and transformation of E. coli were performed as described in SI Materials and Methods. Oligonucleotides are listed in Table S2.
In Vitro Phosphorylation Analysis.
Expression and purification of recombinant proteins.
The expression strains E. coli BL21 harboring plasmids pETPhos-1511 and pEX-StkP-T (26) were cultivated at 37 °C until midlog phase in LB medium. Expression of His-tagged FtsA or His-tagged StkP-KD (kinase domain) was induced with 1 mM isopropyl thio-β-d-galactoside, and cells were harvested after 3 h. Recombinant proteins were purified at room temperature by Ni-nitrilotriacetic acid metal affinity resin (Qiagen) according to the manufacturer's instructions. Purified proteins were dialyzed against buffer containing 25 mM Tris⋅HCl (pH 7.5), 100 mM NaCl, and 10% (vol/vol) glycerol.
In vitro protein phosphorylation.
The in vitro protein kinase reaction mixture contained 0.4 μg of recombinant substrate protein and 0.4 μg of purified StkP-KD in kinase buffer (25 mM Tris⋅HCl [pH 7.5], 25 mM NaCl, 5 mM MnCl2, 10 μM ATP). The reaction was started by the addition of ATP and was terminated after 15 min of incubation in 37 °C by the addition of 5× SDS sample buffer. Samples were separated by SDS/PAGE and electrotransferred to a PVDF membrane. Proteins were detected either with anti-phosphothreonine polyclonal antibody (Cell Signaling) or by amidoblack staining (Sigma).
Microscopy.
Fluorescence microscopy was performed basically as described (62). Where relevant, Nile red (Invitrogen) was added to a final concentration of 8 ng/mL Phase contrast images were segmented automatically and analyzed using Microbetracker (63), and cell-length distributions were plotted using MATLAB. Immunofluorescence experiments were carried out as previously described (43). Van-FL staining was performed as recently described (58). Time-lapse microscopy was basically performed as described (64). For more details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Harma Karsens for excellent technical support and for the construction of strains HK95 and HK96 and Waldemar Vollmer and Michael B. Whalen for critically reading the manuscript and making useful suggestions. K.B. is supported by an Ubbo Emmius Scholarship from the Graduate School of Science of the University of Groningen. Work in the Laboratory of Cell Signaling (P.B. and L.N.) was supported by Grant Agency of the Academy of Sciences of the Czech Republic Project IAA600200801 (to P.B.), by Czech Science Foundation Grants P302/12/0256 (to L.N.) and P207/12/1568 (to P.B.), by Grant LH 12055 of the Ministry of Education, Youth and Sports of the Czech Republic, and by Institutional Research Concept Grant AV0Z50200510. D.F. was supported by a fellowship funded by the Italian Ministry of Instruction, University and Research (MIUR). Work in the laboratory of O.M. was supported by University of Cagliary Research Funds. Work in the J.-W.V. laboratory is supported by a European Union Marie-Curie Reintegration Fellowship, by a VENI fellowship from the Netherlands Organization for Scientific Research, Earth and Life Sciences (NWO-ALW), by a Sysmo2 Grant from NWO-ALW, and by a Horizon Grant from the Netherlands Organization for Health Research and Development (NWO-ZonMW).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.M. is a guest editor invited by the Editorial Board.
See Author Summary on page 5566 (volume 109, number 15).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119172109/-/DCSupplemental.
