Abstract
To ensure genome stability, DNA must be replicated once and only once during each cell cycle. Cdt1 is tightly regulated to make sure that cells do not rereplicate their DNA. Multiple regulatory mechanisms operate to ensure degradation of Cdt1 in S phase. However, little is known about the positive regulators of Cdt1 under physiological conditions. Here we identify FOXO3 as a binding partner of Cdt1. FOXO3 forms a protein complex with Cdt1, which in turn blocks its interaction with DDB1 and PCNA. Conversely, FOXO3 depletion facilitated the proteolysis of Cdt1 in unperturbed cells. Intriguingly, FOXO3 deficiency resulted in impaired S-phase entry and reduced cell proliferation. We provide data that FOXO3 knockdown mimics Cdt1 down-regulation and affects G1/S transitions. Our results demonstrate a unique role of FOXO3 in binding to Cdt1 and maintaining its level required for cell cycle progression.
In eukaryotic cells, DNA replication initiates from thousands of replication origins. Each origin acquires replication competence through the assembly of a prereplication complex (pre-RC) occurring in late mitosis and early G1 (1–3). Pre-RCs are assembled at the origins of DNA replication through the sequential loading of the initiation factors ORC, Cdc6, Cdt1, and MCM2-7 (4). In S phase, pre-RCs are sequentially acted on by two protein kinases, Cdc7 and Cdk2, which promote recruitment of proteins required for helicase activation and replisome assembly, leading to origin unwinding and DNA synthesis. To ensure that no replication origin fires more than once, the assembly of the replication apparatus at origins is tightly regulated by the cell cycle machinery. Among the most important of these regulatory mechanisms are the degradation of Cdt1 during S phase and the sequestration of Cdt1 by the geminin protein (5–7). Phosphorylation of Cdt1 by Cdk2 promotes its binding to SCF-Skp2 E3 ubiquitin ligase (8–10), which results in its degradation in S phase. In addition to the Skp2 pathway, PCNA/DDB1/Cul4-dependent signaling was found to degrade Cdt1 during S phase via the interaction of Cdt1 with PCNA (11–15). Recently, APC/CCdh1 was proposed as a third ubiquitin ligase regulating Cdt1 degradation (16). Cdt1 is also targeted for degradation after DNA damage to stop licensing of new origins until after DNA repair. Both the SCF-Skp2 complex and the Cul4-DDB1 complex have been reported to induce degradation of Cdt1 after UV irradiation (17, 18).
FOXO transcription factors are critical for the regulation of cell cycle arrest, cell death, and DNA damage repair. Ample evidence has suggested that FOXO exerts a negative effect on cell cycle progression. In dividing cells, overexpression of the active form of FOXO family members promotes cell cycle arrest at the G1/S boundary. Target genes that mediate FOXO-induced cell cycle arrest are the Cdk inhibitors p27KIP1 and p21 (in the presence of TGF-β), the Rb family member p130, and cyclin D1 and D2. The ectopically expressed active form of FOXO factors can cause G1 arrest both by up-regulating cell cycle inhibitors (p21 and p27) and by repressing cell cycle activators (cyclin D1/D2). FOXO factors also regulate other cell cycle checkpoints. Cells expressing the constitutively active form of FOXO3 in the S phase display a delay in their progression through the G2 phase of the cell cycle. Two targets were identified that may mediate the effect of FOXOs at the G2/M boundary: cyclin G2 and GADD45. Thus, FOXO factors mediate cell cycle arrest at the G1/S and G2/M transitions, two checkpoints that are critical in the cellular response to stress. Notably, these previous reports characterizing the biological functions of FOXO in cell cycle regulation were largely, if not all, based on overexpression of constitutively active form of FOXO members. In contrast to these previous reports, here we provide evidence that depleting FOXO3 reduced G1/S transition and cell proliferation. Cdt1 was identified as a binding partner of FOXO3. FOXO3 is crucial for maintaining Cdt1 basal levels. Our data suggest a unique biological function of FOXO3 in cell cycle progression.
Results
Cdt1 Interacts with FOXO3.
