Abstract
The structure and dynamics of bacterial communities in the airways of persons with cystic fibrosis (CF) remain largely unknown. We characterized the bacterial communities in 126 sputum samples representing serial collections spanning 8–9 y from six age-matched male CF patients. Sputum DNA was analyzed by bar-coded pyrosequencing of the V3–V5 hypervariable region of the 16S rRNA gene, defining 662 operational taxonomic units (OTUs) from >633,000 sequences. Bacterial community diversity decreased significantly over time in patients with typically progressive lung disease but remained relatively stable in patients with a mild lung disease phenotype. Antibiotic use, rather than patient age or lung function, was the primary driver of decreasing diversity. Interpatient variability in community structure exceeded intrapatient variability in serial samples. Antibiotic treatment was associated with pronounced shifts in community structure, but communities showed both short- and long-term resilience after antibiotic perturbation. There was a positive correlation between OTU occurrence and relative abundance, with a small number of persistent OTUs accounting for the greatest abundance. Significant changes in community structure, diversity, or total bacterial density at the time of pulmonary exacerbation were not observed. Despite decreasing community diversity in patients with progressive disease, total bacterial density remained relatively stable over time. These findings show the critical relationship between airway bacterial community structure, disease stage, and clinical state at the time of sample collection. These features are the key parameters with which to assess the complex ecology of the CF airway.
Keywords: microbial ecology, microbiome, antibiotics
Chronic infection and inflammation of the airways are the leading causes of morbidity and mortality in cystic fibrosis (CF). Therefore it is reasonable to expect that a better understanding of airway infection in CF could provide opportunities for novel strategies to maintain lung health in this population. Recently, the microbiota of the CF airways has been assessed through the application of culture-independent methods that leverage the phylogenetic information of the bacterial 16S rRNA gene. By using a variety of techniques, including Sanger sequencing of clone libraries (1, 2), terminal restriction fragment length polymorphism analysis (3, 4), microarray hybridization (5, 6), or pyrosequencing (7), these studies have revealed the presence of a far more complex bacterial community in the airways of CF patients than previously appreciated with culture-based methods.
Most of these studies have been cross-sectional, analyzing samples collected at a single time point from individual patients. Correlations between microbial communities and disease progression in cross-sectional studies can be confounded by a host of variables, including patient age, sex, lung disease stage, and antibiotic use. Longitudinal studies that analyze serial samples obtained from individual patients over long periods of time allow better assessment of the impact of these potentially confounding variables in constructing tractable models of the relationship between the dynamics of the lung microbial community and disease progression (4, 8).
In this study, we used a culture-independent approach to investigate intensively the dynamics of the CF airway bacterial communities over the course of 8–9 y in six age- and sex-matched CF patients. The findings reveal a dynamic ecological system that is influenced markedly by a patient's rate of disease progression as well as by disease stage and clinical state at the time of sampling. These features have significant bearing on measures of diversity and community structure, reinforcing the power of longitudinal studies in relating the status of the airway microbial community to CF disease activity.
Results
Patients and Lung Microbiota Measures.
Multiple sputum samples were collected from six adult male patients with CF during the course of 8–9 y. Percent predicted forced expiratory volume in 1 s (%FEV1) was used as a measure of lung function. Three patients (S1, S2, and S3) showed a mild phenotype of lung disease, maintaining relatively stable lung function over several years. The other three patients (P1, P2, and P3) had a more typical course of CF lung disease, exhibiting a progressive decline in lung function during the period of observation. Hereafter these two groups of patients are referred to as “stable” and “progressing” patients, respectively (Table 1 and Fig. 1A).
Table 1.
Study subjects
| Stable patients |
Progressing patients |
|||||
| S1 | S2 | S3 | P1 | P2 | P3 | |
| CFTR genotype | Unknown* | ΔF508/1717–1G > A | ΔF508/ΔF508 | ΔF508/ΔF508 | ΔF508/ΔF508 | ΔF508/ΔF508 |
| Age range | 18.9–28.0 | 21.2–30.1 | 19.8–28.1 | 21.1–29.2 | 18.7–27.3 | 22.0–30.1 |
| Observation period (y)† | 9.1 | 8.9 | 8.3 | 8.1 | 8.6 | 8.1 |
| %FEV1 range | 81–83 | 83–88 | 91–82 | 68–30 | 65–41 | 77–25 |
| Avg. %FEV1 change‡ | 0.11 | 0.56 | −1.11 | −4.69 | −2.79 | −6.42 |
| No. specimens | 23 | 14 | 13 | 20 | 30 | 26 |
*No mutations identified from the 70 mutation sequencing panel, which includes ΔF508. CF diagnosis based on positive sweat chloride test.
