Abstract
Histone lysine methylation is an important component of the epigenetic system demarcating transcriptionally active and inactive chromatin domains. It is of primary importance in understanding how different histone lysine methylation marks and a specific combination of them are read and interpreted by chromatin proteins to regulate gene expression. In this paper, we report that the rice CHD3 protein CHR729 that was required for many aspects of plant development can interact with dimethylated histone H3 lysine 4 (H3K4me2, a mark associated with moderately expressed or repressed genes) and with trimethylated histone H3 lysine 27 (H3K27me3, a mark associated with repressed genes), respectively, through the chromodomains and the plant homeodomain (PHD) finger of the protein. A mutation or down-regulation of the gene provoked a decrease of H3K27me3 and H3K4me3 (a mark associated with active genes). Genome-wide analysis revealed that H3K27me3 and H3K4me3, respectively, were lost from about 56 and 23% of marked loci, which correspond mostly to under-expressed or repressed genes. In the mutant, a higher-than-expected proportion of down-regulated genes lost H3K4me3, among which many encode DNA-binding transcription factors. These results suggest that the rice CHD3 protein is a bifunctional chromatin regulator able to recognize and modulate H3K4 and H3K27 methylation over repressed or tissue-specific genes, which may be associated with regulation of a gene transcription program of plant development.
Keywords: Oryza sativa, epigenomics, epigenome, epigenetics, chromatin remodeling
Dynamic regulation of chromatin structure is of primary importance for modulating genome activities in higher eukaryotes. Histone lysine methylation is recognized as an important epigenetic modification that plays essential roles in chromatin remodeling and gene expression. Histone lysine residues can be mono, di-, and trimethylated, and the different methylation states have distinct function in gene expression (1, 2). For example, trimethylated histone H3 lysine 27 (H3K27me3) is associated with gene silencing and provides cellular memory to maintain gene repression during plant development (3, 4). Trimethylated histone lysine 4 (H3K4me3) is generally correlated with gene activation (5, 6), whereas dimethylated histone lysine 4 (H3K4me2) is suggested to be implicated in fine tuning of tissue-specific gene expression (7, 8).
Histone modification marks are recognized by specific chromatin proteins having activities that remodel chromatin structure to regulate DNA accessibility for transcription and other activities. However, a repertoire of chromatin-remodeling proteins capable of reading and interpreting different epigenomic marks, or a specific combination of them, is not yet established. Chromatin-remodeling complexes are compositionally and functionally diverse, but they all share the presence of a subunit that belongs to the sucrose non-fermenting (SNF2)-like family of ATPases. The chromodomain, helicase/ATPase, and DNA-binding domain (CHD) proteins belong to this protein superfamily (9). Several CHD members have been identified in a variety of higher eukaryotic organisms and are divided into three subfamilies. In addition to the double chromodomains and the helicase/ATPase, CHD3 subfamily proteins contain one or two plant homeodomain (PHD) fingers at the N terminus. CHD3 members in Drosophila and mammalian cells are the central components of the nucleosome remodeling and histone deacetylase complexes regulating transcriptional repression (10). However, CHD3 members have also been shown to be implicated in gene activation (11–14). The Arabidopsis CHD3 protein PICKLE (PKL) was initially found to be a repressor of embryonic traits in seedlings (15). Recent results suggest that this protein may be a transcriptional activator required for expression of many H3K27me3-marked genes (11). The chromatin mechanism of CHD3 proteins in gene regulation remains unclear.
In this work, we studied the developmental and chromatin function of a rice CHD3 protein, namely, CHR729. We show that this protein that is essential for rice plant development not only interacted with H3K4me2 and H3K27me3 but also was required for H3K27me3 and H3K4me3 over a large number of targets that are mostly under-expressed or repressed genes, many of which encode transcription factors. These data provide insights into the mechanisms of recognition and modulation of histone methylation that regulate gene expression in plants.
Results
Loss of CHR729 Affected Many Aspects of Plant Development.