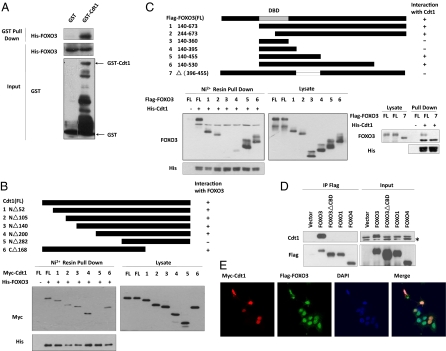
Our laboratory is interested in identifying binding partners of FOXO3. To this end, epitope-tagging strategy and affinity chromatography on M2 (anti-Flag antibody) agarose beads was used to isolate protein complexes containing Flag-tagged FOXO3 from nuclear extracts of HEK293T cells. After SDS/PAGE fractionation and silver staining, we identified a major protein band of ∼64 kDa that copurified with FOXO3 and that mass spectrometry revealed to be Cdt1. We next showed that purified recombinant His-FOXO3 was able to interact with GST-Cdt1 under cell-free conditions (Fig. 1A), suggesting a direct interaction between FOXO3 and Cdt1. To determine the region of Cdt1 required for FOXO3 association, we generated a series of Cdt1 deletion mutants. Mutants lacking the Cdt1 N-terminal domain [amino acids (aa) 1–282] failed to bind to His-FOXO3, whereas those lacking aa 1–200 exhibited robust interaction with FOXO3 (Fig. 1B). These data suggest that the middle region (aa 200–282) of Cdt1 mediates FOXO3-Cdt1 association. Reciprocal mapping using Flag-FOXO3 deletion constructs pinpointed FOXO3 aa 396–455 as a critical region for FOXO3-Cdt1 interaction (Fig. 1C), prompting us to designate this region as the Cdt1-binding domain (CBD). To assess if other FOXO factors could bind to Cdt1, HEK293T cells were transfected with vectors overexpressing Flag-tagged FOXO1, FOXO3, or FOXO4. A Flag-tagged mutant lacking FOXO3 aa 395–455 (FOXO3ΔCBD) served as a negative control for immunoprecipitations. Surprisingly, only FOXO3 was able to bring down and stabilize endogenous Cdt1, and Cdt1 did not accumulate in cells overexpressing FOXO3ΔCBD, FOXO1, or FOXO4 (Fig. 1D). Interestingly, exogenous FOXO3 had no effect on Cdt1 mRNA levels, and geminin protein was unchanged in cells overexpressing FOXO proteins (Fig. S1). To determine the subcellular localization of the FOXO3-Cdt1 complex, we cotransfected MCF-7 cells with vectors expressing Myc-Cdt1 and Flag-FOXO3. Immunofluorescent staining revealed that FOXO3 and Cdt1 colocalized in the nucleus (Fig. 1E). Taken together, these results show that FOXO3 is unique among FOXOs in its interaction with Cdt1 in vivo.
Fig. 1.
Cdt1 is a unique FOXO3-interacting protein. (A) Cdt1 directly interacts with FOXO3. Purified FOXO3 was incubated with GST or GST-Cdt1 coupled to GSH-Sepharose. Cell lysates (“Input”) and the eluates (“GST pull down”) were subjected to SDS/PAGE, followed by immunoblot analysis with the indicated antibodies. (B and C) Mapping of the binding domains for FOXO3 and Cdt1. 293T cells were transfected with the indicated constructs, and lysates were incubated with His-FOXO3 (B) or His-Cdt1 (C). Proteins retained on nickel resin were subjected to Western blotting with the indicated antibodies. (D) Cdt1 interacts specifically with FOXO3. 293T cells transfected with indicated plasmids were subjected to immunoprecipitation with M2 beads, followed by immunoblotting with anti-Cdt1 and anti-Flag antibodies. The asterisk indicates a nonspecific band. (E) Cdt1 and FOXO3 form a complex in the nucleus. MCF-7 cells cotransfected with Myc-Cdt1 and Flag-FOXO3 were subjected to immunofluorescent staining with anti-Myc (red) and anti-Flag (green) antibodies. For A–E, results shown are representative of at least three independent experiments.
FOXO3 Regulates Cdt1 Protein Stability via Interfering PCNA-DDB1-Cul4–Mediated Ubiquitination of Cdt1.
