Abstract
During apoptosis, hundreds of proteins are cleaved by caspases, most of them by the executioner caspase-3. However, caspase-7, which shares the same substrate primary sequence preference as caspase-3, is better at cleaving poly(ADP ribose) polymerase 1 (PARP) and Hsp90 cochaperone p23, despite a lower intrinsic activity. Here, we identified key lysine residues (K38KKK) within the N-terminal domain of caspase-7 as critical elements for the efficient proteolysis of these two substrates. Caspase-7's N-terminal domain binds PARP and improves its cleavage by a chimeric caspase-3 by ∼30-fold. Cellular expression of caspase-7 lacking the critical lysine residues resulted in less-efficient PARP and p23 cleavage compared with cells expressing the wild-type peptidase. We further showed, using a series of caspase chimeras, the positioning of p23 on the enzyme providing us with a mechanistic insight into the binding of the exosite. In summary, we have uncovered a role for the N-terminal domain (NTD) and the N-terminal peptide of caspase-7 in promoting key substrate proteolysis.
Keywords: caspase substrate, molecular recognition, enzymology
Apoptosis employs a family of cysteinyl peptidases, the caspases, to integrate and propagate various signals to cause cell demise. The latter step is governed by a subgroup of caspases named executioners that are responsible for the cleavage of a plethora of cellular proteins. The cleavage of some of these proteins provokes the associated hallmarks of apoptosis (1, 2). A survey of the published literature suggests that, among executioners, caspase-3 performs the bulk of the cleavage events. These data are in agreement with biochemical studies suggesting that this caspase is highly active and is present at the highest concentration among all caspases in most cells (3–5). For these reasons, caspase-3 supersedes other caspases in most biochemical readouts (4).
Caspase-3 and -7 share 57% sequence identity throughout their catalytic domains. Additionally, they have the same substrate preference based on studies using peptide substrate libraries (6, 7). Despite this apparent redundancy, these two capases have an overlapping but nonidentical substrate repertoire. For example, Rho kinase ROCK1, α-fodrin, and Rho-GDI are cleaved efficiently by both caspases (8, 9). However, inhibitor of caspase-activated Dnase (ICAD) (9), the X-linked inhibitor of apoptosis protein (XIAP) (10), and initiator caspase-9 (11) are preferred by caspase-3, whereas Nogo-B (12), ataxin-7 (13), and the p23 cochaperone (9) are cleaved more efficiently by caspase-7. Poly(ADP ribose) polymerase 1 (PARP), the first caspase substrate identified (14), is a particularly interesting death substrate, and its cleavage is now recognized as a hallmark of apoptosis. PARP proteolysis is essential for an adequate energetic balance during apoptosis and protects against necrosis (15, 16). Previous work has suggested that caspase-7 is responsible for PARP inactivation during apoptosis (17), but no mechanism for such selectivity has been proposed. However, many caspases and the immune-derived granzymes have been suggested to cleave PARP (14, 17–21). Notably, whereas PARP-1 is cleaved at a canonical caspase-3/7 site (DEVD↓G), p23 is cleaved at a less suitable site (PEVD↓G). Indeed, studies have shown that a proline instead of an aspartate in the P4 position reduces the catalysis of caspase-7 by 16,500-fold (7).
In the present study, we hypothesized that if caspase-7 is better at cleaving certain death substrates, such as PARP and p23, despite the fact that it is intrinsically less active than caspase-3, it must use exosites to improve catalysis. This attractive possibility has been raised several times in the past decade (22–25), but never confirmed. Our results show that the N-terminal domain (NTD) of caspase-7 contains a conserved basic patch that greatly improves substrate recognition and proteolysis of both PARP and p23. This study provides a unique demonstration of the use of a substrate exosite by a caspase.
Results
Caspase-7 Cleaves PARP and p23 More Efficiently than Caspase-3.
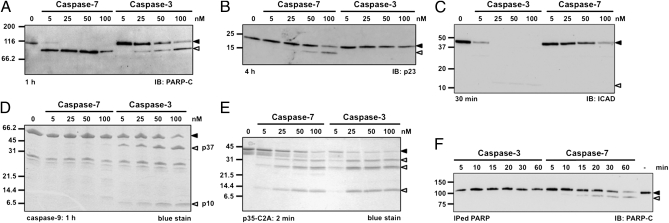
To determine whether caspase-7 or caspase-3 is the more efficient protease at cleaving death substrates, we performed assays in which active site-titrated recombinant caspases were incubated with cell extracts from MCF-7 cells (which lack caspase-3) expressing a short-hairpin RNA against caspase-7 (Fig. S1A). In these conditions, the ectopic caspase faces a mixture of substrates, akin to the cellular context. Furthermore, at the concentration used caspase-7 cannot activate other caspases (26). When various concentrations of caspase-7 or -3 were incubated with the extracts, the 116-kDa full-length PARP was cleaved by both enzymes to yield the typical 89-kDa fragment detected by our antibody (Fig. 1A). However, whereas 5 nM caspase-7 was sufficient to process > 85% of PARP, 50 nM caspase-3 resulted in less than 40% conversion. We also examined the cleavage of p23 and found that caspase-7 cleaves this protein more efficiently than caspase-3, albeit with less efficacy than PARP (Fig. 1B). To ensure that our assays were unbiased toward caspase-7, we analyzed ICAD and recombinant caspase-9 proteolysis and found that caspase-3 is better at cleaving both proteins (Fig. 1 C and D). These results demonstrate that each of these executioner caspases prefers a different set of death substrates.
