Abstract
By commandeering cellular translation initiation factors, or destroying those dispensable for viral mRNA translation, viruses often suppress host protein synthesis. In contrast, cellular protein synthesis proceeds in human cytomegalovirus (HCMV)-infected cells, forcing viral and cellular mRNAs to compete for limiting translation initiation factors. Curiously, inactivating the host translational repressor 4E-BP1 in HCMV-infected cells stimulates synthesis of the cellular poly(A) binding protein (PABP), significantly increasing PABP abundance. Here, we establish that new PABP synthesis is translationally controlled by the HCMV-encoded UL38 mammalian target of rapamycin complex 1-activator. The 5′ UTR within the mRNA encoding PABP contains a terminal oligopyrimidine (TOP) element found in mRNAs, the translation of which is stimulated in response to mitogenic, growth, and nutritional stimuli, and proteins encoded by TOP-containing mRNAs accumulated in HCMV-infected cells. Furthermore, UL38 expression was necessary and sufficient to regulate expression of a PABP TOP-containing reporter. Remarkably, preventing the rise in PABP abundance by RNAi impaired eIF4E binding to eIF4G, thereby reducing assembly of the multisubunit initiation factor eIF4F, viral protein production, and replication. This finding demonstrates that viruses can increase host translation initiation factor concentration to foster their replication and defines a unique mechanism whereby control of PABP abundance regulates eIF4F assembly.
Keywords: human cytomegalovirus replication, translational control
Among the interactions between viruses and their cellular hosts are those that ensure the protein components required to assemble new infectious progeny will be produced. This process represents a critical step in the productive growth of all viruses, regardless of nucleic acid content or family classification. Despite encoding diverse functions in their genomes, including those capable of template-directed nucleic acid synthesis, viruses remain exquisitely dependent on cellular translation factors and ribosomes to meet their intense protein synthetic needs (1). Given the importance of mRNA translation to their replication, it is surprising that more viruses do not encode translation-factor components to ensure access to this machinery. Instead, viral strategies to access the host translation apparatus have focused on capturing and controlling this machinery, rather than replacing it (1, 2).
Rapid viral replication cycles often inhibit host protein synthesis to short-circuit host defenses and allow intensive viral protein accumulation over limited time. Indeed, poliovirus (PV) proteinases cleave translation initiation factors eIF4G and poly(A) binding protein (PABP) to inhibit cap-dependent mRNA translation (3). Host protein synthesis is suppressed, as host mRNAs are capped and require eIF4G and PABP, whereas uncapped PV mRNA translation proceeds via a cis-acting internal ribosome entry site (4). Other viruses produce capped, polyadenylated mRNAs, including HSV-1, Kaposi's sarcoma-associated herpesvirus (KSHV), and Vaccinia virus (VV), and rely on cellular cap-binding functions to recruit ribosomes. Despite suppressing host protein synthesis, each virus stimulates assembly of the cellular multisubunit, cap-binding translation initiation factor eIF4F (5–7). HSV-1, KSHV, and VV suppress host protein synthesis via global mRNA metabolism changes caused in part by viral factors that accelerate mRNA turnover (8–11). Viral mRNAs remain relatively immune to this by virtue of their sheer abundance. Unlike PV, these viruses do not reduce translation initiation factor levels. Instead, these viruses use existing levels and modulate subunit assembly into functional complexes.
Additional obstacles confront human cytomegalovirus (HCMV) mRNAs because host protein synthesis is not suppressed during the protracted viral lifecycle (12). Viral and host mRNAs coexist and compete for the same machinery. Like other herpesviruses, HCMV stimulates eIF4F assembly (13, 14). However, unlike other viruses, HCMV increases the abundance of select translation initiation factors during its productive replication cycle (14–16). One of these factors, PABP, increases via a rapamycin-sensitive pathway requiring 4E-BP1 inactivation (17). However, the underlying mechanism and its significance to productive viral replication remain unknown. Here, we establish that the HCMV-induced PABP increase was controlled translationally and dependent upon UL38, the virus-encoded mammalian target of rapamycin complex 1 (mTORC1) activator. UL38 expression was necessary and sufficient to regulate expression of a reporter gene containing a 32-nt terminal oligopyrimidine (TOP) element from the PABP mRNA 5′ UTR. We also demonstrate that eEF2 and rpS6, other canonical TOP mRNA-encoded proteins, accumulated in HCMV-infected cells. Surprisingly, virus-induced PABP accumulation was required to stimulate eIF4F assembly viral protein production and replication. This finding represents a unique example of a virus that elevates host translation initiation factor abundance to promote eIF4F assembly and foster its own replication. Moreover, this finding defines a strategy for regulating eIF4F assembly based upon translational control of PABP abundance.
Results
Induction of PABP mRNA Translation in HCMV-Infected Cells Requires the Viral UL38 mTORC1 Activator.
The HCMV-induced increase in PABP abundance was sensitive to the mTORC1-selective inhibitor rapamycin (17). mTORC1 is activated by PI3-kinase signaling and typically is Akt-responsive (reviewed in ref. 18). Indeed, ectopic HCMV immediate-early (IE) protein expression activated Akt in uninfected cells (19). Although published reports agree that HCMV activates Akt, time points beyond 48 h were not investigated and disagreements regarding activation kinetics remain (13, 20). To analyze Akt activation in parallel with PABP accumulation, total protein was isolated from mock or HCMV-infected cells and the overall abundance plus activation state of the indicated proteins evaluated (Fig. 1A). Whereas total Akt levels remained fairly constant following infection, the abundance of phospho-T308 and -S473, markers for Akt activation, increased substantially by 6 h postinfection (hpi) relative to mock-infected cells, declined by 9 hpi in agreement with Kudchodkar et al. (13), and remained undetectable through 72 hpi (Fig. 1A). Notably, elevated PABP levels were evident by 24 hpi, but activated Akt was not detected (Fig. 1A). Thus, either a transient Akt activity burst through 6 hpi was harnessed to activate mTORC1 later in infection, or a different strategy, perhaps involving a virus-encoded gene product, was operative.