†Years between collection dates of first and last sputum samples.
‡Calculated as the difference between %FEV1 values when first and last sputum samples were collected divided by the observation period.
Fig. 1.
Changes in lung function and community diversity. (A) Percent predicted FEV1 values (y axis) are plotted against patient age (x axis). Patients S1, S2, and S3 have relatively stable disease; patients P1, P2, and P3 have progressive disease. Solid circles indicate the ages when the first and last sputum samples were obtained. (B) Temporal changes in community diversity in stable (Upper Row) and progressing (Lower Row) patients. In each plot, community diversity measured by Shannon index is plotted against patient age when sputum was collected. (C) Boxplot comparison of microbial diversity in sputum samples obtained during different BETR clinical states. The top and bottom boundaries of each box indicate 75th and 25th quartile values, respectively, and black lines inside each box represent 50th quartile (median) values. Ends of the whiskers mark the lowest and highest diversity index in each BETR category. The letters above the boxes indicate comparison results in group means; groups with the same letter do not have significant differences in means, but those indicated by different letters are significantly different. (See SI Results for details.)
The bacterial communities in 126 sputum samples obtained from these six patients were profiled using pyrosequencing of the 16S rRNA V3–V5 hypervariable region. A total of 633,746 high-quality reads were generated from these samples with an average of 5,030 reads per sample. From these sequences, 662 operational taxonomic units (OTUs) (based on 97% DNA sequence similarity) were identified. On average, 36 OTUs per sample were detected; however, the range was large (5–114 OTUs per sample). After normalization to the sample with the smallest number of reads (1,152), 259 OTUs were included in further analyses. Among these, 47 OTUs each had a relative abundance of more than 1% in at least one sputum sample (Table S1).
Community Diversity Relative to Disease Progression and Clinical State.
The diversity of the bacterial community, based on calculation of the Shannon index, fluctuated in serial samples from all patients but remained relatively constrained in the three stable patients (S1, S2 and S3). In contrast, serial samples from the three progressing patients (P1, P2 and P3) exhibited statistically significant decreases in community diversity during the course of several years (Fig. 1B).
CF is characterized by periods of relatively stable pulmonary function punctuated by episodic exacerbations of respiratory symptoms and decreased lung function which usually warrant antibiotic treatment. To examine the impact of these changes in clinical state on bacterial diversity, each of the 126 samples was assigned to one of four “BETR” categories: baseline (B); pulmonary exacerbation before initiation of episodic antibiotic therapy (E); antibiotic treatment of pulmonary exacerbation (T); and recovering from pulmonary exacerbation (R) (Table 2). Eight samples with insufficient medical record information were labeled as “unknown” (U). Not unexpectedly, a comparison of the diversity of bacterial communities revealed that treatment samples, collected when patients were receiving antibiotics for exacerbation, were significantly less diverse than baseline, exacerbation, or recovering samples (Fig. 1C). No significant differences in diversity levels were observed between baseline and exacerbation samples, whereas diversity levels from recovering samples were significantly lower than those from exacerbation samples.
Table 2.
BETR categories
| Clinical state | Description |
| Baseline (B) | Well or mild increase in pulmonary symptoms |
| Not a doctor defined pulmonary exacerbation | |
| Not hospitalized for increase in pulmonary symptoms | |
| Not on episodic antibiotics for >30 d | |
| May or may not be on maintenance antibiotics | |
| Exacerbation (E) | Doctor defined pulmonary exacerbation or increased pulmonary symptoms fitting definition of exacerbation* |
| Before start of episodic IV or oral antibiotics | |
| Not on episodic antibiotics for >30 d | |
| May or may not be on maintenance antibiotics | |
| Treatment (T) | On IV or oral episodic antibiotics for treatment of doctor defined exacerbation or increased pulmonary symptoms fitting definition of exacerbation* |
| Recovering (R) | Off episodic antibiotics ≤30 d |
| May or may not be on maintenance antibiotics | |
| May or may not be back to baseline clinical state |
*See SI Materials and Methods for details.