References
- 1.Higgins ML, Shockman GD. Model for cell wall growth of Streptococcus faecalis. J Bacteriol. 1970;101:643–648. doi: 10.1128/jb.101.2.643-648.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Higgins ML, Shockman GD. Study of cycle of cell wall assembly in Streptococcus faecalis by three-dimensional reconstructions of thin sections of cells. J Bacteriol. 1976;127:1346–1358. doi: 10.1128/jb.127.3.1346-1358.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomasz A. In: Streptococcus pneumoniae: Molecular Biology and Mechanisms of Disease. Tomasz A, editor. New York: Mary Ann Liebert; 2000. pp. 9–21. [Google Scholar]
- 4.Zapun A, Vernet T, Pinho MG. The different shapes of cocci. FEMS Microbiol Rev. 2008;32:345–360. doi: 10.1111/j.1574-6976.2007.00098.x. [DOI] [PubMed] [Google Scholar]
- 5.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 6.Av-Gay Y, Everett M. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 2000;8:238–244. doi: 10.1016/s0966-842x(00)01734-0. [DOI] [PubMed] [Google Scholar]
- 7.Inouye S, et al. A large family of eukaryotic-like protein Ser/Thr kinases of Myxococcus xanthus, a developmental bacterium. Microb Comp Genomics. 2000;5:103–120. doi: 10.1089/10906590050179783. [DOI] [PubMed] [Google Scholar]
- 8.Bakal CJ, Davies JE. No longer an exclusive club: Eukaryotic signalling domains in bacteria. Trends Cell Biol. 2000;10:32–38. doi: 10.1016/s0962-8924(99)01681-5. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Sun YP, Chen WL, Li JH, Zhang CC. Genomic analysis of protein kinases, protein phosphatases and two-component regulatory systems of the cyanobacterium Anabaena sp. strain PCC 7120. FEMS Microbiol Lett. 2002;217:155–165. doi: 10.1111/j.1574-6968.2002.tb11469.x. [DOI] [PubMed] [Google Scholar]
- 10.Petrícková K, Petrícek M. Eukaryotic-type protein kinases in Streptomyces coelicolor: Variations on a common theme. Microbiology. 2003;149:1609–1621. doi: 10.1099/mic.0.26275-0. [DOI] [PubMed] [Google Scholar]
- 11.Cozzone AJ. Role of protein phosphorylation on serine/threonine and tyrosine in the virulence of bacterial pathogens. J Mol Microbiol Biotechnol. 2005;9:198–213. doi: 10.1159/000089648. [DOI] [PubMed] [Google Scholar]
- 12.Young TA, Delagoutte B, Endrizzi JA, Falick AM, Alber T. Structure of Mycobacterium tuberculosis PknB supports a universal activation mechanism for Ser/Thr protein kinases. Nat Struct Biol. 2003;10:168–174. doi: 10.1038/nsb897. [DOI] [PubMed] [Google Scholar]
- 13.Pereira SF, Goss L, Dworkin J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol Mol Biol Rev. 2011;75:192–212. doi: 10.1128/MMBR.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajagopal L, Clancy A, Rubens CE. A eukaryotic type serine/threonine kinase and phosphatase in Streptococcus agalactiae reversibly phosphorylate an inorganic pyrophosphatase and affect growth, cell segregation, and virulence. J Biol Chem. 2003;278:14429–14441. doi: 10.1074/jbc.M212747200. [DOI] [PubMed] [Google Scholar]
- 15.Kang CM, et al. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: Substrate identification and regulation of cell shape. Genes Dev. 2005;19:1692–1704. doi: 10.1101/gad.1311105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin H, Pancholi V. Identification and biochemical characterization of a eukaryotic-type serine/threonine kinase and its cognate phosphatase in Streptococcus pyogenes: Their biological functions and substrate identification. J Mol Biol. 2006;357:1351–1372. doi: 10.1016/j.jmb.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 17.Fiuza M, et al. From the characterization of the four serine/threonine protein kinases (PknA/B/G/L) of Corynebacterium glutamicum toward the role of PknA and PknB in cell division. J Biol Chem. 2008;283:18099–18112. doi: 10.1074/jbc.M802615200. [DOI] [PubMed] [Google Scholar]
- 18.Beltramini AM, Mukhopadhyay CD, Pancholi V. Modulation of cell wall structure and antimicrobial susceptibility by a Staphylococcus aureus eukaryote-like serine/threonine kinase and phosphatase. Infect Immun. 2009;77:1406–1416. doi: 10.1128/IAI.01499-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banu LD, et al. The Streptococcus mutans serine/threonine kinase, PknB, regulates competence development, bacteriocin production, and cell wall metabolism. Infect Immun. 2010;78:2209–2220. doi: 10.1128/IAI.01167-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silvestroni A, et al. Identification of serine/threonine kinase substrates in the human pathogen group B Streptococcus. J Proteome Res. 2009;8:2563–2574. doi: 10.1021/pr900069n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeats C, Finn RD, Bateman A. The PASTA domain: A beta-lactam-binding domain. Trends Biochem Sci. 2002;27:438. doi: 10.1016/s0968-0004(02)02164-3. [DOI] [PubMed] [Google Scholar]