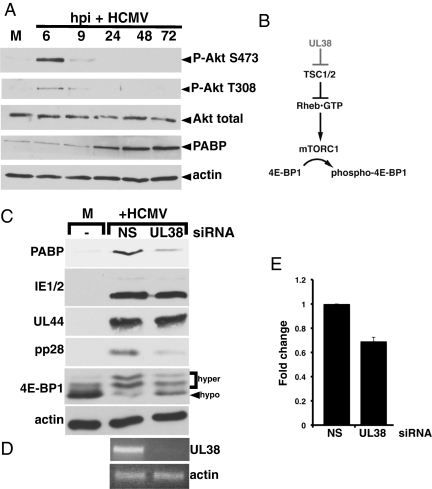
Fig. 1.
Inhibition of UL38 expression reduces PABP accumulation in HCMV-infected cells. (A) Growth-arrested NHDFs were either mock-infected (M) or infected with HCMV (MOI = 5). At the indicated hours postinfection, total protein was isolated, fractionated by SDS/PAGE, and analyzed by immunoblotting using the indicated antibodies. (B) In the absence of Akt activity, mTORC1 is inactive and unable to phosphorylate the translational repressor 4E-BP1 because TSC1/2 inhibits rheb by promoting GTP hydrolysis and rheb•GDP accumulation. By binding to TSC2, UL38 inhibits TSC and allows mTORC1 activation by rheb•GTP (22). (C) NHDFs transiently transfected with control, nonsilencing siRNA (NS), UL38 siRNA (UL38), or no siRNA (−) were either mock-infected (M) or infected with HCMV (MOI = 5). Total protein was harvested at 48 hpi, fractionated by SDS/PAGE, and analyzed by immunoblotting with the indicated antisera (Left). Hyperphosphorylated (hyper) and hypophosphorylated (hypo) 4E-BP1 forms are indicated (Right). (D) Total RNA was isolated at 48 hpi and analyzed by RT-PCR using primers specific for UL38 or actin. (E) As in D, except pp28 mRNA abundance was quantified by real-time PCR and normalized to actin mRNA.
As an early gene, UL38 expression is dependent upon the IE1/2 transactivators (21). Moreover, UL38 expression in uninfected cells prevents mTORC1 inhibition by stress (Fig. 1B) (22). To determine if UL38 was required for PABP accumulation in HCMV-infected cells, total protein was harvested from infected normal human dermal fibroblasts (NHDFs) treated with a nonsilencing control (NS) or UL38-silencing siRNA and analyzed by immunoblotting. Although PABP levels increased in HCMV-infected cultures treated with NS siRNA, UL38 siRNA treatment inhibited the virus-induced rise in PABP abundance in growth-arrested (Fig. 1C) primary cells. Whereas viral IE (IE1/2) and early protein (UL44) levels remained relatively unaffected by the siRNAs, accumulation of pp28, a representative late viral protein, was selectively reduced by the UL38 siRNA. Under these conditions, pp28 mRNA levels only decreased by 30% (Fig. 1E). This finding suggests that UL38-depletion predominately acts posttranscriptionally to regulate late pp28 protein levels and that IE and E protein accumulation proceeds normally. UL38 siRNA also interfered with UL38 protein function by reducing 4E-BP1 hyperphosphorylation (Fig. 1C), in agreement with studies using a UL38-deficient virus (22) and consistent with the partial sensitivity of 4E-BP1 hyperphosphorylation to rapamycin (13, 14). Furthermore, this finding established that partially preventing 4E-BP1 hyperphosphorylation substantially reduced PABP accumulation. Finally, UL38 siRNA, but not NS siRNA, reduced UL38 mRNA abundance to below detection without affecting actin mRNA, establishing the effectiveness and specificity of the UL38 siRNA (Fig. 1D).
To distinguish between effects of UL38 on PABP accumulation vs. synthesis, lysates from metabolically labeled, siRNA-treated cultures were used to immunoprecipitate PABP. Besides altering ongoing protein synthesis, HCMV globally stimulated 35S-amino acid incorporation into protein. Silencing UL38 modestly reduced the intensity with which many proteins in infected cells were radiolabeled, suggesting that its impact was not limited to PABP (Fig. 2 A and B). Whereas new PABP synthesis was stimulated in NS siRNA-treated cultures, UL38 siRNA effectively reduced HCMV-induced new PABP synthesis (Fig. 2B). Thus, the HCMV UL38 gene product was required to stimulate new PABP synthesis in infected cells. This finding suggests that the rapamycin-sensitive nature of HCMV-induced new PABP synthesis can be explained by UL38-mediated mTORC1 activation.
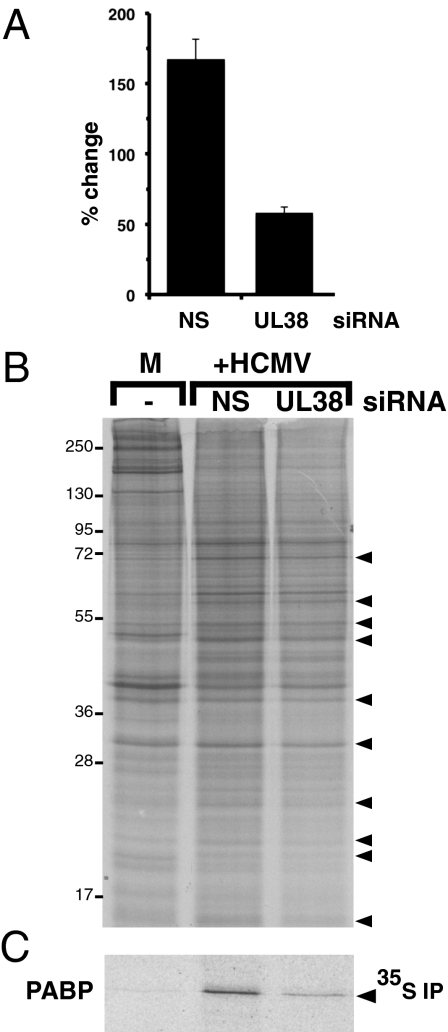
Fig. 2.