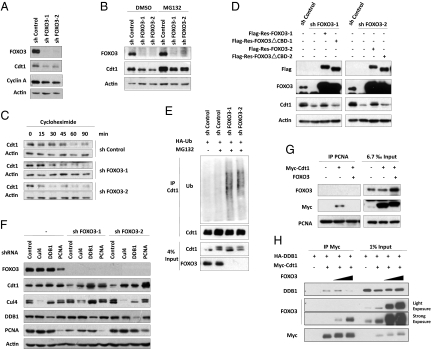
To further confirm the role of FOXO3 in regulating Cdt1, we depleted FOXO3 in MCF-7 cells using two independent FOXO3-specific short hairpin RNAs (shRNAs). Intriguingly, we found that FOXO3 knockdown significantly decreased Cdt1 protein levels (Fig. 2A). Cdt1 mRNA levels were also slightly reduced (Fig. S2A). E2F-dependent transactivation of Cdt1 has been reported previously (19). Indeed, we detected partial blockage of the cell cycle at G1 phase, which was indicated by a decrease in cyclin A levels (Fig. 2A) and by fluorescence-activated cell sorting (FACS) analysis (Fig. 3D). We also observed decreased Cdt1 protein in the human lung cancer cell line H1299 and in the osteosarcoma cell line U2OS upon FOXO3 knockdown (Fig. S2B). Importantly, the degree of reduction in Cdt1 protein was tightly correlated with FOXO3 knockdown efficiency, in that MCF-7 cells infected with various amounts of FOXO3 shRNA lentiviral soup showed dose-dependent Cdt1 protein down-regulation (Fig. S2C). Given that Cdt1 protein is subject to DNA damage-induced degradation (17, 18), we speculated that our shRNA lentiviral soup may have been slightly cytotoxic and induced a DNA damage-like insult that triggered Cdt1 down-regulation. To address this possibility, we examined cell cycle checkpoint pathways in FOXO3-depleted cells. In contrast to the ready activation of Chk2-p53 signaling in response to UV irradiation, FOXO3 knockdown failed to activate these crucial checkpoint regulators (Fig. S2D). Thus, Cdt1 down-regulation in response to FOXO3 depletion is a primary event and not a secondary effect of a DNA damage-like insult. Importantly, the decrease in Cdt1 levels induced by FOXO3 knockdown could be largely reversed by the addition of the proteasome inhibitor MG132 (Fig. 2B), indicating that FOXO3 regulates Cdt1 levels in a proteasome-dependent manner. To confirm that FOXO3 influences Cdt1 stability, we treated control cells or cells stably expressing FOXO3 shRNA with cycloheximide and examined the half-life of Cdt1 protein. A significant decrease in Cdt1 stability occurred in FOXO3 knockdown cells compared with controls (Fig. 2C). Thus, FOXO3 is required for the maintenance of steady-state levels of Cdt1 protein. To determine the specificity of the FOXO3 knockdown-induced Cdt1 destabilization, we performed a rescue experiment. Expression plasmids encoding rescue forms of FOXO3 or FOXO3ΔCBD were generated by introducing silent mutations in the cDNA encoding the FOXO3 proteins designed to render them resistant to FOXO3 shRNA (Res-FOXO3 or Res-FOXO3ΔCBD). Intriguingly, only Res-FOXO3, but not Res-FOXO3ΔCBD, largely blocked Cdt1 decrease mediated by FOXO3 depletion (Fig. 2D), indicating that Cdt1 destabilization in FOXO3-deficient cells is the result of specific knockdown of FOXO3. Furthermore, FOXO3-Cdt1 interaction is essential for maintaining basal Cdt1 levels by FOXO3.
Fig. 2.
FOXO3 regulates Cdt1 protein stability via attenuating PCNA/DDB1/Cul4-mediated ubiquitination of Cdt1. (A) MCF-7 cells infected with the indicated shRNA lentivirus were lysed with Laemmli lysis buffer, and lysates were analyzed by immunoblotting with anti-FOXO3, anti-Cdt1, anti-cyclin A, and anti–β-actin antibodies. (B) Proteasome inhibitor blocks Cdt1 degradation in FOXO3-deficient cells. MCF-7 cells infected with lentivirus expressing the indicated shRNAs were left untreated or treated with 20 μM MG132 for 4 h, and cell lysates were then extracted and subjected to Western blotting. (C) Cdt1 half-life is decreased upon FOXO3 depletion. Control and FOXO3-depleted MCF-7 cells were chased with cycloheximide. Cells were then harvested at indicated time points and analyzed by immunoblotting. (D) Rescue of FOXO3 depletion-induced Cdt1 degradation by Res-FOXO3 but not by Res-FOXO3△CBD. MCF-7 cells were coinfected with indicated lentiviral-expressing constructs. Cell lysates were subjected to Western blotting analysis with the indicated antibodies. (E) FOXO3 depletion results in increased Cdt1 ubiquitylation. MCF-7 cells expressing HA-ubiquitin (Ub) were infected with the indicated shRNA lentivirus in the presence or absence of MG132 for the final 4 h. Cells were harvested and immunoprecipitated with anti-Cdt1 antibody. Western blotting was then performed using indicated antibodies. (F) PCNA and the Cul4-DDB1 complex are responsible for FOXO3 knockdown-mediated Cdt1 destabilization. MCF-7 cells were infected with the indicated shRNA lentivirus. Cell extracts were prepared and analyzed by immunoblotting with indicated antibodies. (G) PCNA-Cdt1 interaction is impaired in the presence of exogenous FOXO3. 293T cells expressing the indicated constructs were subjected to immunoprecipitation with anti-PCNA antibody, followed by Western blotting for anti-FOXO3, anti-Myc, and anti-PCNA antibodies. (H) Ectopic FOXO3 abolishes DDB1-Cdt1 interaction. 293T cells ectopically expressing the indicated constructs were immunoprecipiated with anti-Myc antibody, followed by immunoblotting with anti-DDB1, anti-FOXO3, and anti-Myc antibodies.