The rice genome contains six CHD-related (CHR) genes (http://www.chromdb.org). Phylogenetic analysis using the full length and the chromodomain sequences revealed that one CHR protein (CHR705) was a member of subfamily I (CHD1) and the other CHR proteins belonged to subfamily II (CHD3) of CHD proteins (Fig. S1). No subfamily III member was found in rice or Arabidopsis. To study the function of rice CHD3 proteins, we analyzed a T-DNA insertion and RNAi plants of CHR702 and CHR729. T-DNA insertion mutation and RNAi of CHR702 did not produce any morphological phenotype. In contrast, the chr729 T-DNA mutation caused a set of morphological and growth defects, including short and narrow leaves, reduced stem elongation, reduced chlorophyll contents, and no secondary panicle branches (Fig. 1 and Table 1). Four independent CHR729 RNAi lines showed similar, albeit less severe, phenotypes (Table 1 and Fig. S2). The pleiotropic phenotype indicated that CHR729 plays an important role in plant development in rice.
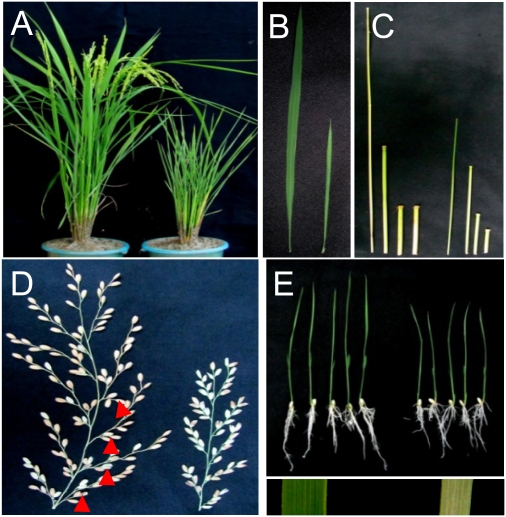
Fig. 1.
The effects of the chr729 mutation on rice plant morphology. (A) Wild type (left) and the T-DNA insertion mutant of chr729 (right) at mature stage. (B) The flag leaf from wild type (left) and chr729 (right). (C) The four uppermost internodes from the main tiller of wild-type (left) and chr729 (right) plants. (D) Panicles from wild type (left) and chr729 (right). Secondary panicle branches indicated by red arrowheads in the wild type are absent from the chr729 panicle. (E) Wild type (left) and chr729 (right) plants at seedling stage (11 d old). Seedling leaf segments are shown at the bottom.
Table 1.
Phenotype comparisons Between chr729 mutant with the wild-type Hwayoung (HY) and Between RNAi lines (21-3-3, 10-17-3, 10-16-3, and 9–2) and the wild type Zhonghua 11 (ZH11)
| Line | No. of panicles | Length of panicle (cm) | No. of PPB | No. of SPB | Plant height (cm) | Length of flag leaf (cm) | Width of flag leaf (mm) | Contents of chlorophyll a (mg/g) |
| HY | 43.1 ± 10.1 | 22.8 ± 1.1 | 10.2 ± 1.3 | 24.6 ± 5.5 | 80.2 ± 2.8 | 35.5 ± 5 | 13.8 ± 1.7 | 3.83 ± 0.24 |
| chr729 | 19.8 ± 4.4** | 15.7 ± 1** | 9 ± 1** | 1.4 ± 1.4** | 45.2 ± 9.4** | 16.8 ± 2.7** | 5.3 ± 0.5** | 2.69 ± 0.19** |
| ZH11 | 10.4 ± 3.8 | 23.1 ± 1.4 | 12.9 ± 2.4 | 18.4 ± 7.1 | 84 ± 5.3 | 32.2 ± 6.8 | 12 ± 2.4 | ND |
| 21–3-3 | 7 ± 1.8** | 18 ± 1.5** | 8.3 ± 1.3** | 10.6 ± 1.5** | 56.4 ± 3.5** | 20.8 ± 2.5** | 6.4 ± 0.6** | ND |
| 10–17-3 | 7 ± 2.5** | 20.7 ± 1.1** | 12.4 ± 1.2 | 12.1 ± 4.5** | 68.5 ± 5** | 28.2 ± 5.5* | 7 0.8 ± 1.2* | ND |
| 10–16-3 | 5.2 ± 1.6** | 19.7 ± 1.7** | 11.9 ± 1.4 | 10.9 ± 2.8** | 58.6 ± 4.4** | 26.3 ± 4.3** | 7.6 ± 0.5* | ND |
| 9–2 | 8.8 ± 4.4 | 21.3 ± 2.3** | 11.5 ± 1.9* | 7.8 ± 5.9** | 62.7 ± 7.3** | 25.7 ± 4** | 7 ± 1.5** | ND |
Data are presented as mean ± SE. Leaf widths were measured through the middle region of the leaves at mature stage. Statistically significant differences from the respective wild type at *P < 0.05 and **P < 0.01 were detected using t tests (n = 20). ND, not determined; PPB, primary panicle branches; SPB, secondary panicle branches.