UL38 depletion inhibits HCMV-induced PABP synthesis. NHDFs were infected and siRNA-treated as described in Fig. 1. After pulse-labeling with 35S Met/Cys for 1 h at 48 hpi, total protein was isolated and samples were either: (A) TCA-precipitated and acid-insoluble radioactivity quantified by counting in liquid scintillant (the amount of 35S incorporation in mock-infected (M) cultures was normalized to 100%); or (B) fractionated by SDS/PAGE, and the proteins synthesized visualized by autoradiography. Arrowheads to the right indicated protein bands, the intensity of which decreases upon UL38 siRNA treatment. Migration of molecular weight standards (in Kd) is indicated on the left. (C) At 48 hpi, cultures were pulse-labeled with 35S Met/Cys and PABP was immunoprecipitated (IP). Immune complexes were fractionated by SDS/PAGE and visualized by autoradiography.
Accumulation of TOP mRNA-Encoded Proteins in HCMV-Infected Cells.
At least two translational control pathways maintain PABP homeostasis in cells. Not only can a heterotrimeric ribonucleoprotein complex, one component of which is PABP, bind to an adenine-rich 5′ UTR sequence and repress PABP mRNA translation (23), but a 5′ TOP stretch in the PABP mRNA 5′ UTR allows it to behave as a TOP mRNA (24). Translation of TOP mRNAs, which include ribosomal proteins and translation factors, is stimulated by mitogenic and nutritional stimuli, such as insulin (24, 25). To determine if HCMV stimulates TOP mRNA-encoded protein accumulation, total protein was isolated from mock-infected or HCMV-infected cells at various times and analyzed by immunoblotting using antibodies specific for eEF2 and ribosomal protein S6 (rpS6), two canonical TOP mRNA-encoded proteins. Whereas actin levels remained relatively constant between 24 and 48 hpi, eEF2 and rpS6 abundance rose considerably (Fig. 3A). In particular, elevated PABP and eEF2 levels were apparent by 24 hpi and continued to increase, along with rpS6 through 48 hpi. In contrast, their mRNA levels remained relatively unchanged (Fig. 3A). To determine if the increase in eEF2 and rpS6 was UL38-dependent, their overall levels were compared in siRNA-treated cultures. Although PABP, eEF2, and rpS6 all increased upon HCMV infection of NS siRNA-treated cultures, their accumulation was reduced by UL38-depletion (Fig. 3B). Thus, steady-state levels of eEF2 and rpS6, two well-characterized TOP mRNA-encoded proteins, together with PABP, all increased via a posttranscriptional, UL38-dependent manner in HCMV-infected cells. Finally, UL38 siRNA treatment inhibited the rise in PABP abundance induced by HCMV not only in growth-arrested cells (Fig. 1C), but in asynchronously growing primary cells as well (Fig. 3B).
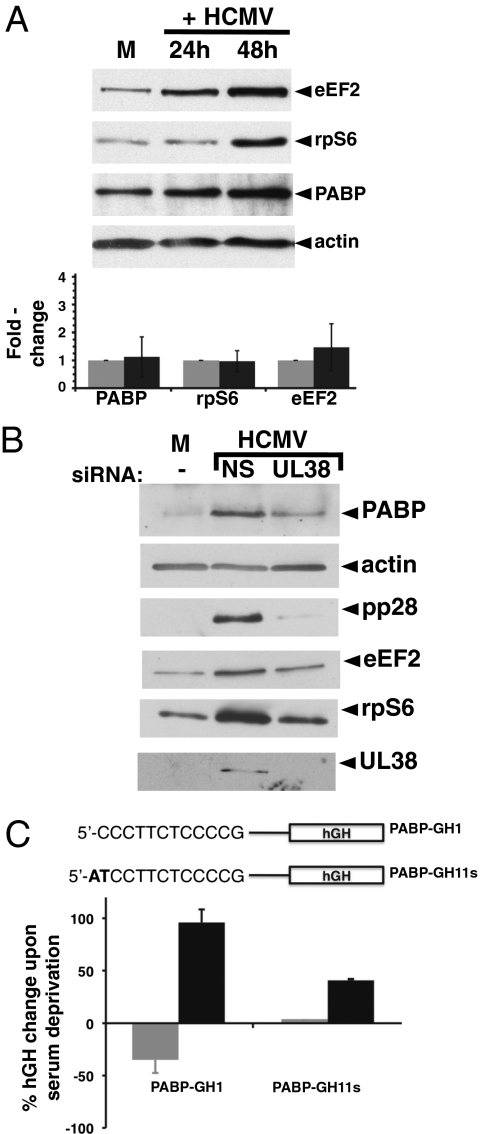
Fig. 3.