Impacts of Age, Antibiotic Therapy, and Lung Function on Diversity.
Having demonstrated that clinical state at the time of sample collection influences measures of community diversity, we next sought to assess the relative impacts of patient age, lung function, and antibiotic therapy on diversity. Antibiotic load was quantified as the sum of the number of days a patient received antibiotics (prescribed for maintenance therapy as well as for acute treatment of pulmonary exacerbations) during the 30 d before specimen collection (see SI Materials and Methods for details). In the three stable patients, the diversity of the community did not decrease significantly with increasing age and mildly decreasing lung function (Fig. 1B); similarly, no significant decrease in diversity was observed after adjustment for antibiotic load. In contrast, in the three progressing patients, where increasing age and decreasing lung function were each associated with decreasing diversity, adjustment for antibiotic load eliminated or diminished the statistical significance of these trends (Fig. S1).
Samples from all six patients were combined to explore further the effects of age, lung function, and antibiotic load on diversity in both univariate and multivariate linear mixed models (Table S2). In the univariate models, all three variables were correlated individually with decreasing diversity. In the multivariate model, however, antibiotic use was the strongest predictor of decrease in diversity, overshadowing the contributions of age and lung function to contribute to a significant trend (P < 0.001 for antibiotic load; P > 0.10 for age and lung function; see SI Results for details). Collectively, these results indicated that antibiotic use, rather than patient age or lung function, was the primary driver of the decreasing bacterial diversity observed in patients with typically progressive CF lung disease.
Community Structure Dynamics.
We compared community structures across patients to determine if interindividual differences in structure exceeded the changes we observed between serial samples from the same patient. Pairwise ecologic distances were calculated for all samples using the Bray–Curtis (BC) distance metric, which takes into account both community membership and relative abundance. These distances then were visualized by principal coordinate analysis (PCoA), which showed considerable overlap between communities from the two sets of stable and progressing patients (Fig. 2A). However, in comparing patient pairs, PCoA revealed appreciably distinct communities in individual patients (Fig. S2). Likewise, analysis of molecular variance (AMOVA) (9) showed significant differences in communities between all patient pairs, except between patients P2 and P3.
Fig. 2.
PCoA of bacterial community structures using BC distances. (A) Community structures of samples from three stable patients (blue) and three progressing patients (red). Distances between symbols on the ordination plot reflect relative similarities in community structures. PC1, principal coordinate 1; PC2, principal coordinate 2. (B) Community structure dynamics in serial samples highlighting the patient's clinical state on day of collection. The scale and percent of variation explained by PC1 and PC2 for each plot are the same as shown in A. Upper row shows stable patients (S1, S2, S3); lower row shows progressing patients (P1, P2, P3). Circles represent individual sputum samples and are color coded to reflect BETR category (B, blue; E, red; T, green; R, yellow; U, gray); lines connect serial samples. Upright and inverted triangles indicate first and last samples, respectively, in each serial set. Samples 10–13 from patient P1 and sample 12 from patient P3 are indicated by black arrows and are described in the text.
BC-based PCoA plots of communities from individual patients also showed the impact of clinical state on the dynamics of community structure (Fig. 2B). The majority of sputum samples from the three stable patients were collected during baseline states, and communities clustered in limited areas of PCoA plots. In contrast, the three progressing patients experienced more exacerbations and more courses of antibiotic treatment. Baseline and exacerbation samples from these patients clustered in the left half of the PCoA plots, whereas samples collected during antibiotic treatment drifted into other areas, particularly in patients P1 and P3. PCoA using the Jaccard distance metric, which takes into account only community membership, revealed similar results (Fig. S3).
The significance of the relationships between the structure of the bacterial community (measured as BC distance), patient age, %FEV1, and antibiotic load was assessed using the Mantel test. The positive correlation between BC distance and antibiotic load was greater (Mantel r = 0.32, P < 0.01) than that between BC distance and age (Mantel r = 0.20, P < 0.01) or between BC distance and %FEV1 (Mantel r = 0.22, P < 0.01). When antibiotic load was used as a control in a partial Mantel test, lesser correlations between BC distance and age (Mantel r = 0.11, P = 0.02) and between BC distance and %FEV1 (Mantel r = 0.12, P < 0.01) were observed.