CHR729 Bound to H3K27me3 and H3K4me2 by the PHD Finger and Chromodomains, Respectively.
CHR729 contains a PHD finger at the N-terminal domain. The PHD finger is a 60-amino-acid module characterized by a conserved C4HC3 sequence and is found in a wide variety of nuclear proteins. The function of the PHD domain of CHD proteins has not been studied. The structure of chromodomain is refined to about 50 amino acids and has been described to recognize different histone modification modules in several chromatin proteins (16). It was not clear whether CHD3 chromodomains bind to histones. The PHD finger and the double chromodomains of CHR729 were produced as GST-fusion proteins in Escherichia coli and incubated with isolated histones (Fig. 2A). Histones that interacted with PHD-GST were eluted and analyzed by Western blots using antibodies against histone H3 and methylated histone lysine modules. Histone H3 was found to be enriched by PHD-GST compared with GST alone. Among the analyzed histone modification modules, only H3K27me3 was found to be clearly enriched by the PHD-GST (Fig. 2B). To refine this observation, biotinylated H3K27me3 and H3K4me2 peptides were incubated with PHD-GST fusion or GST alone. After elution from streptavidine-coupled beads, PHD-GST was found to be retained only by the H3K27me3 peptide (Fig. 2C), confirming the specific interaction between H3K27me3 and the PHD finger. In addition, mutant versions of the PHD finger were generated by PCR-directed mutagenesis. Substitution of the first two cysteine residues of the finger abolished the interaction with H3K27me3, whereas substitution of two nonconserved residues had no clear effect on the binding (Fig. 2D), indicating that the CHR729 PHD finger specifically interacted with H3K27me3.
Fig. 2.
Binding of PHD finger and chromodomains of CHR729 to H3K27me3 and H3K4me2, respectively. (A) Schematic representation of CHR729 protein structure. The amino acid positions of the different motifs are indicated. (Lower) Sequence alignment of different PHD fingers. Amino acid sequence substitutions in mutated versions m1, m2, and m3 of the CHR729 PHD finger are indicated. (B) PHD-binding specificity to methylated H3. Histones were used to interact with GST alone with fusions with PHD (PHD-GST), or with double chromodomains (2Chromo-GST) of CHR729 and analyzed by Western blots using the antibodies indicated on the left. Input histones (10%) were loaded as controls. (C) Analysis of PHD interactions with histone H3K27me3 peptide. GST or PHD-GST proteins were selected by biotinylated H3K27me3 peptide coupled to streptavidin beads and detected by Western blots with anti-GST. (D) Tests of PHD mutants for binding to H3K27me3. The pull-down assays performed as in B were analyzed by SDS/PAGE (Upper) and by Western blots using anti-H3K27me3 (Lower). (E) Preferential interaction of chromodomains with H3K4me2. The histone-binding assays with GST, PHD-GST, PHDm3-GST, and 2Chromo-GST were performed as in B and analyzed by Western blots using the antibodies indicated on the left.
Histone H3 was found to be enriched also by the double chromodomain-GST fusion (Fig. 2B). The above-tested trimethylated lysine modules seemed not to interact with the double chromodomains (Fig. 2B). Tests with antibodies against additional methylated lysine modules revealed that the chromodomains preferentially interacted with H3K4me2 (Fig. 2E). Collectively, the data indicated that CHR729 was associated with H3K27me3 and H3K4me2 through the two different motifs of the protein.
CHR729 Mutation Reduced H3K27me3 and H3K4me3 from Many Target Genes.
To test whether the chr729 mutation affected methylation of H3K27 and H3K4, histones isolated from wild type, the chr729 mutant, and two CHR729 RNAi lines were analyzed by Western blots. H3K27me3 and H3K4me3 showed a decrease in the mutant and RNAi plants compared with wild type, whereas H3K4me2 and H3K4me1 were not clearly changed (Fig. 3A).