Fig. 1.
Caspase-7 is better than caspase-3 at cleaving PARP and p23. (A–C) MCF-7sh7 extracts were incubated for the indicated period in the presence of 0, 5, 25, 50, or 100 nM recombinant caspase-7 or -3 in caspase buffer, then samples were analyzed by immunoblotting (IB) using the antibody indicated. (D and E) Recombinant full-length caspase-9 (1 μM) or p35-C2A (400 nM) was treated as in A. Samples were TCA-precipitated and analyzed by SDS/PAGE. (F) One nanomolar recombinant caspase-3 or -7 was incubated with immunoprecipitated flag-tagged PARP for the indicated period. Samples were analyzed as in A. Closed arrowhead, full-length protein; open arrowhead, cleaved fragment.
Comparison of the activity of these enzymes using the small fluorogenic substrate AcDEVD-Afc yielded catalytic specificity constants (kcat/KM) for caspase-7 and -3 of 1.1 × 105 and 5.9 × 105 M-1⋅s−1, respectively (Table S1), which are in agreement with previously published work (4, 26, 27). To determine whether the observed difference was an artifact of using a peptidic substrate, we used the baculovirus caspase inhibitor p35 with the inactivating C2A mutation that turns this protein into a substrate (28, 29). Similar to AcDEVD-Afc, p35-C2A is cleaved approximately five times more efficiently by caspase-3 than by caspase-7 (Fig. 1E), suggesting that caspase-3 is indeed a more proficient enzyme. We also tested the activity of caspase-3 and -7 on AcPEVD-Afc, which features the same cleavage site motif as that in p23, and found that caspase-3 is ∼500-times more active than caspase-7 (Table S1). Finally, to ensure that the preference for PARP we observed is not caused by the presence of competing substrates or endogenous caspase inhibitors, such as XIAP, we performed assays using antibody-purified PARP (Fig. 1F). However, we still found that caspase-7 was more efficient than caspase-3 at cleaving purified PARP, demonstrating the robustness of our assays. Taken together, these results suggest that caspase-7 likely uses an exosite to promote PARP and p23 cleavage.
Mature NTD of Caspase-7 Promotes PARP Cleavage.
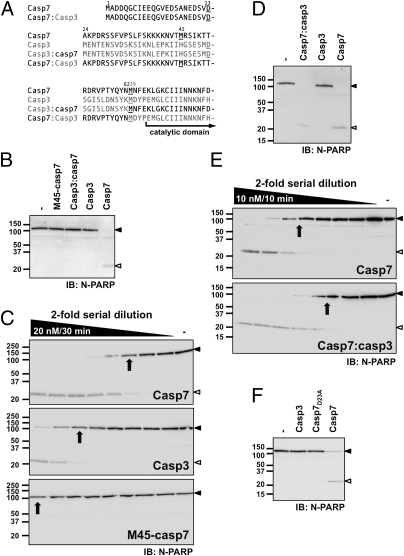
The catalytic domains of caspase-3 and -7 are highly similar. However, the N-terminal region that precedes the catalytic domain is much longer in caspase-7; this region, therefore, is a good exosite candidate. Consequently, we tested the ability of a caspase-7 variant that lacked part of its NTD to cleave PARP. We took advantage of an alternate translation initiation site within the NTD (Met45) (Fig. 2A) to generate an active enzyme (26). M45-caspase-7 is less efficient at cleaving PARP than WT enzyme at identical concentrations (Fig. 2B), even though it is as active on the peptidic substrate (Table S1). The percentage of PARP cleavage was estimated using imaging software as described in SI Materials and Methods and reported in Fig. S2. Specifically, we compared the intensity of the full-length 116-kDa protein because the cleaved fragments have variable detectability and stability, as suggested for many caspase substrate cleavage products, including PARP (30). By readjusting the caspase concentration and incubation time, we were able to estimate that proteolysis of PARP by caspase-7 is nearly eight-times higher than that by caspase-3, and that the cleavage rate by truncated M45-caspase-7 was less than 2 × 104 M-1⋅s−1, which is ∼30-times lower than the rate by WT caspase-7 (Fig. 2C). We then took advantage of the conserved methionine residue (Met62 in caspase-7 and Met39 in caspase-3) located just N-terminal to the catalytic domain to swap the NTD of caspase-7 with that of caspase-3 (casp3:casp7) (Fig. 2B). This transfer failed to restore PARP cleavage, demonstrating that the ability is specific to caspase-7's NTD. Notably, casp3:casp7 is fully active and processes its N-peptide (Fig. S3 and Table S1). Conversely, we tested the ability of the NTD of caspase-7 to confer PARP cleavage proficiency to caspase-3 (gain-of-function) by designing a casp7:casp3 chimera (Fig. 2D). In our assay, this chimeric caspase was as efficient as caspase-7 at cleaving PARP. Kinetic analysis demonstrated that casp7:casp3 was 3.6- and 30-folds more efficient than caspase-7 and WT caspase-3, respectively (compare Fig. 2 E vs. C). Furthermore, no alteration in the proteolysis of ICAD, a caspase-3-preferred substrate, was observed for this caspase-3 chimera (Fig. S4).