Communities in samples obtained after recovery from treatment of exacerbation were closer to the baseline and exacerbation (preantibiotic) communities in the ordination than were samples obtained during antibiotic treatment, suggesting resiliency of communities after antibiotic perturbation. For example, sample 10 from patient P1 (Fig. 2B) was obtained at baseline, and sample 11 was obtained 15 wk later during treatment for an exacerbation. Sample 12 was collected after return to baseline 11 wk thereafter and resided close to sample 10 on the PCoA plot. Resiliency over the course of several years also was suggested by PCoA. In patient P2, for example, the community in the last collected sample, obtained during recovery from treatment for exacerbation, clustered close to the first sample, collected during baseline 8.5 y earlier. Similarly, the last samples obtained during periods without antibiotic treatment in patient P1 (sample 13, collected 7.5 y after the first sample) and in patient P3 (sample 12, collected 6.5 y after the first sample) also resided close to their respective first samples (Fig. 2B).
OTU Occurrence and Relative Abundance.
The occurrence and relative abundance of OTUs were dynamic in serial samples from both stable and progressing patients (Fig. 3, Fig. S4, and Table S1), but differences between these groups were noted. The most pronounced difference was with Staphylococcus, which occurred in 67% of samples from progressing patients but in only 6% of samples from stable patients.
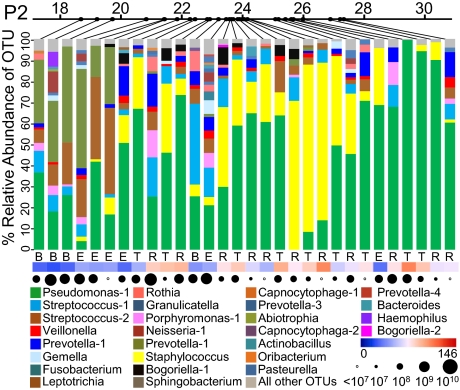
Fig. 3.
Relative abundance of OTUs in serial sputum samples from patient P2. Relative abundances of OTUs, each accounting for >1% of the total bacterial community, are shown in each sample. Relative abundances of all remaining OTUs, each representing ≤1% of the community, are shown in gray at the top of each bar. Top line indicates patient age at time of sample collection. Letters beneath each bar indicate BETR category (Table 2). Heat map indicates antibiotic load (Materials and Methods) at the time of sputum sample collection. Circles indicate total bacterial density (16S rRNA copies/mL sputum) based on quantitative PCR. Similar graphs for patients S1, S2, S3, P1, and P3 are shown in Fig. S4. Pseudomonas-1 represents Pseudomonas aeruginosa. Streptococcus-1 represents species within the Streptococcus mitis group. Streptococcus-2 represents species within the Streptococcus milleri group.
Within each patient's set of serial samples, certain OTUs were characterized as persistent, being defined as detected with a relative abundance of >1% in at least 50% of the samples obtained from that individual throughout the 8–9 y of observation. Pseudomonas was detected in at least 92% of samples in each patient (Table S1). Species in the S. milleri group were found in at least 58% of samples from each patient, and species in the S. mitis group were detected in at least 65% of samples from all patients except patient P3. A positive relationship was found between OTU occurrence and relative abundance. Although only 10 (3.9%) of the 259 total OTUs detected in the 126 sputum samples were defined as persistent (i.e., were found in more than 50% of samples in at least one of the six patients), these taxa accounted for 83–90% of the total relative abundance in each patient (Table S3).
Clinical state, particularly treatment of exacerbation, affected the occurrence and relative abundance of OTUs. In patient P3, for example, S. mitis group species were detected in 46% of all samples; however, they were detected in only 15% of samples obtained during treatment for exacerbation and in 86% of samples obtained at exacerbation before initiation of antibiotic therapy. In all three progressing patients, more OTUs were characterized as persistent when only samples obtained at baseline or exacerbation were considered (Table S3). In other words, in the absence of antibiotic treatment or recovery from treatment, more OTUs (primarily anaerobic species) met the criteria for being considered persistent in these patients.