Fig. 3.
The effect of the chr729 mutation on H3K4me3 and H3K27me3. (A) Decrease of H3K4me3 and H3K27me3 in the mutant and RNAi lines. Histones isolated from wild type, chr729 mutant, and two RNAi lines were analyzed by Western blots using antibodies of the indicated H3K27 and H3K4 methylation modules and histone H3. Two independent repeats for the mutant are shown. Relative signals (with wild type set at 1) are indicated. (B) Venn diagram representing the number of genes marked by H3K27me3 in wild type (blue) and in chr729 (red) and by H3K4me3 in wild type (green) and chr729 (yellow). (C) Overlaps of genes marked by H3K27me3 and H3K4me3 in wild type (Upper) and chr729 (Lower). (D) Distribution of H3K27me3 and H3K4me3 ChIP-seq reads along 2 kb upstream, gene body and 2-kb downstream regions of 56,797 rice genes in wild type, and chr729 mutant. Each gene was divided into 20 intervals (5% each interval), and the 2-kb regions upstream and downstream of each gene were divided into 100-bp intervals. Transcriptional start site is at the position 0. The y axis is the average read coverage of intervals divided by total reads and by the length of the intervals.
To obtain a high-resolution map of genome-wide distribution of H3K27me3 and H3K4me3 in chr729 versus wild type, chromatin immunoprecipitation (ChIP) coupled with high-throughput sequencing (ChIP-seq) was performed on 11-d-old seedlings. The ChIP-seq data and analysis are presented in Fig. S3. About 6% of annotated genes were found to be modified by H3K27me3 in wild-type rice seedlings (Fig. 3B), which showed a 60% overlap with previously published H3K27me3-marked rice genes and is comparable to the percentage of H3K27me3-marked genes in Arabidopsis (3, 4, 17). In the mutant, 1,932 (about 56%) marked genes lost H3K27me3, whereas 754 genes displayed ectopic H3K27me3 (Fig. 3B). ChIP-PCR verifications of 12 genes confirmed the ChIP-seq data (Fig. S4).
About 37.3% (21,166 of 56,797) of the genes were marked with H3K4me3 in wild-type rice seedlings (Fig. 3B), which indicates a 78% overlap with previously published H3K4me3-marked rice genes and is also comparable to the percentage of H3K4me3-marked genes in Arabidopsis (6, 17). In chr729, H3K4me3 was lost from about 23% (4,884 of 21,166) of marked genes. In addition, 724 genes gained ectopic H3K4me3 in chr729. Validation by ChIP-PCR with a selection of 14 genes confirmed the ChIP-seq data (Fig. S5).
In wild type, a set of 1,125 genes were found to be comodified with both H3K27me3 and H3K4me3 (Fig. 3C). Because the starting materials for ChIP-seq were seedlings, the number of comodified genes most likely represents an overestimate, as some genes might be marked with H3K4me3 in one cell type/tissue and modified by H3K27me3 in another. Due to the decrease of both marks, a smaller number of genes were found to be comarked in chr729. H3K27me3 and H3K4me3 are suggested to be mutually repulsive and antagonist marks (6, 8, 18). However, gain or loss of H3K27me3 did not generally influence H3K4me3, and vice versa (Table S1), suggesting that CHR729 function in H3K4me3 and H3K27me3 might be independent.
H3K27me3 was found to be distributed all over the gene bodies, whereas H3K4me3 was found highly enriched in the 5′ of the genes (Fig. 3D), corroborating previous data in Arabidopsis and rice. The chr729 mutation reduced H3K4me3 preferentially at the 5′ end of the genes and H3K27me3 all over the gene body (Fig. 3D).
Higher Proportion of Down-Regulated Genes Lost H3K4me3 in chr729.
Analysis of chr729 transcripts compared with wild type revealed that 345 and 253 genes, respectively, were down- and up-regulated more than two-fold (q-value ≤0.05) in 11-d-old seedlings (Fig. 4). About 9.6% of down-regulated and 7.1% of up-regulated genes were marked by H3K27me3, which were higher than the overall rate of H3K27me3-marked genes in chr729. About half of the marked genes lost H3K27me3 in both up-regulated and down-regulated groups, which was close to the overall H3K27me3 loss rate in the mutant, indicating that there was no clear correlation between H3K27me3 loss and gene up-regulation in chr729. Loss of H3K27me3 in chr729 might be not sufficient for gene activation, as observed for most H3K27me3-marked genes that are not activated upon loss of H3K27me3 in Arabidopsis mutants (18, 19).