Fig. 2.
Caspase-7's N-terminal domain contains a transferable exosite to promote PARP cleavage. (A) The amino acid sequence of the N-terminal domain of caspase-7 and -3 are presented. Asp23 of caspase-7 or Asp28 of caspase-3 are the P1 cleavage site residue of the N-peptide that is removed during apoptosis. Mature caspase-7 and -3 start at Ala24 and Ser29, respectively. Key residues are underlined including the conserved methionine used as a convenient location for NTD deletion or chimera design. (B, D, and F) MCF-7sh7 detergent extracts were incubated for 30 min in the presence of 1 nM of the indicated caspases. Samples were analyzed by immunoblotting using an antibody recognizing the N-terminus of PARP. Quantification of PARP proteolysis is presented in Fig. S2. (C and E) Detergent extracts were incubated for the indicated period with twofold serial dilution of the indicated recombinant enzyme in caspase buffer starting at the indicated concentration. PARP hydrolysis rates were estimated as described in Materials and Methods. The enzyme concentration at which 50% of PARP is cleaved (arrow) was used to estimate rates. In C, the calculated rates are 6.2 × 105, <0.2, and 0.8 × 105 M-1⋅s−1 for caspase-7, M45-caspase-7, and caspase-3, respectively. In E, the calculated rates are 6.5 and 22.1 × 105 M-1⋅s−1 for caspase-7 and caspase-7:caspase-3 chimera, respectively.
Because removal of caspase-7's N-peptide precedes its activation (31), we asked if its removal was necessary to the function of the exosite. Thus, we compared a caspase-7 mutant that cannot process its N-peptide because of a D23A mutation (26) and found that this mutant is unable to cleave PARP in our standard assay (Fig. 2F). Importantly, caspase-7D23A does not impair catalytic activity (Table S1 and ref. 26). These results demonstrate that the NTD of caspase-7 contains molecular determinants that promote efficient PARP proteolysis.
Basic Patch in Caspase-7's NTD Is Critical to Promote PARP Cleavage.
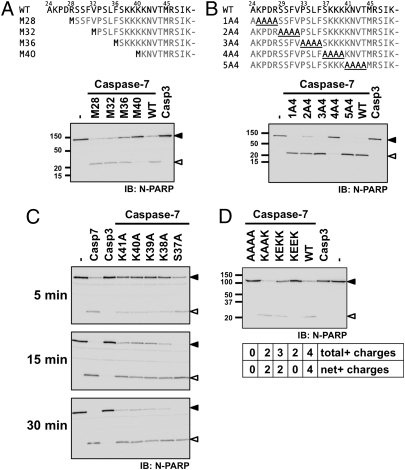
We wanted to identify the critical residues within the NTD that constitute the exosite. To this end, we tested the ability of deletion mutants to cleave PARP. Caspase-7 deletion mutants that lacked NTD residues downstream of Phe36 failed to process PARP in the assay conditions used (Fig. 3A). Conversely, a caspase-7 variant missing residues up to Val32 was still able to cleave the substrate. We also tested tetra-alanine substitution mutants (Fig. 3B) and found that changing residues 37–40 to alanine (mutant 4A4) had a dramatic effect on PARP proteolysis. These two sets of results suggest that the exosite is present in the basic sequence contained within the NTD. We then fine-tuned our assay using shorter incubation time to detect the influence of individual residues and scanned the residues contained in the 4A4 mutant (Ser37-Lys40) plus the following Lys41 (Fig. 3C). Unlike the mutation of Ser37, mutation of any lysine hampered, but did not abrogate, PARP cleavage, and K39A and K40A mutations had the greatest effect. The latter result suggests that charges within the exosite are the main determinant. Finally, to further characterize the motif, we designed four mutants with altered charges: K38KKK→AAAA (different from the 4A4 mutant), →KAAK, →KEKK, and →KEEK (Fig. 3D). From now on, all K38KKK mutations are referred to using subscript of the four replacing amino acids (WT is caspase-7KKKK). Compared with WT caspase-7, caspase-7KAAK was as efficient at cleaving PARP, whereas a decrease in efficacy was observed as the net charge decreased and negative charges were added. Indeed, caspase-7KEKK was worse than caspase-7KAAK, although both have the same net charge. These results demonstrate that the exosite relies on at least two lysine residues and does not tolerate negative charges. Importantly, all tested mutants displayed proteolytic activity against the peptide substrate that was either similar to the WT enzyme or, if different, could not account for the change in PARP proteolysis (Table S1).
Fig. 3.
The K38KKK motif is critical to the exosite function. (A–D) MCF-7sh7 detergent extracts were incubated for 30 min or the indicated time in the presence of 1 nM of the indicated caspases. Samples were analyzed by immunoblotting using an anti-PARP antibody recognizing the N terminus. Proteolysis quantification is presented in Fig. S2.
With the ability to transfer the exosite on caspase-3, these latest results suggest that the exosite is fully contained within the NTD. To further support this conclusion, we performed GST pull-down assays using only caspase-7 residues 24–62. Although GST alone and the tetra-alanine NTD mutant (NTDAAAA) fused to GST failed to bind PARP, the WT NTD readily precipitated PARP (Fig. S5). Indeed, quantification of the immunoblot signal revealed that 38% of PARP was pulled down. This result was also confirmed by analysis of the postpull-down supernatant that showed > 50% depletion of PARP. These results show that the NTD contains a complete exosite for PARP.