TOP mRNA-encoded protein accumulation in HCMV-infected cells and regulation of a PABP TOP-containing reported in uninfected cells are UL38-dependent. (A) (Upper) Growth-arrested NHDFs were either mock-infected (M) or infected with HCMV (MOI = 5). At 24 or 48 hpi, total protein was isolated, fractionated by SDS/PAGE, and analyzed by immunoblotting using the indicated antibodies. (Lower) Total RNA was isolated at 0 hpi (gray bars) or 48 hpi (black bars) and PABP, rpS6, and eEF2 mRNA levels analyzed by real-time RT-PCR (normalized to actin). (B) Asynchronous NHDFs transiently transfected with control, NS siRNA (NS), UL38 siRNA (+), or no siRNA (−) were either mock-infected (M) or infected with HCMV AD169 (MOI = 5). Total protein was harvested at 48 hpi, fractionated by SDS/PAGE, and analyzed by immunoblotting with the indicated antisera (Right). (C) Asynchronous NHDFs in media + 5% FBS were transiently transfected with a functional TOP-containing hGH reporter plasmid (PABP-GH1) or a hGH reporter containing a nonfunctional, mutated TOP element (PABP-GH11s) in the presence of a UL38 or GFP-expression plasmid. (Upper) The 5′ UTR of PABP-GH1 contains the 32 nt TOP element (+1 to +32) upstream of the hGH reporter ORF. PABP-GH11s is identical to GH1 except for the first 2 nt (24). (Lower) Secreted hGH in the media was measured by ELISA at 72 h posttransfection, and the cells subsequently transferred to media containing 0.2% FBS. Secreted hGH was again measured in the same samples 72 h after serum-deprivation. The percent change in hGH levels after serum-deprivation is shown. Gray bars, +GFP; black bars, +UL38.
PABP mRNA is translationally controlled in a growth-dependent manner that requires the TOP element within the first 32 nt of the 5′ UTR (24). To determine if the PABP TOP element was UL38-responsive, the behavior of a human growth hormone (hGH) reporter containing or lacking a functional PABP 32-nt TOP sequence was evaluated in the presence of a control GFP or UL38 expression plasmid following transient transfection into asynchronous NHDFs. Identical reporters, one containing the wild-type PABP 32-nt TOP (PABP-GH1) or a mutant variant (PABP-GH11s) that abrogates TOP translational control, were used to originally define the PABP TOP sequence and its growth-dependent activity (24). Secreted hGH levels were quantified by ELISA 72 h posttransfection, and again from the same sample following an additional 72 h of serum-deprivation. As reported, hGH levels declined by 40% upon serum-deprivation, and this decline was not detected using the PABP-GH11s reporter lacking TOP function (Fig. 3C) (24). UL38 not only suppressed the hGH decline induced by serum-deprivation, but increased secreted hGH levels by ∼100% (Fig. 3C). Thus, UL38 antagonized the suppression of hGH expression induced by growth-arrest, which required the functional PABP 32-nt TOP sequence. Thus, UL38 expression is necessary and sufficient to regulate expression of a PABP TOP reporter. Although the magnitude of the UL38-mediated change was greatest using a TOP reporter, a more modest increase in non-TOP reporter expression was also observed, consistent with a general stimulation of cap-dependent mRNA translation resulting from 4E-BP1 inactivation.
Preventing the Increase in PABP Abundance Inhibits eIF4E Binding to eIF4G, Impairing eIF4F Assembly, Viral Protein Accumulation, and Replication.
Although HCMV stimulated new PABP synthesis via a cellular translational control pathway, the contribution of increased PABP to productive viral replication remained unknown. To determine if increased PABP abundance contributed to HCMV replication, the HCMV-induced PABP increase was prevented using RNAi without reducing PABP levels below the threshold amount in uninfected cells (Fig. 4A). Mock-infected and HCMV-infected growth-arrested (Fig. 4A) or asynchronously growing (Fig. S1A). NHDFs were transfected with NS siRNA or PABP-specific siRNA, and protein accumulation monitored (Fig. 4A). Remarkably, selectively preventing the HCMV-induced PABP increase impaired representative viral protein accumulation (IE1/2, pp28). Indirect immunofluorescence confirmed that all cells showed both reduced PABP and pp28 (Fig. 4B). The pp28 levels were also modestly reduced by interfering with the HCMV-induced accumulation of eIF4A, an eIF4F component. In contrast, silencing DAP5, an eIF4G family member important for cap-independent translation—the abundance of which was increased to a lesser extent by HCMV—did not detectably reduce pp28 accumulation (Fig. 4C).
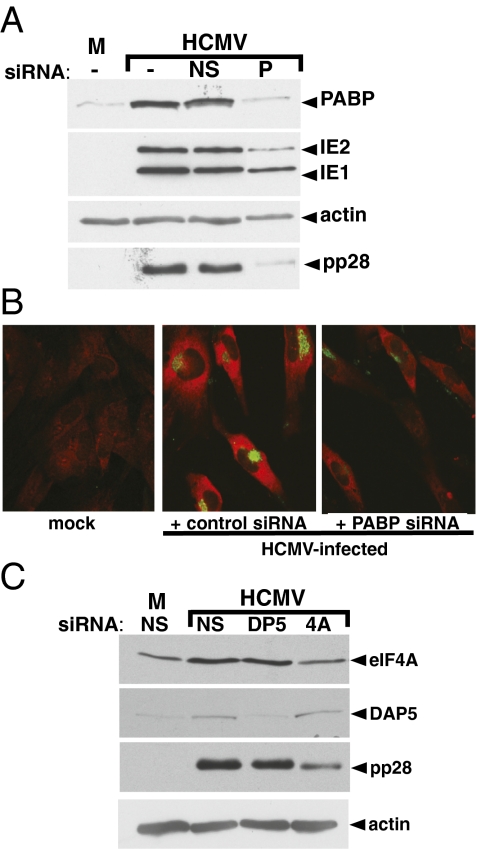
Fig. 4.