The effects of repeated courses of antibiotic therapy for exacerbation were especially evident in later stages of lung disease, in which communities were heavily dominated by one or two OTUs (Fig. 3 and Fig. S4). In patient P1, communities late in the disease course were heavily dominated by Achromobacter and Pseudomonas, with these two OTUs accounting for up to 99.9% of the relative abundance in some samples. In patients P2 and P3, communities in later samples were dominated by Pseudomonas, nearly to the exclusion of other OTUs.
Quantitation of Bacterial Load.
Bacterial densities in samples were calculated by quantitative PCR analysis using a universal 16s rRNA primer-probe assay. No significant differences in total bacterial densities were detected in sputum samples from individual patients or from the three stable patients compared with the three progressing patients (Fig. S5A; P > 0.05, linear mixed model). Similarly, bacterial density did not differ significantly between samples based on clinical BETR state (Fig. S5B; P > 0.05, linear mixed model).
Discussion
Previous cross-sectional studies have reported that bacterial community diversity in CF is correlated positively with pulmonary function and inversely with patient age, suggesting a role for community composition in the pathophysiology of CF airway disease (5, 6). In our longitudinal study, we, too, found a positive correlation between diversity and lung function; however, at the individual patient level, decreasing community diversity was not necessarily correlated with increasing age. In the three patients with a mild lung disease phenotype, community diversity remained quite stable throughout time spans of 8–9 y. Furthermore, in patients S1 and S2, community diversity levels were comparable to those observed late, during periods of advanced lung disease, in the three progressing patients (Fig. 1B). Thus, community diversity alone is not a sufficient indicator of disease status. Of particular note is patient S3, who was homozygous for the ΔF508 CF transmembrane conductance regulator allele, a genotype more commonly associated with a more aggressive lung disease phenotype. Despite having relatively mild lung disease, this patient had airway bacterial communities that were significantly more diverse than those in the other two stable patients and comparable to those observed in early samples from the three progressing patients (Fig. 1B and Fig. S6). This apparent correlation between community diversity and CF genotype raises intriguing questions about the role the ΔF508 allele plays in susceptibility to airways infection.
Schluchter and colleagues (10) characterized CF lung disease phenotype by using longitudinal pulmonary function data, demonstrating the advantages of this approach over the use of a single FEV1 measurement to describe the severity of a patient's lung disease at a particular point in time. More recently, Konstan and colleagues (11) used these same principles to clarify the distinction between a patient's overall CF lung disease severity phenotype (lung disease aggressiveness) and that patient's current state of lung disease (lung disease stage). The rate at which %FEV1 declines over a period is an indicator of the aggressiveness of a patient's lung disease, independent of the stage of the patient's lung disease, which is defined as “early” when serial %FEV1 measures are >70; “intermediate” when %FEV1 measures range between 70 and 40; and “advanced” when %FEV1 measures are <40. In this context, the three stable patients included in our study can be described as having a mild lung disease-aggressiveness phenotype, and the three progressing patients represent a more typical moderate lung disease-aggressiveness phenotype (Fig. S7).
Our study demonstrates that among patients with moderately aggressive lung disease, disease stage (early, intermediate, or advanced), as indicated by %FEV1 measures, has significant bearing on estimates of the diversity of the lung bacterial community. As the three (stable) patients with mildly aggressive lung disease ultimately progress from early to intermediate stages of disease, and as antibiotic therapy intensifies, a similar decrease in diversity likely would be observed; however, continued long-term follow-up of these patients will be necessary to demonstrate this decrease. More importantly, our results show that clinical state (BETR category) at the time of sample collection has a pronounced effect on measures of diversity. It remains to be determined how finer-scale demarcation of clinical state (e.g., subcategories taking into account the exact number of days on or off antibiotic therapy relative to sample collection) might further impact measures of community diversity.