NTD of Caspase-7 Promotes Death Substrate Cleavage in Cells.
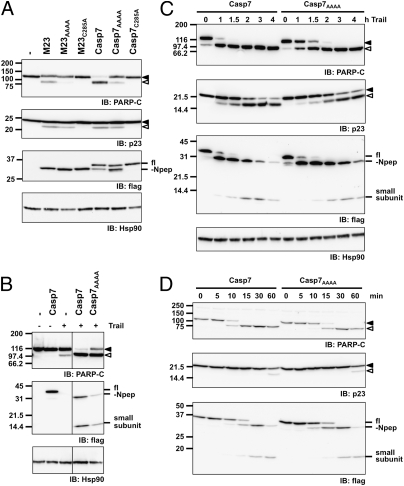
To demonstrate that the exosite supports efficient cleavage of PARP in living cells, we transfected AD-293 cells stably expressing a short-hairpin RNA (293sh7) against caspase-7 (Fig. S1B) with cDNAs encoding short-hairpin RNA-resistant flag-tagged caspase-7, caspase-7AAAA, or a caspase-7 baring the catalytic mutation C285A. In this paradigm, caspase-7 self-activates and induces cell death without the contribution of any other caspase (26, 32). These constructs also feature the deletion of the N-terminal peptide (M23) to mimic its removal by caspase-3. In agreement with our in vitro assays, the K38KKK->AAAA mutation decreases PARP proteolysis (Fig. 4A). Albeit less pronounced than for PARP cleavage, quantification of cleavage products did revealed a decrease of 39% and 26% in the processing of p23 by M23AAAA and caspase-7AAAA, respectively, compared with the corresponding protein bearing an intact exosite. Importantly, the capacity of caspase-7AAAA to self-activate was assessed by labeling the active enzyme with the activity-based probe biotinyl-VAD-fmk before lysis (Fig. S6). Using this approach, we showed that mutation of the exosite did not impede M23-caspase-7AAAA to self-activate, but did alter full-length processing, which suggests an interaction between the N-terminal peptide and the exosite.
Fig. 4.
Efficient PARP proteolysis in apoptotic cells depends on the exosite. (A) 293sh7 cells were transfected with the indicated flag-tagged short-hairpin RNA-resistant caspase-7 cDNAs. Lysates were analyzed 24 h posttransfection with the indicated antibodies. (B) Trail (200 ng/mL)-induced PARP cleavage in cells expressing nonlethal levels of WT or caspase-7AAAA. Samples were harvested after 3 h. All lanes were from the same blot. (C) Time-course of Trail-induced PARP cleavage in cells expressing nonlethal levels of WT or caspase-7AAAA. (D) Hypotonic extracts from 293sh7 cells reconstituted with low amounts of WT or caspase-7AAAA and supplemented with PARP-containing extracts were activated with cytc and dATP. fl, full-length; -Npep, caspase-7 lacking the N-peptide.
We also used the death ligand Trail to activate the extrinsic pathway in 293sh7 cells expressing caspase-7 or caspase-7AAAA at protein levels that do not provoke their autoactivation (Fig. 4B). Using this physiologic death stimulus, cells expressing caspase-7AAAA did process PARP to a lesser extent compared with cells expressing the WT caspase. Furthermore, time-course of Trail-induced apoptosis showed a 30- to 60-min delay in PARP cleavage, but no change in caspase-7 cleavage profile (Fig. 4C). Processing of p23 was also negatively affected by the exosite mutation.
Finally, we used cell-free extracts from 293sh7 cells expressing C-terminally flag-tagged WT and caspase-7AAAA, in which we induced caspase activation by the addition of cytochrome c (cytc) and dATP, leading to the initiator caspase-9 activation. As a source of PARP, we added dialyzed detergent extract from MCF-7sh7 cells. Time-course experiments showed a delay in the processing of PARP in extracts reconstituted with caspase-7AAAA (Fig. 4D).
Efficient Cleavage of p23 Requires the Exosite.
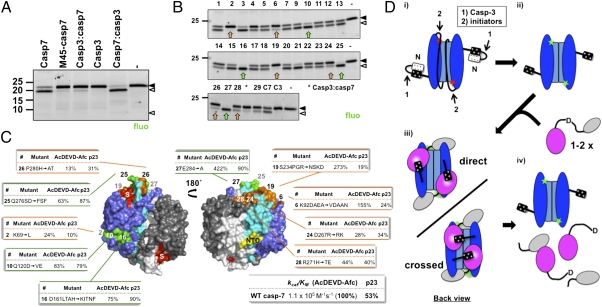
In Fig. 1 we showed that p23 is preferentially cleaved by caspase-7. Consequently, we wanted to extend our findings to p23 by analyzing the ability of key mutants of both executioner caspases to cleave fluorescein-labeled recombinant p23. Importantly, p23 labeling on free amines did not impair p23 cleavage by caspase-7 (Fig. S7). In an assay in which WT caspase-7 cleaves > 80% of p23, neither M45-caspase-7 nor caspase-3 processed p23 to the cleaved fragments (Fig. 5A). As we have shown for PARP, the casp7:casp3 chimera more efficiently cleaves p23. A decrease in p23 cleavage was also observed in cellular experiments expressing caspase-7AAAA (Fig. 4 A and C). These latest results demonstrate that caspase-7 also uses the exosite for p23 proteolysis. However, unlike PARP, p23 failed to precipitate with GST-NTD, suggesting that the interaction between the NTD and p23 is much weaker.