Regulation of viral protein accumulation by an HCMV-induced increase in PABP concentration. (A) Precluding the HCMV-induced PABP increase reduces viral polypeptide accumulation. Quiescent NHDFs were transiently transfected with control nonsilencing siRNA (NS), PABP siRNA (P), or no siRNA (−). After 48 h posttransfection, cells were either mock-infected (M) or infected with HCMV (MOI = 5). Total protein was harvested at 48 hpi, fractionated by SDS/PAGE and analyzed by immunoblotting using the indicated antisera. (B) Indirect immunofluorescence of mock or HCMV-infected siRNA-treated cultures. Red, PABP; green, pp28. Images were captured using identical acquisition settings and scaled using the same brightness/contrast settings. (Magnification, 20×.) (C) As in A, using the indicated siRNAs (DP5 = DAP5; 4A = eIF4A) and antisera.
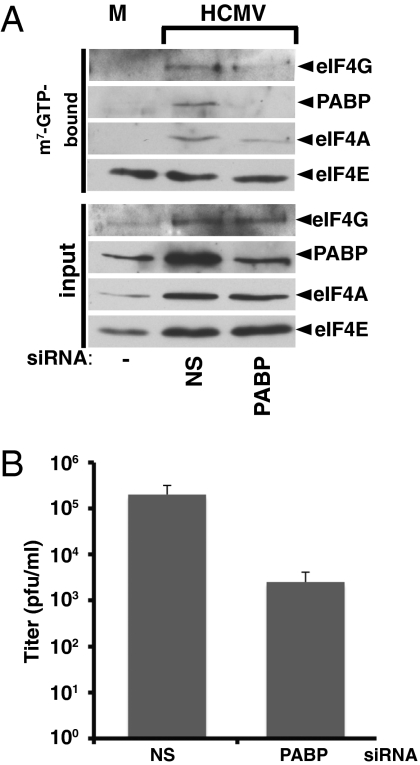
To evaluate if increased PABP concentration contributed to eIF4E, eIF4G, and eIF4A assembly into a multisubunit eIF4F complex, cell-free extracts from mock or HCMV-infected cultures treated with NS or PABP-siRNA were subjected to batch chromatography on methyl-7 (m7) GTP Sepharose. After extensive washing, proteins associated with m7GTP cap-bound eIF4E were analyzed (Fig. 5A). Notably, HCMV stimulated eIF4F assembly, as measured by enhanced retention of eIF4G, eIF4A, and the eIF4F-associated protein PABP on cap-bound eIF4E (Fig. 5A). Interfering with the virus-triggered PABP increase inhibited association of PABP with eIF4F (Fig. 5A) and prevented virus-induced eIF4F assembly, as evidenced by reduced binding of eIF4G and eIF4A to eIF4E (Fig. 5A). Overall, eIF4G, eIF4E, and eIF4A amounts were not detectably reduced by PABP-silencing (Fig. 5A). Finally, preventing the PABP increase reduced viral replication by 80-fold (Fig. 5B). This finding demonstrates that the increased abundance of at least one cellular translation initiation factor (PABP) in HCMV-infected cells is biologically significant and important for virus-induced remodeling of host translation-factor complexes, viral protein accumulation, and replication. Although reducing translation initiation factor levels in virus-infected cells (ie: eIF4G and PABP cleavage by virus-encoded proteases) is a well-accepted translational control strategy (reviewed in refs. 1 and 3), this finding establishes that a virus-triggered increase in host translation factor abundance can contribute to viral replication by regulating eIF4F assembly and viral protein accumulation.
Fig. 5.
Preventing the HCMV-induced increase in PABP concentration inhibits eIF4F assembly and virus replication. (A) At 48 hpi, cell extracts were prepared from NHDFs treated as described in Fig 4. Extract samples (input) and proteins bound to m7GTP-Sepharose 4B (m7GTP-bound) were fractionated by SDS/PAGE, immunoblotted, and visualized with the indicated antibodies. (B) NHDFs treated or untreated with the indicated siRNAs were infected with GFP-expressing wild-type HCMV AD-GFP (MOI = 0.1). After 10 d, cell-free lysates were prepared and infectious virus quantified. NS = control, nonsilencing siRNA. The experiment was performed three times and error bars indicate SEM.
Discussion
In HCMV-infected cells, host protein synthesis is not suppressed, requiring viral and cellular mRNAs to compete for limiting translation initiation factors. Although one of these factors, the host PABP, increases via a rapamycin-sensitive translational control pathway, the underlying mechanism and its significance to the viral lifecycle remained unexplored (17). Here, we establish that the HCMV-induced increase in PABP abundance was dependent upon the UL38 mTORC1 activator. TOP mRNA-encoded proteins eEF2 and rpS6 also accumulated in a UL38-dependent manner. Furthermore, UL38 expression was sufficient to stimulate translation of a PABP TOP-containing reporter in uninfected cells. Finally, preventing the increase in PABP abundance impaired eIF4F assembly, reduced viral protein production, and inhibited replication. This finding demonstrates that eIF4F assembly in HCMV-infected cells requires new PABP synthesis and defines a mechanism for regulating eIF4F based upon translational control of PABP abundance. Thus, although viruses typically do not encode translation initiation factors, they can direct their hosts to increase translation-factor levels to foster their replication.
Although Akt typically activates mTORC1, UL38-dependent PABP accumulation was rapamycin-sensitive and proceeded without detectable Akt activation. Activation of mTORC1 by UL38 involves binding to TSC2 and mTOR activity is required for viral replication (22, 26). By intervening with PI3-kinase signaling at this downstream juncture, S6K1-mediated feedback inhibition of upstream components is precluded and constitutive mTORC1 activation is ensured in infected cells. Although the UL38 protein is nuclear by 8 hpi, it appears in the cytoplasm between 24 and 48 hpi (27), during which time PABP accumulation is evident and suggests that changes in UL38 subcellular distribution play a role in activating PABP mRNA translation. A mutant UL38 protein that only accumulates in nuclei would test this hypothesis.