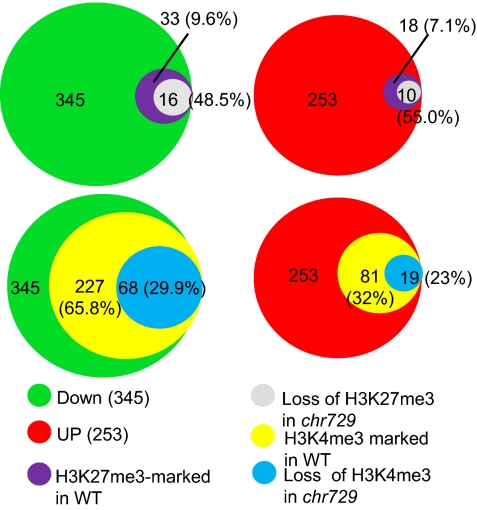
Fig. 4.
Proportions of down-regulated genes that lost H3K4me3 in chr729. (Upper) Numbers of genes marked by H3K27me3 in wild type and numbers of down- and up-regulated genes that lost H3K27me3 in chr729. (Lower) Numbers of genes marked by H3K4me3 in wild type and numbers of down- and up-regulated genes that lost H3K4me3 in chr729.
Among the 345 down-regulated genes, 277 (65.8%) were marked with H3K4me3, much higher than the overall rate. In contrast, a much lower percentage (32%) of up-regulated genes was marked by H3K4me3 (Fig. 4). About 30% (68) of down-regulated genes lost H3K4me3, higher than the overall loss rate of this mark in chr729 and that in up-regulated genes, which were about 23%. This analysis revealed some correlation between H3K4me3 loss and gene down-regulation in chr729.
Many Transcription Factor Genes Were Down-Regulated in chr729.
Among the down-regulated genes, about 15% (52) were DNA-binding transcription factor genes (Table S2). This percentage was about three to four times that of up-regulated transcription factor genes and that of overall transcription factor genes in the rice genome (Table S2) (20). Quantitative PCR validation confirmed the down-regulation of the AP2-domain protein genes (Fig. S6). Many of the transcription factor genes have been shown to be involved in signaling and plant development in rice (Table S2). The pleiotropic phenotype of the mutant and the down-regulation of a relatively large number of transcription factor genes suggest that CHR729 may be involved primarily in gene expression programs of plant growth and development control.
Loss of H3K4me3 in chr729 Mostly Affects Under-Expressed or Repressed Genes.
Comparison of the microarray and the ChIP-seq data, which were generated from rice seedlings grown and harvested under the same conditions, revealed that more than 70% of the H3K27me3-marked genes were not expressed (Fig. 5). Conversely, almost 70% of the H3K4me3-marked genes were expressed. These data supported the notion that H3K4me3 is generally associated with gene activation, whereas H3K27me3 is mostly correlated to gene repression. Genes that lost H3K27me3 in chr729 displayed a similar distribution of expression levels as that of H3K27me3-marked genes in wild type (Fig. 5). However, a higher percentage of repressed or under-expressed genes were found to lose H3K4me3 in chr729 compared with wild type (Fig. 5). This analysis revealed that CHR729 modulates H3K4me3 and H3K27me3 mostly on repressed or under-expressed genes.
Fig. 5.
Expression levels of H3K27me3- and H3K4me3-marked genes in wild type and chr729. Percentages of H3K27me3- and H3K4me3-marked genes in wild type and those of genes that lost H3K27me3 or H3K4me3 (y axis) in chr729 with different expression levels revealed by microarray analysis (x axis) are shown.
Discussion
Chromatin Function of CHR729.