Fig. 5.
The exosite promotes cleavage of p23. (A) Fluorescein-labeled p23 (50 nM) was incubated with 40 nM of the indicated caspase for 30 min and was analyzed by fluorescence imaging of gels. (B) Same as in A, with caspases from the caspase-7:caspase-3 library (Fig. S8). Mutants showing decrease (orange arrows) or increase (green arrows) in p23 processing are indicated. Mut8 was inactive, therefore not analyzed. (C) Surface model of caspase-7 [PDB: 1F1J (33)] presenting the 10 sets of residues identified in B. The dimeric caspase contains two catalytic units, each of them is constituted of a large (blue or gray) and a small (cyan or white) subunits. The active site (S) is identified in red. For each mutant, the relative specific activity (kcat/KM) vs. WT for the fluorogenic substrate and the percentage of p23 cleavage are presented. Mut2 lies in a crevasse on the side of the caspase. The NTD is attached at the yellow region. (D) Proposed model for the exosite mechanism. (i) Caspase-3 primes caspase-7 zymogen for activation by removing the negatively charged N-terminal peptide, simultaneously uncovering the basic exosite. (ii) Then, an initiator caspase activates caspase-7 by cleaving the linker that separates the large and small subunits. (iii) Up to two molecules of substrates bind the caspase dimer, either in a direct or in a crossed configuration. (iv) Exosite binding promotes cleavage of the aspartate-containing motif by the catalytic site.
To build a model for the interaction of the caspase-7's exosite with a substrate, we sought to identify regions for which mutations interfere with the cleavage of p23. We used p23 because (i) it is a smaller protein than PARP; (ii) cleavage is much slower, which allows us to easily fine-tune the assay; and (iii) the exosite is not as dominant as it is the case for PARP, which allows us to better appreciate interference by other regions. We built a rational caspase-7 mutant library that incorporated groups of residues that were different in caspase-3 (Fig. S8). This mutagenesis strategy is very conservative because both executioners share high-sequence identity and have identical fold, thus limiting potential adverse effect of mutations. Indeed, all but one (mut8) of the designed mutants are active and self-activate in bacteria (Table S1). We tested this series of mutants under conditions in which only partial p23 cleavage is obtained. We identified six mutants (mut2, -6, -19, -24, -26, and -28) with diminished proteolysis of p23 (Fig. 5B, orange arrows). Mutants 6 (K92DAEA→VDAAN) and 19 (S234PGR→NSKD) are particularly interesting because they are ∼0.5 and ∼3 times, respectively, more active than WT caspase-7 on the fluorogenic substrate, yet ineffective at processing p23. Remarkably, these two mutants map to the back of the P4 substrate-binding pocket (Fig. 5C). We also identified mutants (mut10, -16, -25, and -27) with higher activity toward p23 (Fig. 5B, green arrows). One of these mutants, mut25 (Q276SD→FSF), mapped to the so-called L4 loop, is even less effective at cleaving the peptidic substrate, yet has improved activity on p23. Two more mutants (mut10 and mut16) with higher cleavage efficiency on p23, localized on the front side of the caspase large subunit enzyme (Fig. 5C). These results suggest that the globular domain of p23 lies close to the catalytic site when binding to the exosite.
Discussion
At a minimum, caspases select their substrates based on the structural accessibility of a P1 aspartate motif. To be efficient, however, more determinants are required to restrict the list of candidate proteins. Concretely, initiator caspases must rapidly activate downstream executioner caspases, which then must select among the much larger pool of death substrates to induce cell death. To this end, cells have evolved proficient enzymes, more discriminatory motifs, and presented optimal cleavage sites in flexible loops. In many proteolytic systems, further proficiency is achieved through the use of exosites.
Our results clearly demonstrate that caspase-7 uses a basic patch located in its NTD to improve the proteolysis of PARP and p23, two death substrates. This motif is conserved in mammals, frog, and chicken but not in other birds and fish. This domain is whole because it is transferable to another caspase and can precipitate PARP from a cell lysate. The improvement bestowed by the NTD is nontrivial because we observed an ∼30-fold decrease in cleavage efficacy when the NTD was removed from caspase-7, and a similar change (∼30-fold), but in the opposite direction, upon its transfer onto caspase-3. Furthermore, the fact that lysine mutants and truncated caspase-7 proteins have similar enzymatic properties on the peptidic substrate compared with the WT enzyme argues against an allosteric model involving the NTD. Consequently, we conclude that the tetra-basic motif fulfills all of the requirements of an exosite. This discovery gives credence to a recent study in which it was demonstrated that caspases could not reach proficiency solely by presenting a good cleavage motif (22). Indeed, engineering the best known cleavage motif (DEVD↓G) into a flexible loop in the small subunit of Escherichia coli carA protein resulted in a cleavage efficiency by caspase-3 of ∼5 × 104 M−1⋅s−1, which is lower than the cleavage rate observed for PARP with caspase-7, an intrinsically less active enzyme. By comparing the pool of evolutionary unrelated substrates found in the above-mentioned study with the higher cleavage rates of some natural caspase substrates, the authors concluded that caspases likely use exosites to increase catalysis. Another important finding of our study is that the exosite works best if the N-peptide of caspase-7 is removed. During apoptosis, this short 23-amino acid segment is clipped by caspase-3 before caspase-7 activation by the initiators, at least in some cells (31), although the involvement of other caspases have not been completely ruled out. We favor a simple model in which the highly acidic N-peptide (9 Asp/Glu residues, no Arg/Lys) shields the basic exosite until it is cleaved.