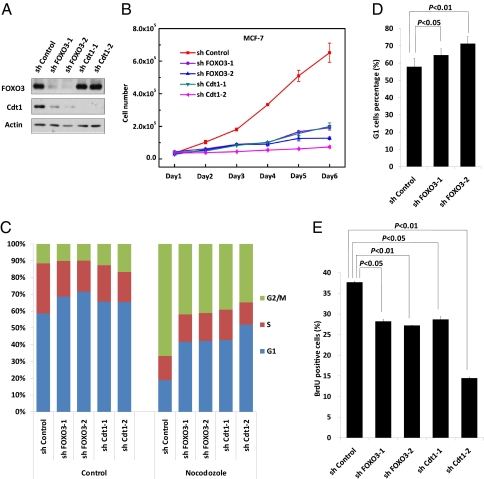
Fig. 3.
FOXO3 depletion impairs G1/S transition and cell proliferation. (A) Depleting FOXO3 and Cdt1 in MCF-7 cells. MCF-7 cells were infected with the indicated shRNA lentivirus, and knockdown efficiency was measured by immunoblotting with anti-FOXO3 and anti-Cdt1 antibodies. Actin, loading control. (B) Impaired cell proliferation upon depleting FOXO3 or Cdt1. MCF-7 cells stably expressing the indicated shRNAs were plated, and cell proliferation was then quantified at the indicated time points. Data presented are the mean ± SD of three independent experiments. (C) Flow cytometry analysis by PI staining of the DNA content of cells in A before and after synchronization in mitosis with nocodazole (100 ng/mL) for 16 h. Percentages of cells in each culture in the G1, S, and G2/M phases are shown. Results are from one trial representative of three independent experiments. (D) MCF-7 cells stably expressing the indicated shRNAs were stained by PI, followed by FACS analysis of cells in G1 phase. Data shown are mean ± SD of six independent experiments. (E) Cells in A were pulsed with BrdU for 1 h and then stained with anti-BrdU antibody followed by FACS analysis for BrdU-positive cells. Data represent mean ± SD of three independent experiments.
To elucidate the mechanism by which FOXO3 protects Cdt1 from proteasomal degradation, we first tested if FOXO3 inhibits Cdt1 ubiquitination. Indeed, endogenous Cdt1 ubiquitination was substantially increased in FOXO3 knockdown cells (Fig. 2E). We next investigated which E3 ligase system is responsible for Cdt1 ubiquitination under conditions of FOXO3 depletion. SCFSkp2 ubiquitin ligase, Cul4 ubiquitin ligase, and APC/CCdh1 ubiquitin ligase all play crucial roles in the proteolytic regulation of Cdt1 in mammalian cells (8, 16–18, 20). When we used specific shRNAs to deplete Cul4, DDB1, PCNA, Cdh1, or Skp2 in MCF-7 cells in the presence or absence of FOXO3 shRNA, we found that silencing of Cul4, DDB1, or PCNA significantly, albeit partially, inhibited Cdt1 down-regulation in FOXO3 knockdown cells (Fig. 2F). In contrast, depletion of Skp2 or Cdh1 in FOXO3 knockdown cells failed to block Cdt1 decrease in FOXO3-deficient cells (Fig. S3). We therefore concluded that Cul4, DDB1, and PCNA all participate in the degradation of Cdt1 induced by FOXO3 depletion. To gain more molecular insights, we then examined the effect of exogenous FOXO3 overexpression on the association of Cdt1 with PCNA/DDB1/Cul4. The ability of ectopically expressed Cdt1 to form a complex with endogenous PCNA (Fig. 2G) or exogenous DDB1 (Fig. 2H) was progressively lost as the cells expressed increasing amounts of FOXO3, suggesting that FOXO3 inhibits Cdt1 ubiquitination by attenuating DDB1-Cdt1 and PCNA-Cdt1 interaction.
FOXO3 Depletion Impairs S-Phase Entry and Cell Proliferation.