The pathophysiologic bases for the intermittent exacerbations that characterize CF are poorly understood. Speculation has focused on presumed acute changes in the composition of the airways bacterial community around the time of exacerbation. Although we detected significantly lower community diversity in samples obtained during antibiotic treatment, we observed no differences in diversity between baseline samples obtained during periods of relatively stable health and samples obtained at the time of exacerbation but before initiation of antibiotic therapy. This lack of a clear microbial signature for the onset of exacerbation suggests that exacerbations (i) may not be associated with specific changes in airway bacterial communities; (ii) may be associated with changes in only a few OTUs and/or very minor changes in several OTUs; or (iii) may result primarily from host-specific factors such as the nature and degree of inflammatory response to some as yet undefined stimulus. We also found no significant changes in total bacterial density by quantitative PCR in samples based on clinical state, suggesting that the reduction in some OTUs with antibiotic treatment may be offset by an increase in other, possibly more resistant, OTUs. It is important to note, however, that our quantitative PCR analysis did not differentiate between live and dead bacterial cells (12). Thus, it is likely that measures of bacterial density, particularly in samples obtained during antibiotic therapy, represent both viable and dead bacteria. Whether this distinction is important with respect to host inflammatory response and disease progression remains unclear.
It is clear from our results and those of others (5, 6) that, as most persons with CF age, lung function typically declines, antibiotic therapy both accumulates and accelerates, and the diversity of the airway community decreases. However, these confounding variables present a challenge to establishing a simple linear cause-and-effect relationship between the progression of lung disease and decreasing community diversity. Although age, lung function (%FEV1), and antibiotic use were all significantly correlated with community diversity in our study, we found that the effects of age and lung function lost significance when controlled for antibiotic use, suggesting that antibiotic therapy is the stronger driver of decreasing diversity. More robust models of antibiotic use in CF that incorporate antibiotic class, spectrum of activity, route of delivery, synergistic or antagonistic interactions, and duration of therapy are needed to provide a more precise understanding of the impact of antibiotic use on microbial community diversity.
Although we found considerable overlap in the community structures from all six patients, pairwise analyses using PCoA and AMOVA showed significant community differences between patient pairs (with the exception of patients P2 and P3), suggesting that interpatient variability in community structures exceeds intrapatient variability in longitudinal sample sets. When pairwise analyses were performed on subsets of samples based on disease stage (Fig. S8 and Table S4), we found that communities from the three progressing patients become more or less distinct in the advanced stage of disease, depending on the dominant species. More specifically, the advanced-stage communities in samples from patient P1, which were dominated by Achromobacter, were quite distinct from those of patients P2 and P3, which were dominated by Pseudomonas. This result suggests that, as community diversity decreases in the advanced stage of disease, community structures adopt a smaller number of distinct profiles, mainly described by the species that dominates the community.
Our analyses of serial samples from individual patients reinforced the impact of antibiotic therapy on community structure dynamics. BC-based PCoA plots showed marked movement of communities in samples obtained during antibiotic therapy for exacerbation relative to other samples, with the correlation between BC distance and antibiotic use exceeding that of either BC distance and age or BC distance and %FEV1. Our results also suggest that CF airways communities are resilient after perturbation by antibiotics. PCoA plots of communities in serial samples from the three progressing patients generally show communities in treatment recovery samples returning to profiles resembling the communities in samples obtained before therapy, indicating that resilience, rather than stochastic reassembly of depleted communities with new species, drives community dynamics in CF. A more complete assessment of the resilience of CF airway communities will require analysis of a larger set of samples from around the time of exacerbations. An important question to address in this regard is whether the dynamic range of communities changes as diversity decreases with disease progression. It may be reasonable to expect that, as airway communities become increasingly constrained by repeated antibiotic use, they may exhibit greater resistance to further perturbation, particularly if the community becomes restricted to a small number of highly antibiotic-resistant species.
Our study reinforces the dominant role of Pseudomonas as a constituent of the CF airway microbiota. Pseudomonas was identified in all six patients and quite often represented the most abundant species in individual samples. Of particular note, the communities in patients S1 and S2 were dominated almost uniformly by Pseudomonas during 8–9 y of clinical stability, indicating that the presence of Pseudomonas, even in relatively high abundance, does not necessarily predict poor clinical outcome. Additional analyses that allow quantitative assessment of potential fluctuations in the absolute density of P. aeruginosa are required to understand better the ecology and clinical impact of this species in CF. Our study also reaffirms that several anaerobic species, including a number of Veillonella and Prevotella species, are common and relatively abundant members of the airway microbiota in CF (13). The facultative anaerobic species of the S. milleri group (S. anginosus, S. constellatus, and S. intermedius) deserve particular attention. Using both culture and culture-independent methods, Sibley and colleagues (4) found that these species are detected frequently in CF respiratory samples and may be associated with clinical decline in health. Our results support these observations: S. milleri species were both persistent (present in most samples) and relatively abundant in all patients included in our study.