Like many proteins, UL38 is multifunctional, potentially executing different functions at specific times in discrete subcellular compartments. Besides antagonizing TSC2, UL38 inhibits apoptosis, suppresses endoplasmic reticulum stress-induced cell death, and associates with many cellular proteins (22, 27, 28). Here, we show that the UL38-dependent PABP increase is critical for wild-type levels of HCMV replication, eIF4F assembly, and late protein accumulation. The importance of posttranscriptional and translational control in regulating late HCMV gene expression has long been recognized (29, 30). However, the mechanisms responsible remained obscure. Although UL38-mediated elevation of PABP abundance is important for late viral gene expression, replication, and eIF4F assembly, additional strategies, including interactions between PABP, eIF4A, and pUL69, may also contribute to translational control of gene expression in HCMV-infected cells (31).
Besides inactivating the translational repressor 4E-BP1, viruses whose mRNAs are capped and polyadenylated like their cellular counterparts use diverse mechanisms to regulate eIF4F assembly in infected cells. Replicating in the cytoplasm, poxviruses exploit preexisting eIF4F levels by concentrating eIF4E and eIF4G within replication compartments (6, 32). Overall levels of eIF4F-core and associated components also remain constant in HSV-1–infected cells, where eIF4G directly binds the viral ICP6 protein, which promotes eIF4F assembly (5, 33). A different strategy drives eIF4F assembly in HCMV-infected cells where rising PABP levels, perhaps in conjunction with eIF4G and eIF4E, contribute to eIF4F assembly, stimulating viral protein accumulation and replication. Indeed, PABP-mediated binding of eIF4E and eIF4A to a capped, polyadenylated reporter mRNA is consistent with a direct role for PABP in eIF4F assembly (34). Regulating subunit abundance to control eIF4F assembly is likely not confined to HCMV biology and has important developmental and pathogenic consequences. Discrete mRNAs better compete for limiting initiation factors when factor concentration increases (35–40). Alternatively, PABP overexpression can reduce polysome recruitment and interfere with select mRNA translation (41). Finally, initiation factor levels contribute to gene-expression profiles in human cancers (42). Abortive infection with HCMV, a fairly ubiquitous human pathogen, could contribute to translational disregulation within preneoplastic cells, perhaps by a hit-and-run mechanism described for IE1/2 (43) or a similar strategy involving PABP accumulation stimulated by UL38.
As obligate intracellular parasites, the reliance of viruses on the cell's protein synthesis machinery has defined many translational control paradigms. Cleavage of eIF4G by picornaviral proteinases provides a stunning example of this, reducing the abundance of a full-length initiation factor necessary for cap-dependent host mRNA translation. Similarly, enterovirus proteinases cleave PABP (3). Viral cap-independent mRNA translation subsequently proceeds via an internal ribosome entry site. Thus, viruses can reduce the abundance of full-length cellular translation initiation factors to foster viral mRNA translation and inhibit most host mRNA translation. Other viruses (HSV, KSHV, VV) that produce capped mRNAs require eIF4F to recruit 40S ribosomes and therefore promote eIF4F assembly, but do not alter eIF4E, eIF4G, or PABP abundance (5–7, 32). A more extreme example involves an eIF4E-like molecule encoded by a virus infecting acanthameoba (mimivirus), although its function and relevance to virus biology remains unknown (44). Instead of encoding translation factors, HCMV forces its host to increase the existing supply. To exploit host translational control pathways, activate mTORC1 signaling, and stimulate production of at least one cellular translation initiation factor critical for its replication, HCMV uses the UL38 protein. By increasing the available pool of initiation factor subunits exemplified by PABP, HCMV drives eIF4F assembly in infected cells, enabling wild-type levels of viral protein accumulation and infectious virus production.
Materials and Methods
Cell Culture, Viruses, and Chemicals.
Primary NHDFs (Clonetics) were propagated in 5% (vol/vol) CO2 incubators with DMEM + 5% (vol/vol) FBS or serum-deprived, as previously described (14). HCMV AD169 strain was obtained from ATCC and propagated as previously described (14). The GFP-expressing wild-type virus AD-GFP was a gift from D. Yu (Washington University, St. Louis, MO) (27). Antibodies and chemical inhibitors were described previously (5, 6, 14), except for the following antibodies from Cell Signaling: eEF2 (# 2332), S6 (# 2217), S6K1/2 (# 9430), Akt (#9272), Akt-pSer473 (#9271s), Akt-pThr 308 (#9275s), DAP5 (#2182).
Metabolic Labeling, Protein Half-Life Determinations, RNA Isolation, Real-Time PCR, m7GTP-Sepharose Chromatography, Immunoprecipitation, Immunofluorescence Microscopy, and Immunoblotting.
These procedures were performed as previously described (14, 17).
RNA Interference.