- 22.Jones G, Dyson P. Evolution of transmembrane protein kinases implicated in coordinating remodeling of gram-positive peptidoglycan: Inside versus outside. J Bacteriol. 2006;188:7470–7476. doi: 10.1128/JB.00800-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee M, et al. Synthetic peptidoglycan motifs for germination of bacterial spores. Chembiochem. 2010;11:2525–2529. doi: 10.1002/cbic.201000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pallová P, Hercík K, Sasková L, Nováková L, Branny P. A eukaryotic-type serine/threonine protein kinase StkP of Streptococcus pneumoniae acts as a dimer in vivo. Biochem Biophys Res Commun. 2007;355:526–530. doi: 10.1016/j.bbrc.2007.01.184. [DOI] [PubMed] [Google Scholar]
- 26.Nováková L, et al. Characterization of a eukaryotic type serine/threonine protein kinase and protein phosphatase of Streptococcus pneumoniae and identification of kinase substrates. FEBS J. 2005;272:1243–1254. doi: 10.1111/j.1742-4658.2005.04560.x. [DOI] [PubMed] [Google Scholar]
- 27.Osaki M, et al. The StkP/PhpP signaling couple in Streptococcus pneumoniae: Cellular organization and physiological characterization. J Bacteriol. 2009;191:4943–4950. doi: 10.1128/JB.00196-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maestro B, et al. Recognition of peptidoglycan and β-lactam antibiotics by the extracellular domain of the Ser/Thr protein kinase StkP from Streptococcus pneumoniae. FEBS Lett. 2011;585:357–363. doi: 10.1016/j.febslet.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Echenique J, Kadioglu A, Romao S, Andrew PW, Trombe MC. Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect Immun. 2004;72:2434–2437. doi: 10.1128/IAI.72.4.2434-2437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giefing C, et al. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J Exp Med. 2008;205:117–131. doi: 10.1084/jem.20071168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasková L, Nováková L, Basler M, Branny P. Eukaryotic-type serine/threonine protein kinase StkP is a global regulator of gene expression in Streptococcus pneumoniae. J Bacteriol. 2007;189:4168–4179. doi: 10.1128/JB.01616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dias R, Félix D, Caniça M, Trombe MC. The highly conserved serine threonine kinase StkP of Streptococcus pneumoniae contributes to penicillin susceptibility independently from genes encoding penicillin-binding proteins. BMC Microbiol. 2009;9:121. doi: 10.1186/1471-2180-9-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nováková L, et al. Identification of multiple substrates of the StkP Ser/Thr protein kinase in Streptococcus pneumoniae. J Bacteriol. 2010;192:3629–3638. doi: 10.1128/JB.01564-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giefing C, Jelencsics KE, Gelbmann D, Senn BM, Nagy E. The pneumococcal eukaryotic-type serine/threonine protein kinase StkP co-localizes with the cell division apparatus and interacts with FtsZ in vitro. Microbiology. 2010;156:1697–1707. doi: 10.1099/mic.0.036335-0. [DOI] [PubMed] [Google Scholar]
- 35.Eberhardt A, Wu LJ, Errington J, Vollmer W, Veening JW. Cellular localization of choline-utilization proteins in Streptococcus pneumoniae using novel fluorescent reporter systems. Mol Microbiol. 2009;74:395–408. doi: 10.1111/j.1365-2958.2009.06872.x. [DOI] [PubMed] [Google Scholar]
- 36.Lanie JA, et al. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol. 2007;189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun X, et al. Phosphoproteomic analysis reveals the multiple roles of phosphorylation in pathogenic bacterium Streptococcus pneumoniae. J Proteome Res. 2010;9:275–282. doi: 10.1021/pr900612v. [DOI] [PubMed] [Google Scholar]
- 38.Ulijasz AT, Falk SP, Weisblum B. Phosphorylation of the RitR DNA-binding domain by a Ser-Thr phosphokinase: Implications for global gene regulation in the streptococci. Mol Microbiol. 2009;71:382–390. doi: 10.1111/j.1365-2958.2008.06532.x. [DOI] [PubMed] [Google Scholar]
- 39.Kahne D, Leimkuhler C, Lu W, Walsh C. Glycopeptide and lipoglycopeptide antibiotics. Chem Rev. 2005;105:425–448. doi: 10.1021/cr030103a. [DOI] [PubMed] [Google Scholar]
- 40.Tipper DJ, Strominger JL. Mechanism of action of penicillins: A proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc Natl Acad Sci USA. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science. 2006;313:89–92. doi: 10.1126/science.1127912. [DOI] [PubMed] [Google Scholar]