Because Cdt1 promotes pre-RC assembly and DNA replication, we examined the proliferation of MCF-7 cells following FOXO3 or Cdt1 knockdown. First, we confirmed the high efficiency of our specific shRNAs (Fig. 3A). Surprisingly, the cell-number counting assay showed that cell proliferation was significantly impaired by FOXO3 depletion, just as observed after Cdt1 knockdown (Fig. 3B). FOXO3 depletion in H1299 and U2OS cells also resulted in profoundly impaired cell proliferation (Fig. S4). To gain more insight into the cell cycle progression defect caused by FOXO3 depletion, we synchronized cells in mitosis using nocodazole, which blocks exit from mitosis in a reversible manner. FACS analysis revealed that nocodazole treatment of MCF-7 cells expressing control shRNA resulted in G2/M arrest (Fig. 3C). Strikingly, depletion of FOXO3 prevented this nocodazole-induced block and substantially increased the fraction of cells in G1 (Fig. 3 C and D). Identical results were obtained in cells expressing FOXO3 shRNA in the absence of nocodazole (Fig. 3D). Furthermore, FOXO3 knockdown blocked DNA synthesis and resulted in fewer S-phase cells, as determined by measurement of DNA content (Fig. 3C) and BrdU incorporation (Fig. 3E). Significantly, the phenotypes of FOXO3-depleted cells were remarkably similar to those of Cdt1 knockdown cells (Fig. 3 B–E). These data suggest that FOXO3 and Cdt1 may operate in a common pathway to control the G1/S transition and DNA replication.
Down-Regulation of Geminin Is Secondary to FOXO3-Mediated Cdt1 Destabilization.
We noted that endogenous geminin levels were also decreased in our FOXO3-depleted cells (Fig. S5A). Evidence from Drosophila and mammalian cells has suggested the existence of an interesting balance between the geminin and Cdt1 proteins, such that geminin depletion leads to Cdt1 down-regulation (6, 21, 22). Accordingly, the reduced geminin protein that we observed could be either a direct consequence of FOXO3 knockdown or a secondary effect of the resulting decrease in Cdt1. To distinguish between these possibilities, we compared the checkpoint activation, DNA content, and proliferation of geminin knockdown cells with FOXO3 knockdown cells. Consistent with a previous report (23), Cdt1 protein was reduced following geminin knockdown (Fig. S5A). However, we observed activation of H2AX and Chk2 only in geminin-depleted cells and not in FOXO3-depleted cells (Fig. S5A). Geminin knockdown also decreased cell proliferation, but to a lesser extent than did FOXO3 knockdown (Fig. S5B). FACS analysis and BrdU incorporation measurements revealed a significant accumulation of S-phase cells following geminin depletion (Fig. S5 C and D). Importantly, geminin knockdown resulted in the accumulation of cells showing >4N DNA content (Fig. S5E), in line with previous reports showing that DNA rereplication occurs in the absence of geminin (23, 24). In contrast, FOXO3-depleted cells showed no signs of DNA rereplication (Fig. S5E). These data suggest that the reduced geminin protein in FOXO3-depleted cells is likely a consequence of Cdt1 destabilization.
Discussion
Genomic integrity requires that chromosomal DNA replicate only once during a single cell cycle. Therefore, the re-establishment of the pre-RC must be suppressed during the S, G2, and M phases. A key aspect of this suppression involves tight and multifaceted regulation of the replication licensing factor Cdt1. Cdt1 is degraded after the initiation of S phase but accumulates during mitosis and G1. However, the signaling pathways and molecular machinery underlying Cdt1 accumulation remain largely unknown. Our study has demonstrated a unique function for FOXO3 in the G1/S transition and during cell proliferation that involves its binding to Cdt1. Surprisingly, endogenous Cdt1 levels were decreased in FOXO3-depleted cells due to impaired Cdt1 protein stability. Although we attempted to determine during which phase FOXO3 regulates Cdt1 levels, we encountered technical difficulties in inducing endogenous Cdt1 down-regulation by FOXO3 knockdown. The knockdown experiment either by transient transfection of siRNA or by lentiviral shRNA infection, in combination with cell cycle synchronization protocol, failed to destabilize Cdt1. This failure may stem from inadequate timing to allow siRNA or shRNA to work properly. Indeed, we found that the gene dose of FOXO3 has a pivotal influence on steady-state levels of Cdt1 and likely on the subsequent biological consequences resulting from FOXO3 knockdown. Our comparison of DNA damage checkpoint regulators (phosphorylated levels of Chk2, p53, and H2AX) in FOXO3-depleted cells with cells exposed to UV damage failed to detect activation of these molecules in FOXO3-depleted cells. However, we cannot exclude the possibility that other unidentified DNA damage signaling may have been up-regulated in our FOXO3 knockdown cells, which in turn could activate signals leading to Cdt1 proteolysis.