Caspase-7 mutants with altered activity toward p23, yet presenting opposite or unchanged activity on the peptidic substrate, provide an important structural constraint on the positioning of the substrate to the enzyme. In our model (Fig. 5D), the core of the substrate sits in close proximity to the back of the P4 substrate binding pocket. A second constraint is the requirement for the cleavage site to occupy the substrate-binding pocket. In the case of p23, the length of the C-terminal region spans 49 residues and is unstructured (34), which theoretically allows it to reach either catalytic center. However, in the model we proposed, the C-terminal binds the closest active site. This arrangement is analogous to the binding of XIAP to caspase-3 (35), with the second baculovirus inhibitory repeat (BIR2) positioning itself behind the catalytic site and extending its N-terminus over the catalytic cleft, but in a reverse orientation in this particular case. Probably the most interesting feature of our model is that it raises the possibility of a crossed substrate presentation (i.e., the NTD of one catalytic unit provides the exosite for the substrate cleaved by the other catalytic unit within a dimer) (Fig. 5D). However, the segment following the basic motif also allows all interactions to take place on the same catalytic unit (direct substrate presentation). It is not certain that the model we have just outlined applies to PARP because this protein has six independently folded domains (reviewed in ref. 36), thus offering several surfaces for exosite binding. Furthermore, Germain et al. demonstrated that caspase-7, but not caspase-3, has affinity for the polyanionic ADP ribose polymer (PAR) synthetized by PARP as part of its auto-modification activity (17). The protocol we used to extract PARP was not designed to preferentially select modified or unmodified PARP, but may have a bias toward a particular form. We did not use inhibitors of PAR formation and PAR glycohydrolase, and the protocol does not explicitly preserve the energy required for PAR synthesis. Consequently, we favor a model of interaction with PARP itself and not PAR. This finding is further supported by the ability of the exosite to promote p23 proteolysis, which is not modified, but we cannot rule out an important contribution by PAR to the interaction. Another difference may come from the screening of the caspase-7 mutant library with PARP that did not resulted in the identification of any determinant other than the NTD, although the strong interaction with the NTD could have masked other determinants. If the proposed model for p23 interaction also holds for PARP, the exosite would bind to the second zinc-finger domain, which precedes the cleavage site.
The interaction of the NTD with p23 is not as strong as it is with PARP. The reason for this difference is unknown, but it is tempting to speculate that the additional interactions provided by the residues we identified on the catalytic domain of caspase-7 also contribute to p23 cleavage efficiency, thus removing the requirement for strong binding to the NTD. Furthermore, we estimated that p23 is cleaved at least 100-times more slowly than PARP, which cast some doubt on the relevance of its cleavage during apoptosis, at least in the early phase. The simplest answer to explain the large difference in proteolysis rates of PARP and p23 probably resides in the more efficient exosite for PARP (present study) compounded with the favorable aspartate residue at the P4 position of the cleavage site compared with a proline in p23 (7). Other determinants, such as secondary and tertiary structures surrounding the cleavage site may also contribute to the difference observed. The p23 promotes ATP hydrolysis and may also specify the client protein repertoire of Hsp90, thus potentially affecting the many activities performed by this system. Furthermore, if many of the roles performed by its yeast ortholog, Sba1, are conserved in humans, p23 would play a pivotal role in DNA repair, which is reminiscent of the crucial role of PARP in genome maintenance. Consequently, p23 proteolysis could have widespread consequences on cellular functions.
Our findings raise the attractive possibility that caspases could use different exosites to achieve their apoptotic and nonapoptotic functions. This multiplicity of exosites is not unheard of, as some proteases, such as thrombin, use many exosites to bind different substrates and inhibitors. It is likely that other caspases have evolved different exosites to achieve proficiency in their roles during apoptosis and the ever-growing list of nonapoptotic roles of these enzymes.
Materials and Methods
Plasmids.
For mammalian cell expression, caspases were expressed with a C-terminal Flag epitope using the pcDNA3 plasmid. The various mutants were generated using standard PCR techniques. A short-hairpin RNA construct in pSuper plasmid targeting caspase-7 was created using the following sequences: 5′-gatccccAGACCGGTCCTCGTTTGTAttcaagagaTACAAACGAGGACCGGTCTttttta-3′ and 5′-agcttaaaaaAGACCGGTCCTCGTTTGTAtctcttgaaTACAAACGAGGACCGGTCTGGG-3′ (capital letters denote the interfering sequence). Caspase-7 resistance to the short-hairpin RNA was provided by silent mutations of the recognized sequence (underlined nucleotides).
Cell Culture and Transfection.