- 42.Drlica K. Mechanism of fluoroquinolone action. Curr Opin Microbiol. 1999;2:504–508. doi: 10.1016/s1369-5274(99)00008-9. [DOI] [PubMed] [Google Scholar]
- 43.Fadda D, et al. Streptococcus pneumoniae DivIVA: Localization and interactions in a MinCD-free context. J Bacteriol. 2007;189:1288–1298. doi: 10.1128/JB.01168-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA. Two-step assembly dynamics of the Bacillus subtilis divisome. J Bacteriol. 2009;191:4186–4194. doi: 10.1128/JB.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barendt SM, et al. Influences of capsule on cell shape and chain formation of wild-type and pcsB mutants of serotype 2 Streptococcus pneumoniae. J Bacteriol. 2009;191:3024–3040. doi: 10.1128/JB.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: Two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 47.Morlot C, Zapun A, Dideberg O, Vernet T. Growth and division of Streptococcus pneumoniae: Localization of the high molecular weight penicillin-binding proteins during the cell cycle. Mol Microbiol. 2003;50:845–855. doi: 10.1046/j.1365-2958.2003.03767.x. [DOI] [PubMed] [Google Scholar]
- 48.Lara B, et al. Cell division in cocci: Localization and properties of the Streptococcus pneumoniae FtsA protein. Mol Microbiol. 2005;55:699–711. doi: 10.1111/j.1365-2958.2004.04432.x. [DOI] [PubMed] [Google Scholar]
- 49.Aarsman ME, et al. Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol. 2005;55:1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- 50.Fadda D, et al. Characterization of divIVA and other genes located in the chromosomal region downstream of the dcw cluster in Streptococcus pneumoniae. J Bacteriol. 2003;185:6209–6214. doi: 10.1128/JB.185.20.6209-6214.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schultz C, et al. Genetic and biochemical analysis of the serine/threonine protein kinases PknA, PknB, PknG and PknL of Corynebacterium glutamicum: Evidence for non-essentiality and for phosphorylation of OdhI and FtsZ by multiple kinases. Mol Microbiol. 2009;74:724–741. doi: 10.1111/j.1365-2958.2009.06897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manteca A, Ye J, Sánchez J, Jensen ON. Phosphoproteome analysis of Streptomyces development reveals extensive protein phosphorylation accompanying bacterial differentiation. J Proteome Res. 2011;10:5481–5492. doi: 10.1021/pr200762y. [DOI] [PubMed] [Google Scholar]
- 53.Domínguez-Escobar J, et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 54.Garner EC, et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Margolin W. Sculpting the bacterial cell. Curr Biol. 2009;19:R812–R822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Massidda O, Anderluzzi D, Friedli L, Feger G. Unconventional organization of the division and cell wall gene cluster of Streptococcus pneumoniae. Microbiology. 1998;144:3069–3078. doi: 10.1099/00221287-144-11-3069. [DOI] [PubMed] [Google Scholar]
- 57.Maggi S, et al. Division protein interaction web: Identification of a phylogenetically conserved common interactome between Streptococcus pneumoniae and Escherichia coli. Microbiology. 2008;154:3042–3052. doi: 10.1099/mic.0.2008/018697-0. [DOI] [PubMed] [Google Scholar]
- 58.Albarracín Orio AG, Piñas GE, Cortes PR, Cian MB, Echenique J. Compensatory evolution of pbp mutations restores the fitness cost imposed by β-lactam resistance in Streptococcus pneumoniae. PLoS Pathog. 2011;7:e1002000. doi: 10.1371/journal.ppat.1002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Land AD, Winkler ME. The requirement for pneumococcal MreC and MreD is relieved by inactivation of the gene encoding PBP1a. J Bacteriol. 2011;193:4166–4179. doi: 10.1128/JB.05245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pérez-Núñez D, et al. A new morphogenesis pathway in bacteria: Unbalanced activity of cell wall synthesis machineries leads to coccus-to-rod transition and filamentation in ovococci. Mol Microbiol. 2011;79:759–771. doi: 10.1111/j.1365-2958.2010.07483.x. [DOI] [PubMed] [Google Scholar]
- 61.Mir M, et al. The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog. 2011;7:e1002182. doi: 10.1371/journal.ppat.1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Minnen A, Attaiech L, Thon M, Gruber S, Veening JW. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol Microbiol. 2011;81:676–688. doi: 10.1111/j.1365-2958.2011.07722.x. [DOI] [PubMed] [Google Scholar]
- 63.Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Jong IG, Beilharz K, Kuipers OP, Veening JW. Live cell imaging of Bacillus subtilis and Streptococcus pneumoniae using automated time-lapse microscopy. J Vis Exp. 2011;53:3145. doi: 10.3791/3145. [DOI] [PMC free article] [PubMed] [Google Scholar]