Our results support the recent observations by van der Gast and colleagues (2) that the CF airway bacterial metacommunity can be partitioned into two groups. One group is comprised of taxa that are prevalent and typically abundant; the other group consists of less commonly found species occurring in low abundance. We, too, found a positive correlation between OTU occurrence and relative abundance; communities in our six patients were dominated most often by Pseudomonas, S. milleri species, and strictly anaerobic species. Again, however, our results highlight the importance of taking clinical state into account in characterizing the airway bacterial community in this regard. We observed that a greater number of OTUs met our definition of persistent, which considers both rate of occurrence and relative abundance, when samples were limited to those obtained in the absence of antibiotic therapy.
Given these observations, it is important to note that the method of DNA extraction may have a significant impact on measures of the relative abundance of specific taxa. For example, we have shown recently that a method of DNA extraction that does not include the use of lysozyme and lysostaphin significantly underrepresents the abundance of Staphylococcus in CF sputum samples (14). Thus, firm conclusions regarding the relative abundance of species in individual specimens must be made with caution. In comparing communities across multiple specimens, however, a given method of DNA extraction should impart equal bias across all samples; therefore, observed intersample differences would not be expected to be the result of the methodology used.
Finally, by using quantitative PCR we have shown, similar to the recent report by Stressmann and colleagues (15), that the total bacterial density in the airway does not increase significantly at the time of pulmonary exacerbation. Our study extends these observations to show further that, as bacterial community diversity decreases with advancing age, progression of lung disease, and increasing antibiotic use, overall bacterial density remains relatively stable over prolonged periods of time. Our data suggest that, as community diversity decreases, the density of the remaining species increases to maintain a stable airway bacterial load. This result is most evident in the terminal treatment samples from patients P1 and P3, wherein bacterial density remained high (108–109 copies/mL) in the context of markedly species-restricted airway communities and intensive antibiotic therapy.
In summary, this study describes the structure and dynamics of airway bacterial communities in persons with CF over periods of several years, relating these measures to changes in patients’ clinical status and treatment. We describe diverse communities that, nevertheless, often are dominated by a few select genera. Our longitudinal analysis shows that high levels of diversity can be maintained for prolonged periods of time in patients with a mildly aggressive disease phenotype but that diversity decreases in patients with more typical progressive lung disease, being driven primarily by antibiotic therapy. We found that interpatient variability in community structures exceeds intrapatient variability in serial samples, indicating that patients harbor distinguishable communities. With decreasing diversity, communities converge to a narrower spectrum of distinct profiles, each being described by the dominant species present. Our analyses suggest that CF airway communities are resilient after antibiotic perturbation and that overall bacterial density remains relatively stable despite markedly decreasing diversity in the advanced stages of lung disease. Significant changes in community structure, diversity, or bacterial density at the time of pulmonary exacerbation were not observed. We have shown that patient clinical state and disease stage are critical in interpreting changes in the diversity and structure of the airway community, underscoring the importance of longitudinal studies in determining patterns of association between the CF airway microbiome and lung disease.
Materials and Methods
Patients, Sputum Specimens, and Medical Record Review.
Sputum sample collection and medical record review were approved by the University of Michigan Institutional Review Board. Each of the 126 sputum specimens included in this study was assigned to one of four clinical states based on medical record review (see SI Materials and Methods for details). Antibiotic load was calculated as the sum of the number of days a patient received an antibiotic within the 30 d before sample collection (see SI Materials and Methods for details).
DNA Extraction and Quantitative PCR.
DNA was extracted from sputum samples by treatment with Sputolysin (EMD Chemicals), mechanical disruption, and processing on an automated nucleic acid purification platform (MagNA Pure Compact System; Roche) (see SI Materials and Methods for details). Total bacterial load was measured by quantitative PCR using the universal primer/probe set of Nadkarni et al. (16) targeting the bacterial 16S rRNA gene (see SI Materials and Methods for details).