AD-293 cells were transfected using Lipofectamine 2000 reagent. High-level expression was achieved using 2–4 μg (35/60-mm dishes) of plasmid DNA, whereas nonlethal levels were obtained using 0.25–0.5 μg of the caspase DNA and empty plasmid to 3–4 μg.
Caspase Expression, Purification, and Characterization.
Recombinant caspase proteins were expressed as C-terminal His6-tagged proteins in the bacteria and purified using immobilized metal affinity chromatography. All enzymes were active site-titrated using the irreversible inhibitor Z-VAD-fmk. The kinetic parameters (kcat and KM) were determined using the fluorogenic substrate AcDEVD-Afc (37).
Cleavage Assays.
Cellular extracts containing PARP, ICAD, or p23 were prepared from clonal MCF-7 cells stably expressing an anti-caspase-7 short-hairpin RNA (see above) to reduce potential interference by endogenous executioner caspases. Cells were grown to confluence, washed, and harvested in PBS/EDTA/EGTA. Extracts were prepared in ice-cold buffer [50 mM Hepes (pH 7.4), 150 mM NaCl, 1% Nonidet P-40] and incubated on ice for 30 min. The soluble material (detergent extract) was recovered by centrifugation at 7,000 × g for 10 min and kept at −80 °C in small aliquots. A 20- to 30-μL reaction mixture containing detergent extract (1.5–3.5 mg⋅mL−1) was used with 0.5–1.0 nM active-site-titrated protease in caspase buffer at 37 °C for 30–60 min. Assays that used recombinant caspase-9 or p35-C2A were performed similarly by substituting the extracts with purified proteins.
Hypotonic Extracts.
Extracts from transfected 293sh7 cells were prepared as described previously (3). Caspases activation was programmed by the addition of 1 μM horse cytc and 1 mM dATP to 40 μL of extracts and 20 μL of extracts as a source of PARP. Ectopically expressed caspase-7 levels were adjusted using extracts from empty plasmid-transfected cells.
Recombinant p23 and Fluorescein Labeling.
The p23 cDNA was cloned from a human fetal brain cDNA library, expressed using the pGEX system. Proteins were cleaved with thrombin. The free amines in p23 were trace labeled using NHS-fluorescein in a 2:1 fluorescein:p23 molar ratio. Unreacted labeling reagent and buffer were removed using a 3,000 molecular weight cutoff spin filter.
Supplementary Material
Acknowledgments
We thank Catherine Duclos and Alexandre Murza for technical assistance; Éric Marsault, Marie-Ève Beaulieu, and Pierre Lavigne (Université de Sherbrooke) for helpful discussion; and Guy Poirier (Université Laval) for the gift of the poly(ADP ribose) polymerase 1. This work was supported by Grant MOP-86563 from the Canadian Institutes of Health Research and Grant 355388-2010 from the Natural Sciences and Engineering Research Council (to J.-B.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200934109/-/DCSupplemental.
References
- 1.Galluzzi L, et al. Molecular definitions of cell death subroutines: Recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor RC, Cullen SP, Martin SJ. Apoptosis: Controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 3.Stennicke HR, et al. Pro-caspase-3 is a major physiologic target of caspase-8. J Biol Chem. 1998;273:27084–27090. doi: 10.1074/jbc.273.42.27084. [DOI] [PubMed] [Google Scholar]
- 4.McStay GP, Salvesen GS, Green DR. Overlapping cleavage motif selectivity of caspases: Implications for analysis of apoptotic pathways. Cell Death Differ. 2008;15:322–331. doi: 10.1038/sj.cdd.4402260. [DOI] [PubMed] [Google Scholar]
- 5.Pop C, et al. Removal of the pro-domain does not affect the conformation of the procaspase-3 dimer. Biochemistry. 2001;40:14224–14235. doi: 10.1021/bi011037e. [DOI] [PubMed] [Google Scholar]
- 6.Thornberry NA, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 7.Stennicke HR, Renatus M, Meldal M, Salvesen GS. Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem J. 2000;350:563–568. [PMC free article] [PubMed] [Google Scholar]
- 8.Sebbagh M, et al. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat Cell Biol. 2001;3:346–352. doi: 10.1038/35070019. [DOI] [PubMed] [Google Scholar]
- 9.Walsh JG, et al. Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc Natl Acad Sci USA. 2008;105:12815–12819. doi: 10.1073/pnas.0707715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slee EA, Adrain C, Martin SJ. Executioner caspase-3, -6, and -7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J Biol Chem. 2001;276:7320–7326. doi: 10.1074/jbc.M008363200. [DOI] [PubMed] [Google Scholar]
- 11.Denault JB, Eckelman BP, Shin H, Pop C, Salvesen GS. Caspase 3 attenuates XIAP (X-linked inhibitor of apoptosis protein)-mediated inhibition of caspase 9. Biochem J. 2007;405:11–19. doi: 10.1042/BJ20070288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schweigreiter R, et al. Phosphorylation-regulated cleavage of the reticulon protein Nogo-B by caspase-7 at a noncanonical recognition site. Proteomics. 2007;7:4457–4467. doi: 10.1002/pmic.200700499. [DOI] [PubMed] [Google Scholar]