The siRNA duplexes targeting PABP1 (sense target 5′-GUGCUUCACCGAAGAAAAA-3′), eIF4A (sense target 5′-GGGAUGGACAUCUUGUCAU-3′), DAP5 (45), and control NS duplexes were composed of synthetic (Qiagen) 21-nt complementary RNAs with 2-nt overhangs (gifts from F. Ramirez-Valle and R. Schneider, New York University School of Medicine, New York). To prevent the HCMV-induced PABP increase, confluent NHDFs in a 12-well plate were serum-starved in DMEM + 0.2% FBS for 24 h and transfected with 2 μL oligofectamine plus 100 nM siRNA per well in opti-MEM, according to the manufacturer's instructions (Invitrogen). After 4 h, 0.4% FBS + DMEM was added for a final FBS concentration of 0.2%. After 24 h, the procedure was repeated, and the cultures infected with HCMV [multiplicity of infection (MOI) = 3–5). Total protein was harvested at the specified time and protein abundance evaluated by immunoblotting. To quantify viral replication, NHDFs (70–80% confluent in DMEM + 5% FBS) on a 12-well plate were transfected with 2 μL RNAi Max + 50 nM siRNA per well according to the manufacturer's instructions (Invitrogen). After 48 h, cells were infected (MOI = 0.1) with AD-GFP (27), lysed after 10 d by freeze-thawing twice, and viral progeny production determined by TCID50, as previously described (13). To interfere with UL38 expression, siRNA transfections were performed as reported (46) and cultures infected with HCMV (MOI = 5). UL38 (sense target: 5′-AGATGGTGGTCGTGGTCTA-3′) and control NS siRNAs were composed of synthetic 21-nt complementary RNAs with 2-nt overhangs (Dharmacon).
TOP-Dependent hGH Reporter Assay.
NHDFs grown to ∼80% confluence in DMEM + 5% FBS on a 12-well plate were cotransfected with 0.5 μg of a UL38 or GFP expression plasmid (provided by N. Moorman and T. Shenk, Princeton University, Princeton, NJ) together with 0.5 μg of an hGH reporter plasmid PABP-GH1 or PABP-GH11s (gift from O. Meyuhas, Hebrew University-Hadassah Medical School, Jerusalem, Israel) (24) using TransIT-2020 (Mirus Bio) per the manufacturer's instructions. After 72 h, the DMEM + 5% FBS media was harvested and replaced with DMEM + 0.2% FBS. Media was harvested a second time following an additional 72 h in 0.2% FBS. Secreted hGH present in the media of samples in 5% FBS and 0.2% FBS was quantified by ELISA (Cat KAQ1081; Invitrogen).
Supplementary Material
Acknowledgments
We thank O. Meyuhas, F. Ramirez, S. Morley, R. Schneider, N. Moorman, D. Yu, and T. Shenk for helpful reagents; C. Arias, A. Wilson, D. Walsh, R. Cuesta-Sanchez, and R. Schneider for many useful discussions and their critical review of the manuscript; and Tobi Maguire for helping us perfect the TCID50 assays. This work was supported by National Institutes of Health (NIH) Grants AI073898 and GM056927 (to I.M.); an American Society for Microbiology Watkins fellowship (to C.P.); and NIH training Grant T32 AI007647 (to C.P. and C.M.). I.M. was a scholar of the Hirshl Trust. Purchase of the confocal microscope was funded by NIH shared instrumentation Grant S10 RR017970.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202829109/-/DCSupplemental.
References
- 1.Walsh D, Mohr I. Viral subversion of the host protein synthesis machinery. Nat Rev Microbiol. 2011;9:860–875. doi: 10.1038/nrmicro2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchkovich NJ, Yu Y, Zampieri CA, Alwine JC. The TORrid affairs of viruses: Effects of mammalian DNA viruses on the PI3K-Akt-mTOR signalling pathway. Nat Rev Microbiol. 2008;6:266–275. doi: 10.1038/nrmicro1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lloyd RE. Translational control by viral proteinases. Virus Res. 2006;119:76–88. doi: 10.1016/j.virusres.2005.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarnow P. Viral internal ribosome entry site elements: Novel ribosome-RNA complexes and roles in viral pathogenesis. J Virol. 2003;77:2801–2806. doi: 10.1128/JVI.77.5.2801-2806.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh D, Mohr I. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev. 2004;18:660–672. doi: 10.1101/gad.1185304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh D, et al. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol Cell Biol. 2008;28:2648–2658. doi: 10.1128/MCB.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arias C, Walsh D, Harbell J, Wilson AC, Mohr I. Activation of host translational control pathways by a viral developmental switch. PLoS Pathog. 2009;5:e1000334. doi: 10.1371/journal.ppat.1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oroskar AA, Read GS. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J Virol. 1989;63:1897–1906. doi: 10.1128/jvi.63.5.1897-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glaunsinger BA, Ganem DE. Messenger RNA turnover and its regulation in herpesviral infection. Adv Virus Res. 2006;66:337–394. doi: 10.1016/S0065-3527(06)66007-7. [DOI] [PubMed] [Google Scholar]
- 10.Parrish S, Moss B. Characterization of a second vaccinia virus mRNA-decapping enzyme conserved in poxviruses. J Virol. 2007;81:12973–12978. doi: 10.1128/JVI.01668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandri-Goldin RM. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front Biosci. 2008;13:5241–5256. doi: 10.2741/3078. [DOI] [PubMed] [Google Scholar]