In this work, we have provided evidence that CHR729 is a bifunctional chromatin protein that can recognize and modulate both activating and repressive histone methylation marks. A subset of PHD fingers from diverse proteins have been shown to bind to H3K4me3 or to unmethylated H3K4 (H3K4me0) (21). The present data show that the single PHD finger of CHR729 interacts instead with H3K27me3. The difference may be due to divergent amino acid sequences among the different PHD motifs (21). It is suggested that PHD fingers have a sophisticated histone sequence reading capacity that may be modulated by the interplay between different histone modifications (21). Therefore, the PHD modules may have functional versatility in epigenome reading to control genome activity and gene expression.
Chromodomains are found in a limited number of proteins, including HETEROCHROMATIN PROTEIN1 (HP1) and LIKE HETEROCHROMATIN PROTEIN1 (LHP1) in addition to the CHD proteins. HP1 chromodomain has been shown to bind to H3K9me2/me3 to induce heterochromatin formation in animal cells (22). However, Arabidopsis LHP1 specifically associates with H3K27me3-marked genes (3). CHD1 proteins associate with active loci and are suspected to function during transcriptional elongation (23, 24). Accordingly, the double chromodomains of mammalian CHD1 proteins interact with di- and trimethylated H3K4 (23, 25). Although CHD3 proteins are primarily identified in transcriptional repressive complexes, the present data suggest that this CHD3 protein may be also involved in gene activation, corroborating previous data showing that CHD3 members also have transcription activation function (11–14).
Polycomb group (PcG)-mediated H3K27me3 is a repressive mark that provides a cellular memory allowing the cell to maintain repressed transcriptional states of target genes. In Arabidopsis, most of the H3K27me3 mark is recognized by LHP1, which may be involved in maintaining the repressive chromatin state of the target genes (3, 26, 27). Loss of function of either PcG proteins or LHP1 impairs gene expression programs and leads to severe developmental defects (28, 29). The present data indicate that H3K27me3 can be recognized by additional chromatin proteins, suggesting that there may be distinct mechanisms of H3K27me3-mediated gene repression. Like lhp1 or PcG gene mutants, CHR729 knockdown and knockout plants display a set of severe developmental defects. Unlike the lhp1 mutation that does not affect H3K27me3 genome-wide (3), the chr729 mutation leads to loss of H3K27me3 from more than 50% of the target genes, indicating that CHR729 and LHP1 may play different roles in H3K27 trimethylation and H3K27me3-mediated gene repression. Previous data show that double mutations of Arabidopsis PKL and PKL-RELATED2 (PKR2) lead to a decrease of H3K27me3 in roots (11). The decrease seems to result from an indirect effect of the mutations and is attributed to the repression of PcG genes in the mutants (11). The expression of the six rice PRC2 genes was not clearly affected in chr729 (30) (Fig. S7). The decrease of H3K27me3 in chr729 may be regulated at a different level. Because loss of H3K27me3 in chr729 is not generally correlated with gene up-regulation, loss of CHR729-dependent H3K27me3 may not be sufficient for gene activation in chr729, as observed for most H3K27me3 target genes in mutants of Arabidopsis PRC2 genes (18, 19). Additional activation signals or activators may be required.
The preferential colocalization of H3K4me3 and H3K27me3 does not appear to occur at a genome-wide level in Arabidopsis (6). The present data suggest also a low frequency of comarking of H3K4me3 and H3K27me3 over rice genes (Fig. 3C). Moreover, recent studies suggest that the two marks are mutually repulsive (18). However, H3K4me2 and H3K27me3 appear to colocalize at a higher-than-expected frequency in Arabidopsis (6). Recent analysis indicates that the majority of H3K4me2 is associated with H3K4me3 within the so-called gene-activating chromatin signature 1 (CS1), and a significant proportion of H3K4me2 is found in combination with H3K27me3 within chromatin signature 2 (CS2), which is mainly associated with repressed or under-expressed genes (8). Because CHR729 can recognize both H3K4me2 and H3K27me3, we speculate that CHR729 may preferentially target genes with CS2 in rice. The observation that a higher proportion of repressed or under-expressed genes showed loss of H3K4me3 in chr729 (Fig. 5) favors the hypothesis that CHR729 may be mostly involved in CS2-associated gene expression. Because the chr729 mutation led to loss of H3K4me3 from about 23% of the target genes and many down-regulated genes showed reduced H3K4me3, it can be suggested that binding of H3K4me2 by CHR729 may recruit activity for H3K4 trimethylation during the target gene activation. Alternatively, CHR729 may activate transcription of a set of genes, resulting in an increase of H3K4me3 over the targets, as recent results suggest that increased H3K4me3 may serve to mark activated genes rather than to activate gene transcription (31).