Our examination of the molecular mechanisms underlying FOXO3-dependent Cdt1 stabilization showed that Cdt1 ubiquitination was increased in FOXO3 knockdown cells. It has been proposed that Cdt1 is targeted for ubiquitination by Cul4-DDB1 in S phase and in response to DNA damage (15, 17, 25) and that PCNA is a cofactor for this process (13, 15, 25). Consistent with these reports, we found that individual depletion of each of these molecules in our FOXO3 knockdown cells impaired Cdt1 proteolysis. In contrast, knockdown of Skp2 or APC/CCdh1 in FOXO3-depleted cells failed to prevent the decrease in Cdt1 protein. Furthermore, the physical association between Cdt1 and DDB1 or PCNA was disrupted in the presence of ectopically expressed FOXO3, suggesting that FOXO3 positively regulates Cdt1 protein stability by attenuating Cdt1 interaction with DDB1 and/or PCNA.
The biological consequences of FOXO3 knockdown were somewhat unanticipated. FOXO3-depleted cells showed significantly decreased proliferation along with a modest but significant increase in G1 cells and a reduction in S-phase cells. Intriguingly, upon nocodazole treatment, FOXO3 knockdown failed to block cells in G2/M and instead caused cells to accumulate in G1. BrdU incorporation studies revealed that FOXO3 knockdown profoundly attenuated DNA synthesis. Most importantly, the cell cycle defects caused by FOXO3 depletion were remarkably similar to those observed after Cdt1 knockdown.
FOXO3 knockdown also diminished geminin expression, most likely because FOXO3 depletion decreases Cdt1 levels. We concluded that FOXO3 had no direct effect on geminin stability because, whereas geminin depletion activated H2AX and Chk2-p53, FOXO3 knockdown did not. Furthermore, although geminin knockdown increased the proportion of cells in S phase and only mildly affected proliferation, FOXO3 knockdown profoundly attenuated cell division. Importantly, geminin depletion results in the accumulation of aneuploid cells, whereas our knockdown of FOXO3 failed to generate any cells with DNA content over 4N. Altogether, FOXO knockdown does not phenocopy geminin knockdown, thus excluding the possibility that FOXO regulates geminin.
In cell culture-based systems, FOXO family members all direct the transcription of common target genes and thus appear to have very similar biological functions. However, characterization of FOXO knockout mice has revealed unique roles for particular FOXO proteins (26). Knockout mice lacking a single FOXO factor are not tumor-prone (except FOXO1 KO, which is embryonic lethal), a surprising result given that PI3K/Akt signaling, which operates upstream of FOXO, plays a crucial role in tumorigenesis (27, 28). Moreover, conditional knockout mice lacking three FOXO family members (FOXO1/3/4; FOXO TKO mice) unexpectedly show only lineage-restricted tumor phenotypes (29). Furthermore, gene expression profiling and in silico promoter analyses have revealed that transcriptional targets of FOXO factors are both cell type-specific and tissue-dependent (26). For example, p27 is a FOXO target that is down-regulated in thymocytes of FOXO TKO mice but not in endothelial cells (30). In our study, shRNA depletion of FOXO3 did not compromise p27 or cyclin D1 expression at either the mRNA or the protein level (Fig. S6 A and B). In fact, both p27 and cyclin D1 were originally identified and characterized as FOXO target genes using FOXO-TM (a constitutively active form of FOXO members), which are deficient in phosphorylation by Akt (30, 31). Data related to the regulation of these genes under nonstress conditions by endogenous FOXO members are absent. Therefore, one has to be cautious when using these targets to interpret the biological functions of FOXO (i.e., as negative regulators of cell cycle progression). In contrast to previous reports where FOXO members have been suggested to be negative cell cycle regulators (28, 30, 31).
Our results are in line with several studies of the effects of FOXOs on tumorigenesis. Elevated FOXO3 expression is associated with an adverse prognosis in cases of acute myeloid leukemia (AML) exhibiting normal cytogenetics (32), and genetic ablation of FoxO3 reduces disease burden in a murine model of chronic myeloid leukemia (33). Another recent study has revealed that FOXO inhibition triggers myeloid cell maturation and AML cell death (34). It will be interesting to elucidate if FOXO-mediated Cdt1 destabilization accounts for the negative impact of FOXO depletion on cell survival.
Materials and Methods
Cell Culture and Reagents.