- 12.Stinski MF. Synthesis of proteins and glycoproteins in cells infected with human cytomegalovirus. J Virol. 1977;23:751–767. doi: 10.1128/jvi.23.3.751-767.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kudchodkar SB, Yu Y, Maguire TG, Alwine JC. Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J Virol. 2004;78:11030–11039. doi: 10.1128/JVI.78.20.11030-11039.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh D, Perez C, Notary J, Mohr I. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J Virol. 2005;79:8057–8064. doi: 10.1128/JVI.79.13.8057-8064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isler JA, Skalet AH, Alwine JC. Human cytomegalovirus infection activates and regulates the unfolded protein response. J Virol. 2005;79:6890–6899. doi: 10.1128/JVI.79.11.6890-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanton RJ, et al. Cytomegalovirus destruction of focal adhesions revealed in a high-throughput Western blot analysis of cellular protein expression. J Virol. 2007;81:7860–7872. doi: 10.1128/JVI.02247-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez C, McKinney C, Chuluunbaatar U, Mohr I. Translational control of cytoplasmic poly A binding protein (PABP) abundance in HCMV- infected cells. J Virol. 2011;85:156–164. doi: 10.1128/JVI.01778-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 19.Yu Y, Alwine JC. Human cytomegalovirus major immediate-early proteins and simian virus 40 large T antigen can inhibit apoptosis through activation of the phosphatidylinositide 3′-OH kinase pathway and the cellular kinase Akt. J Virol. 2002;76:3731–3738. doi: 10.1128/JVI.76.8.3731-3738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson RA, Wang X, Ma XL, Huong SM, Huang ES. Human cytomegalovirus up-regulates the phosphatidylinositol 3-kinase (PI3-K) pathway: Inhibition of PI3-K activity inhibits viral replication and virus-induced signaling. J Virol. 2001;75:6022–6032. doi: 10.1128/JVI.75.13.6022-6032.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tenney DJ, Colberg-Poley AM. Expression of the human cytomegalovirus UL36-38 immediate early region during permissive infection. Virology. 1991;182:199–210. doi: 10.1016/0042-6822(91)90663-v. [DOI] [PubMed] [Google Scholar]
- 22.Moorman NJ, et al. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe. 2008;3:253–262. doi: 10.1016/j.chom.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J, Bag J. Negative control of the poly(A)-binding protein mRNA translation is mediated by the adenine-rich region of its 5′-untranslated region. J Biol Chem. 1998;273:34535–34542. doi: 10.1074/jbc.273.51.34535. [DOI] [PubMed] [Google Scholar]
- 24.Hornstein E, Git A, Braunstein I, Avni D, Meyuhas O. The expression of poly(A)-binding protein gene is translationally regulated in a growth-dependent fashion through a 5′-terminal oligopyrimidine tract motif. J Biol Chem. 1999;274:1708–1714. doi: 10.1074/jbc.274.3.1708. [DOI] [PubMed] [Google Scholar]
- 25.Hamilton TL, Stoneley M, Spriggs KA, Bushell M. TOPs and their regulation. Biochem Soc Trans. 2006;34:12–16. doi: 10.1042/BST20060012. [DOI] [PubMed] [Google Scholar]
- 26.Moorman NJ, Shenk T. Rapamycin-resistant mTORC1 kinase activity is required for herpesvirus replication. J Virol. 2010;84:5260–5269. doi: 10.1128/JVI.02733-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terhune S, et al. Human cytomegalovirus UL38 protein blocks apoptosis. J Virol. 2007;81:3109–3123. doi: 10.1128/JVI.02124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xuan B, Qian Z, Torigoi E, Yu D. Human cytomegalovirus protein pUL38 induces ATF4 expression, inhibits persistent JNK phosphorylation, and suppresses endoplasmic reticulum stress-induced cell death. J Virol. 2009;83:3463–3474. doi: 10.1128/JVI.02307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geballe AP, Leach FS, Mocarski ES. Regulation of cytomegalovirus late gene expression: Gamma genes are controlled by posttranscriptional events. J Virol. 1986;57:864–874. doi: 10.1128/jvi.57.3.864-874.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerry JA, et al. Translational regulation of the human cytomegalovirus pp28 (UL99) late gene. J Virol. 1997;71:981–987. doi: 10.1128/jvi.71.2.981-987.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aoyagi M, Gaspar M, Shenk TE. Human cytomegalovirus UL69 protein facilitates translation by associating with the mRNA cap-binding complex and excluding 4EBP1. Proc Natl Acad Sci USA. 2010;107:2640–2645. doi: 10.1073/pnas.0914856107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katsafanas GC, Moss B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe. 2007;2:221–228. doi: 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh D, Mohr I. Assembly of an active translation initiation factor complex by a viral protein. Genes Dev. 2006;20:461–472. doi: 10.1101/gad.1375006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lodish HF. Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature. 1974;251:385–388. doi: 10.1038/251385a0. [DOI] [PubMed] [Google Scholar]
- 36.McKeehan WL. Regulation of hemoglobin synthesis. Effect of concentration of messenger ribonucleic acid, ribosome subunits, initiation factors, and salts on ratio of alpha and beta chains synthesized in vitro. J Biol Chem. 1974;249:6517–6526. [PubMed] [Google Scholar]
- 37.Kabat D, Chappell MR. Competition between globin messenger ribonucleic acids for a discriminating initiation factor. J Biol Chem. 1977;252:2684–2690. [PubMed] [Google Scholar]
- 38.Koromilas AE, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 1992;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mamane Y, et al. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS ONE. 2007;2:e242. doi: 10.1371/journal.pone.0000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma S, Bhattacharjee RB, Bag J. Expression of poly(A)-binding protein is upregulated during recovery from heat shock in HeLa cells. FEBS J. 2009;276:552–570. doi: 10.1111/j.1742-4658.2008.06803.x. [DOI] [PubMed] [Google Scholar]
- 41.Wormington M, Searfoss AM, Hurney CA. Overexpression of poly(A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes. EMBO J. 1996;15:900–909. [PMC free article] [PubMed] [Google Scholar]
- 42.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 43.Shen Y, Zhu H, Shenk T. Human cytomagalovirus IE1 and IE2 proteins are mutagenic and mediate “hit-and-run” oncogenic transformation in cooperation with the adenovirus E1A proteins. Proc Natl Acad Sci USA. 1997;94:3341–3345. doi: 10.1073/pnas.94.7.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raoult D, et al. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 45.Ramírez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J Cell Biol. 2008;181:293–307. doi: 10.1083/jcb.200710215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiebusch L, Truss M, Hagemeier C. Inhibition of human cytomegalovirus replication by small interfering RNAs. J Gen Virol. 2004;85:179–184. doi: 10.1099/vir.0.19453-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.