- 13.Young JE, et al. Proteolytic cleavage of ataxin-7 by caspase-7 modulates cellular toxicity and transcriptional dysregulation. J Biol Chem. 2007;282:30150–30160. doi: 10.1074/jbc.M705265200. [DOI] [PubMed] [Google Scholar]
- 14.Tewari M, et al. Yama/CPP32 beta, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 15.Los M, et al. Activation and caspase-mediated inhibition of PARP: A molecular switch between fibroblast necrosis and apoptosis in death receptor signaling. Mol Biol Cell. 2002;13:978–988. doi: 10.1091/mbc.01-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oliver FJ, et al. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis. Lesson from an uncleavable mutant. J Biol Chem. 1998;273:33533–33539. doi: 10.1074/jbc.273.50.33533. [DOI] [PubMed] [Google Scholar]
- 17.Germain M, et al. Cleavage of automodified poly(ADP-ribose) polymerase during apoptosis. Evidence for involvement of caspase-7. J Biol Chem. 1999;274:28379–28384. doi: 10.1074/jbc.274.40.28379. [DOI] [PubMed] [Google Scholar]
- 18.Malireddi RK, Ippagunta S, Lamkanfi M, Kanneganti TD. Cutting edge: Proteolytic inactivation of poly(ADP-ribose) polymerase 1 by the Nlrp3 and Nlrc4 inflammasomes. J Immunol. 2010;185:3127–3130. doi: 10.4049/jimmunol.1001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandes-Alnemri T, Litwack G, Alnemri ES. Mch2, a new member of the apoptotic Ced-3/Ice cysteine protease gene family. Cancer Res. 1995;55:2737–2742. [PubMed] [Google Scholar]
- 20.Van Damme P, et al. The substrate specificity profile of human granzyme A. Biol Chem. 2010;391:983–997. doi: 10.1515/BC.2010.096. [DOI] [PubMed] [Google Scholar]
- 21.Quan LT, et al. Proteolytic activation of the cell death protease Yama/CPP32 by granzyme B. Proc Natl Acad Sci USA. 1996;93:1972–1976. doi: 10.1073/pnas.93.5.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Timmer JC, et al. Structural and kinetic determinants of protease substrates. Nat Struct Mol Biol. 2009;16:1101–1108. doi: 10.1038/nsmb.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.López-Otín C, Overall CM. Protease degradomics: A new challenge for proteomics. Nat Rev Mol Cell Biol. 2002;3:509–519. doi: 10.1038/nrm858. [DOI] [PubMed] [Google Scholar]
- 25.Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu Rev Biochem. 2011;80:1055–1087. doi: 10.1146/annurev-biochem-061809-121639. [DOI] [PubMed] [Google Scholar]
- 26.Denault JB, Salvesen GS. Human caspase-7 activity and regulation by its N-terminal peptide. J Biol Chem. 2003;278:34042–34050. doi: 10.1074/jbc.M305110200. [DOI] [PubMed] [Google Scholar]
- 27.Denault JB, et al. Engineered hybrid dimers: Tracking the activation pathway of caspase-7. Mol Cell. 2006;23:523–533. doi: 10.1016/j.molcel.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 28.Xu G, et al. Covalent inhibition revealed by the crystal structure of the caspase-8/p35 complex. Nature. 2001;410:494–497. doi: 10.1038/35068604. [DOI] [PubMed] [Google Scholar]
- 29.Riedl SJ, Renatus M, Snipas SJ, Salvesen GS. Mechanism-based inactivation of caspases by the apoptotic suppressor p35. Biochemistry. 2001;40:13274–13280. doi: 10.1021/bi010574w. [DOI] [PubMed] [Google Scholar]
- 30.Dix MM, Simon GM, Cravatt BF. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell. 2008;134:679–691. doi: 10.1016/j.cell.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X, et al. Granzyme B mimics apical caspases. Description of a unified pathway for trans-activation of executioner caspase-3 and -7. J Biol Chem. 1998;273:34278–34283. doi: 10.1074/jbc.273.51.34278. [DOI] [PubMed] [Google Scholar]
- 32.Duan H, et al. ICE-LAP3, a novel mammalian homologue of the Caenorhabditis elegans cell death protein Ced-3 is activated during Fas- and tumor necrosis factor-induced apoptosis. J Biol Chem. 1996;271:1621–1625. doi: 10.1074/jbc.271.3.1621. [DOI] [PubMed] [Google Scholar]
- 33.Wei Y, et al. The structures of caspases-1, -3, -7 and -8 reveal the basis for substrate and inhibitor selectivity. Chem Biol. 2000;7:423–432. doi: 10.1016/s1074-5521(00)00123-x. [DOI] [PubMed] [Google Scholar]
- 34.Weaver AJ, Sullivan WP, Felts SJ, Owen BA, Toft DO. Crystal structure and activity of human p23, a heat shock protein 90 co-chaperone. J Biol Chem. 2000;275:23045–23052. doi: 10.1074/jbc.M003410200. [DOI] [PubMed] [Google Scholar]
- 35.Riedl SJ, et al. Structural basis for the inhibition of caspase-3 by XIAP. Cell. 2001;104:791–800. doi: 10.1016/s0092-8674(01)00274-4. [DOI] [PubMed] [Google Scholar]
- 36.Krishnakumar R, Kraus WL. The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denault JB, Salvesen GS. In: Apoptotic Caspase Activation and Activity. Apoptosis and Cancer, Methods in Molecular Biology. Mor G, Rutherford TJ, editors. Clifton, NJ: Humana Press; 2007. p. 272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.