MCF-7, HEK293T, H1299, and U2OS cells were cultured in Dulbecco's modified Eagle's medium (GIBCO) supplied with 10% (vol/vol) FBS, 100 units/mL penicillin G sodium, and 100 μg/mL streptomycin (GIBCO). Cells were grown in an atmosphere of 5% CO2 and at 37 °C. Other reagents were obtained from the following sources: MG132 (Boston Biochem), BrdU (Sigma), anti-BrdU (Becton Dickinson), nocodazole (Sigma), imidazole (Sigma), and glutathione-agarose (Sigma). Deletion forms of Myc-Cdt1 constructs and the GST-Cdt1 construct for Cdt1 antibody generation were gifts from Xiaohua Wu (Department of Molecular and Experimental Medicine, The Scripps Research Institute, La Jolla, CA). The HA-DDB1 construct was kindly provided by Yue Xiong (Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, NC).
Western Blotting.
For Western blotting, whole-cell lysates were prepared with Triton lysis buffer [1% or 0.5% triton, 50 mM Tris (pH 7.4), 100 mM NaCl, 2 mM EDTA, 10 mM NaF, 40 mM β-glycerophosphate, 1 mM Na4VO3] or Laemmli lysis buffer [2% SDS, 0.1 M DTT, 0.06 M Tris⋅HCl (pH 6.8), and 10% glycerol].
Immunoprecipitation.
293T cells expressing indicated constructs were cultured for 48 h, and whole-cell lysates were prepared with triton lysis buffer. Anti-Flag M2 affinity gel (Sigma) and anti–c-Myc mouse monoclonal antibody (9B11; Cell Signaling) were used for immunoprecipitation. The bound proteins were eluted by boiling in SDS sample buffer and were detected by Western blotting.
Propidium Iodide Staining and BrdU Pulse Staining.
For cell cycle analyses, MCF-7 cells were fixed with 70% ethanol and then stained with 20 μg/mL propidium iodide (PI) and 200 μg/mL RNase A (Sigma), followed by FACS analysis. Pulse labeling of cells with BrdU (10 μM) was performed following the manufacturer's recommended protocol (Beckton Dickinson; 347580). BrdU-positive cells were detected according to standard protocol.
RNA Interference.
We used the following shRNA sequences: control—5′-GCAAAGAAGGCCACTACTATA-3′; FOXO3-1—5′-GCACAACCTGTCACTGCAT-3′; FOXO3-2—5′-GGAGCTTGGAATGTGACAT-3′; Cdt1-1—5′-GCGCAATGTTGGCCAGATCAA-3′; Cdt1-2—5′-GTACCCCCGAGGCCCCAGA-3′; geminin—5′-TGCCAACTCTGGAATCAAA-3′; Cul4A (35); Cul4B—5′-GCAGCAGTGGATCGAATATAT-3′; DDB1 (36); PCNA (37); Cdh1 (16); and Skp2 (38). shRNA oligos were cloned into the pLV-H1-EF1α lentiviral vector. Lentiviruses were generated according to the manufacturer's protocol (Biosettia).
Statistical Analysis.
Results are reported as mean ± SD of three or more independent experiments. Comparisons were performed with a two-tailed paired Student's t test.
Supplementary Material
Acknowledgments
We thank Dr. Xiaohua Wu (Department of Molecular and Experimental Medicine, The Scripps Research Institute) and Dr. Yue Xiong (Lineberger Comprehensive Cancer Center, University of North Carolina) for reagents and discussions; Jinhong Gao for excellent technical support; and Dr. Mary Saunders for insightful scientific editing. This study was supported by China-Canada Collaborative Research National Natural Science Foundation of China Grant 30700130 (to H.Y.); a Canadian Institutes of Health Research grant (to T.W.M.); the National Basic Research Program of China 973 Program Grant 2009CB522202 (to H.Y.); and Ministry of Education of China 111 Project B06016.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203210109/-/DCSupplemental.