DNA Sequencing and Data Analyses.
Bar-coded pyrosequencing of the 16S rRNA V3–V5 hypervariable region was performed by the Human Genome Sequencing Center at Baylor College of Medicine using protocols developed for the Human Microbiome Project (http://www.hmpdacc.org/tools_protocols/tools_protocols.php). The software package mothur v. 1.21 (17) was used to process sequences and to calculate alpha and beta diversities. Statistical analyses were performed in PASW Statistics 18 and R packages vegan and ade4 (see SI Materials and Methods for details).
Supplementary Material
Acknowledgments
We thank Richard Gibbs, Donna Muzny, and the production team at the Human Genome Sequencing Center at Baylor College of Medicine for their effort on this project. We thank Courtney Robinson and Christine Bassis for helpful discussions. This work was funded by National Institutes of Health Clinical and Translational Science Award UL1RR024986 and National Heart, Lung, and Blood Institute Grant 1RC1HL100809-01 (to J.J.L., V.B.Y., and J.Z.L.). V.B.Y. also was supported by National Institutes of Health Grants U01HL098961, R01HG004906, and P30DK034933. P.D.S. was supported by National Institutes of Health Grant 1R01HG005975-01 and National Science Foundation Grant 0743432. J.J.L. also was supported by the Cystic Fibrosis Foundation. Additional support was provided by the Charles Woodson Pediatric Research Fund.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence data reported in this paper have been deposited in the NCBI Short Read Archive database data base (accession no. SRA050467.1).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120577109/-/DCSupplemental.
References
- 1.Harris JK, et al. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci USA. 2007;104:20529–20533. doi: 10.1073/pnas.0709804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Gast CJ, et al. Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J. 2011;5:780–791. doi: 10.1038/ismej.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogers GB, et al. characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16s ribosomal DNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol. 2004;42:5176–5183. doi: 10.1128/JCM.42.11.5176-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sibley CD, et al. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc Natl Acad Sci USA. 2008;105:15070–15075. doi: 10.1073/pnas.0804326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox MJ, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS ONE. 2010;5:e11044. doi: 10.1371/journal.pone.0011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klepac-Ceraj V, et al. Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ Microbiol. 2010;12:1293–1303. doi: 10.1111/j.1462-2920.2010.02173.x. [DOI] [PubMed] [Google Scholar]
- 7.Guss AM, et al. Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. ISME J. 2011;5:20–29. doi: 10.1038/ismej.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogers GB, Stressmann FA, Walker AW, Carroll MP, Bruce KD. Lung infections in cystic fibrosis: Deriving clinical insight from microbial complexity. Expert Rev Mol Diagn. 2010;10:187–196. doi: 10.1586/erm.09.81. [DOI] [PubMed] [Google Scholar]
- 9.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 10.Schluchter MD, Konstan MW, Drumm ML, Yankaskas JR, Knowles MR. Classifying severity of cystic fibrosis lung disease using longitudinal pulmonary function data. Am J Respir Crit Care Med. 2006;174:780–786. doi: 10.1164/rccm.200512-1919OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konstan MW, Wagener JS, VanDevanter DR. Characterizing aggressiveness and predicting future progression of CF lung disease. J Cyst Fibros. 2009;8(Suppl 1):S15–S19. doi: 10.1016/S1569-1993(09)60006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogers GB, et al. Assessing the diagnostic importance of nonviable bacterial cells in respiratory infections. Diagn Microbiol Infect Dis. 2008;62:133–141. doi: 10.1016/j.diagmicrobio.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Tunney MM, et al. Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. Am J Respir Crit Care Med. 2008;177:995–1001. doi: 10.1164/rccm.200708-1151OC. [DOI] [PubMed] [Google Scholar]
- 14.Zhao J, et al. Impact of enhanced Staphylococcus DNA extraction on microbial community measures in cystic fibrosis sputum. PLoS ONE. 2012;7(3):e33127. doi: 10.1371/journal.pone.0033127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stressmann FA, et al. Does bacterial density in cystic fibrosis sputum increase prior to pulmonary exacerbation? J Cyst Fibros. 2011;10:357–365. doi: 10.1016/j.jcf.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- 17.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.