Developmental Function of Plant CHD3 Proteins.
The observation that the chr729 mutation preferentially affected the expression of transcription factor genes suggests that CHR729 may be a higher hierarchical regulator of transcriptional cascades in plant developmental regulation. However, mutation of the Arabidopsis homolog PICKLE-RELATED 1 (PKR1) does not induce any morphological phenotype (11). In addition, a significant overlap of genes with reduced expression in pkl mutants and genes marked by H3K27me3 was not observed in chr729, as about the same percentages of H3K27-marked genes were up- or down-regulated in the mutant (Fig. 4). Phenotypic and transcriptomic data suggest that CHR729 is unlikely to play a similar role as PKL/PKR2 in rice. Collectively, the developmental function of CHD proteins may not be conserved between rice and Arabidopsis. Although CHD3 proteins are generally considered as transcriptional repressors, CHD3 proteins including Arabidopsis PKL also show transcriptional activation function. Because CHR729 can recognize both the activating and the repressing histone methylation marks and is required for H3K4me3 and H3K27me3 over apparently different subsets of genes, it is possible that CHR729 may have both activating and repressive functions on different targets.
Materials and Methods
Plant Materials.
The rice variety Zhonghua 11 (Oryza sativa spp. japonica) was used for transformation in this study. The T-DNA insertion mutant line 3D-02766 of CHR729 and the wild-type variety (Hwayoung) were obtained from the Pohang, South Korea (32). The T-DNA insertion lines of CHR702 were obtained from the Rice Mutant Database. The insertions were confirmed using gene-specific primers and a T-DNA right primer for the Korean mutant and a left primer for Rice Mutant Database mutants (33, 34).
Vector Construction and Rice Transformation.
To construct RNAi vectors, 381 and 414 bp of CHR702 and CHR729 cDNA fragments were amplified respectively from Zhonghua 11 and were inserted into the KpnI and BamH I sites (for the forward insert) and the SacI and SpeI sites (for the reverse insert) of the pDS1301 vector (35). The RNAi constructs were used to transform Zhonghua11 plants using Agrobacterium tumefaciens (strain EHA105).
Chromatin Immunoprecipitation.
The ChIP experiment was essentially performed as described in ref. 36. Two grams of 11-d-old seedlings were harvested and crosslinked in 1% formaldehyde under vacuum. Chromatin was extracted and fragmented to 200–2,000 bp by sonication, and ChIP was performed using two antibodies: H3K4me3 (Millipore; DAM1731494) and H3K27me3 (Millipore; DAM166204). The precipitated and input DNAs were then analyzed by real-time PCR with gene-specific primers.
ChIP-seq and Microarray Analysis.
The high-throughput data analysis is described in SI Materials and Methods. The ChIP-seq data from this publication have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE30490). The microarray data from this publication have been deposited in the GEO database (accession no. GSE25073; http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE25073).
Supplementary Material
Acknowledgments
We thank Prof. Caiguo Xu for culturing the rice plants and Prof. Xianghua Li for technical management. This work was supported by the plant transgenic program of the Chinese Ministry of Agriculture (2009ZX08009-078B); the “863” projects of the Ministry of Science and Technology (2012AA10A303 and 2012AA10A304); National Natural Science Foundation of China 30921091; and the French Agence Nationale de la Recherche (program “blanc” BLAN08-3_330480).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE30490 and GSE25073).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203148109/-/DCSupplemental.