References
- 1.Diffley JF. Once and only once upon a time: Specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert DM. Making sense of eukaryotic DNA replication origins. Science. 2001;294:96–100. doi: 10.1126/science.1061724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bielinsky AK. Replication origins: Why do we need so many? Cell Cycle. 2003;2:307–309. [PubMed] [Google Scholar]
- 4.Wyrick JJ, et al. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: High-resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- 5.McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 6.Wohlschlegel JA, et al. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 7.Tada S, Li A, Maiorano D, Méchali M, Blow JJ. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Zhao Q, Liao R, Sun P, Wu X. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Biol Chem. 2003;278:30854–30858. doi: 10.1074/jbc.C300251200. [DOI] [PubMed] [Google Scholar]
- 9.Liu E, Li X, Yan F, Zhao Q, Wu X. Cyclin-dependent kinases phosphorylate human Cdt1 and induce its degradation. J Biol Chem. 2004;279:17283–17288. doi: 10.1074/jbc.C300549200. [DOI] [PubMed] [Google Scholar]
- 10.Sugimoto N, et al. Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding. J Biol Chem. 2004;279:19691–19697. doi: 10.1074/jbc.M313175200. [DOI] [PubMed] [Google Scholar]
- 11.Ishii T, et al. Proliferating cell nuclear antigen-dependent rapid recruitment of Cdt1 and CRL4Cdt2 at DNA-damaged sites after UV irradiation in HeLa cells. J Biol Chem. 2010;285:41993–42000. doi: 10.1074/jbc.M110.161661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sansam CL, et al. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 2006;20:3117–3129. doi: 10.1101/gad.1482106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higa LA, et al. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle. 2006;5:1675–1680. doi: 10.4161/cc.5.15.3149. [DOI] [PubMed] [Google Scholar]
- 14.Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Senga T, et al. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J Biol Chem. 2006;281:6246–6252. doi: 10.1074/jbc.M512705200. [DOI] [PubMed] [Google Scholar]
- 16.Sugimoto N, et al. Identification of novel human Cdt1-binding proteins by a proteomics approach: Proteolytic regulation by APC/CCdh1. Mol Biol Cell. 2008;19:1007–1021. doi: 10.1091/mbc.E07-09-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- 18.Kondo T, et al. Rapid degradation of Cdt1 upon UV-induced DNA damage is mediated by SCFSkp2 complex. J Biol Chem. 2004;279:27315–27319. doi: 10.1074/jbc.M314023200. [DOI] [PubMed] [Google Scholar]
- 19.Yoshida K, Inoue I. Regulation of geminin and Cdt1 expression by E2F transcription factors. Oncogene. 2004;23:3802–3812. doi: 10.1038/sj.onc.1207488. [DOI] [PubMed] [Google Scholar]
- 20.Takeda DY, Parvin JD, Dutta A. Degradation of Cdt1 during S phase is Skp2-independent and is required for efficient progression of mammalian cells through S phase. J Biol Chem. 2005;280:23416–23423. doi: 10.1074/jbc.M501208200. [DOI] [PubMed] [Google Scholar]
- 21.Saxena S, Dutta A. Geminin–Cdt1 balance is critical for genetic stability. Mutat Res. 2005;569(1–2):111–121. doi: 10.1016/j.mrfmmm.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 22.Ballabeni A, et al. Human geminin promotes pre-RC formation and DNA replication by stabilizing CDT1 in mitosis. EMBO J. 2004;23:3122–3132. doi: 10.1038/sj.emboj.7600314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melixetian M, et al. Loss of geminin induces rereplication in the presence of functional p53. J Cell Biol. 2004;165:473–482. doi: 10.1083/jcb.200403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu W, Chen Y, Dutta A. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol Cell Biol. 2004;24:7140–7150. doi: 10.1128/MCB.24.16.7140-7150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J, Xiong Y. An evolutionarily conserved function of proliferating cell nuclear antigen for Cdt1 degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA damage. J Biol Chem. 2006;281:3753–3756. doi: 10.1074/jbc.C500464200. [DOI] [PubMed] [Google Scholar]
- 26.Hosaka T, et al. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci USA. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzivion G, Hay N. PI3K-AKT-FoxO axis in cancer and aging. Biochim Biophys Acta. 2011;1813:1925. doi: 10.1016/j.bbamcr.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 28.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 29.Paik JH, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stahl M, et al. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J Immunol. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt M, et al. Cell cycle inhibition by FoxO forkhead transcription factors involves downregulation of cyclin D. Mol Cell Biol. 2002;22:7842–7852. doi: 10.1128/MCB.22.22.7842-7852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santamaría CM, et al. High FOXO3a expression is associated with a poorer prognosis in AML with normal cytogenetics. Leuk Res. 2009;33:1706–1709. doi: 10.1016/j.leukres.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 33.Naka K, et al. TGF-beta-FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature. 2010;463:676–680. doi: 10.1038/nature08734. [DOI] [PubMed] [Google Scholar]
- 34.Sykes SM, et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 2011;146:697–708. doi: 10.1016/j.cell.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lovejoy CA, Lock K, Yenamandra A, Cortez D. DDB1 maintains genome integrity through regulation of Cdt1. Mol Cell Biol. 2006;26:7977–7990. doi: 10.1128/MCB.00819-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishitani H, et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 2006;25:1126–1136. doi: 10.1038/sj.emboj.7601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abbas T, et al. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Méndez J, et al. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol Cell. 2002;9:481–491. doi: 10.1016/s1097-2765(02)00467-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.