References
- 1.Mosammaparast N, Shi Y. Reversal of histone methylation: Biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 2.Liu C, Lu F, Cui X, Cao X. Histone methylation in higher plants. Annu Rev Plant Biol. 2010;61:395–420. doi: 10.1146/annurev.arplant.043008.091939. [DOI] [PubMed] [Google Scholar]
- 3.Turck F, et al. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:e86. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X, et al. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell. 2008;20(2):259–276. doi: 10.1105/tpc.107.056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pekowska A, Benoukraf T, Ferrier P, Spicuglia S. A unique H3K4me2 profile marks tissue-specific gene regulation. Genome Res. 2010;20:1493–1502. doi: 10.1101/gr.109389.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roudier F, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lusser A, Kadonaga JT. Chromatin remodeling by ATP-dependent molecular machines. Bioessays. 2003;25:1192–1200. doi: 10.1002/bies.10359. [DOI] [PubMed] [Google Scholar]
- 10.Bowen NJ, Fujita N, Kajita M, Wade PA. Mi-2/NuRD: Multiple complexes for many purposes. Biochim Biophys Acta. 2004;1667(1-3):52–57. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Aichinger E, et al. CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 2009;5:e1000605. doi: 10.1371/journal.pgen.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murawska M, et al. dCHD3, a novel ATP-dependent chromatin remodeler associated with sites of active transcription. Mol Cell Biol. 2008;28:2745–2757. doi: 10.1128/MCB.01839-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimono Y, et al. Mi-2 beta associates with BRG1 and RET finger protein at the distinct regions with transcriptional activating and repressing abilities. J Biol Chem. 2003;278:51638–51645. doi: 10.1074/jbc.M309198200. [DOI] [PubMed] [Google Scholar]
- 14.Srinivasan S, et al. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development. 2005;132:1623–1635. doi: 10.1242/dev.01713. [DOI] [PubMed] [Google Scholar]
- 15.Ogas J, Kaufmann S, Henderson J, Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:13839–13844. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yap KL, Zhou MM. Structure and mechanisms of lysine methylation recognition by the chromodomain in gene transcription. Biochemistry. 2011;50:1966–1980. doi: 10.1021/bi101885m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He G, et al. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell. 2010;22(1):17–33. doi: 10.1105/tpc.109.072041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouyer D, et al. Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 2011;7:e1002014. doi: 10.1371/journal.pgen.1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinhofer I, Hehenberger E, Roszak P, Hennig L, Köhler C. H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 2010;6:e1001152. doi: 10.1371/journal.pgen.1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goff SA, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296(5565):92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez R, Zhou MM. The PHD finger: A versatile epigenome reader. Trends Biochem Sci. 2011;36:364–372. doi: 10.1016/j.tibs.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vermaak D, Malik HS. Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu Rev Genet. 2009;43:467–492. doi: 10.1146/annurev-genet-102108-134802. [DOI] [PubMed] [Google Scholar]
- 23.Flanagan JF, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438(7071):1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 24.Hall JA, Georgel PT. CHD proteins: A diverse family with strong ties. Biochem Cell Biol. 2007;85:463–476. doi: 10.1139/O07-063. [DOI] [PubMed] [Google Scholar]
- 25.Sims RJ, III, et al. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, et al. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol. 2007;14:869–871. doi: 10.1038/nsmb1283. [DOI] [PubMed] [Google Scholar]
- 27.Hennig L, Derkacheva M. Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet. 2009;25:414–423. doi: 10.1016/j.tig.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Gaudin V, et al. Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development. 2001;128:4847–4858. doi: 10.1242/dev.128.23.4847. [DOI] [PubMed] [Google Scholar]
- 29.Goodrich J, et al. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature. 1997;386(6620):44–51. doi: 10.1038/386044a0. [DOI] [PubMed] [Google Scholar]
- 30.Luo M, Platten D, Chaudhury A, Peacock WJ, Dennis ES. Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol Plant. 2009;2:711–723. doi: 10.1093/mp/ssp036. [DOI] [PubMed] [Google Scholar]
- 31.Hu Y, Shen Y, Conde E Silva N, Zhou DX. The role of histone methylation and H2A.Z occupancy during rapid activation of ethylene responsive genes. PLoS ONE. 2011;6:e28224. doi: 10.1371/journal.pone.0028224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.An S, et al. Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol. 2003;133:2040–2047. doi: 10.1104/pp.103.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C, et al. Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 2003;35:418–427. doi: 10.1046/j.1365-313x.2003.01808.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, et al. RMD: A rice mutant database for functional analysis of the rice genome. Nucleic Acids Res. 2006;34(Database issue):D745–D748. doi: 10.1093/nar/gkj016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu Z, et al. Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 2006;20:1250–1255. doi: 10.1101/gad.1416306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang L, et al. Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 2007;144:1508–1519. doi: 10.1104/pp.107.099473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.