Abstract
During sexual reproduction, one-half of the genetic material is deposited in gametes, and a complete set of chromosomes is restored upon fertilization. Reduction of the genetic information before gametogenesis occurs in meiosis, when cross-overs (COs) between homologous chromosomes secure an exchange of their genetic information. COs are not evenly distributed along chromosomes and are suppressed in chromosomal regions encompassing compact, hypermethylated centromeric and pericentromeric DNA. Therefore, it was postulated that DNA hypermethylation is inhibitory to COs. Here, when analyzing meiotic recombination in mutant plants with hypomethylated DNA, we observed unexpected and counterintuitive effects of DNA methylation losses on CO distribution. Recombination was further promoted in the hypomethylated chromosome arms while it was inhibited in heterochromatic regions encompassing pericentromeric DNA. Importantly, the total number of COs was not affected, implying that loss of DNA methylation led to a global redistribution of COs along chromosomes. To determine by which mechanisms altered levels of DNA methylation influence recombination—whether directly in cis or indirectly in trans by changing expression of genes encoding recombination components—we analyzed CO distribution in wild-type lines with randomly scattered and well-mapped hypomethylated chromosomal segments. The results of these experiments, supported by expression profiling data, suggest that DNA methylation affects meiotic recombination in cis. Because DNA methylation exhibits significant variation even within a single species, our results imply that it may influence the evolution of plant genomes through the control of meiotic recombination.
Keywords: epigenetic, chromatin, epigenetic recombinant inbred lines, met1-3
Regulation of meiotic recombination, as with other essential chromosomal activities like transcription and replication, depends on both DNA sequence and chromatin properties (1, 2). Although regulatory aspects of meiotic recombination have been studied in great detail, it is still not well understood how chromatin structure influences the frequency and distribution of recombination events, as reflected by the final number and distribution of cross-overs (COs) along chromosomes. Biased chromosomal positioning of COs has been recognized for many years; indeed, COs are most likely to occur in euchromatic chromosomal arms, distal to the recombinationally suppressed pericentromeric heterochromatin. These two chromatin compartments are characterized by differences in the abundance of genes and transposable elements (TEs). TEs accumulate in pericentromeric regions, whereas genes are enriched in distal euchromatin. Because suppressive epigenetic marks are primarily directed at silencing TEs, these two chromatin types also differ in their epigenetic signatures. Pericentromeric chromatin is enriched in the methylation of histone H3 at lysine 9 (H3K9me) and encompasses hypermethylated DNA. In contrast, distal chromatin exhibits active marks such as acetylation of histones, methylation of histone H3 at lysine 4 (H3K4me), and low levels of DNA methylation. For example in Arabidopsis, DNA methylation at centromeric regions can reach up to 80% of cytosines compared with <20% in distal euchromatin (3, 4). Noticeably, the meiotic recombination rate (MRR) is lower in highly condensed, transcriptionally inert heterochromatin than in actively transcribed and structurally relaxed euchromatin (5, 6), and it typically increases with distance from the centromere (7). This observation led to the hypothesis that double-strand breaks (DSBs) occur more readily in open euchromatic regions (8, 9). Euchromatic marks have been reported recently to have an impact on meiotic recombination. For example, H3K4me was associated with the presence of hot spots for recombination in mammalian chromosomes (10). In Arabidopsis, elevated histone H3 acetylation, triggered by overexpression of a histone acetyltransferase or by an inhibitor of histone deacetylases, was associated with defects in meiosis and changes in CO number (11).
These results imply a regulatory role for chromatin in meiotic recombination. However, it is not known whether DNA methylation—the best-characterized epigenetic mark and an essential factor for stabilization of mammalian and plant heterochromatin—is responsible for the suppression of recombination in heterochromatic pericentromeres. Therefore, it remains to be determined whether specific, well-characterized changes in DNA methylation would affect recombination and whether these effects would differ in euchromatic vs. heterochromatic chromosomal regions. The model plant Arabidopsis thaliana is well suited to address these questions because regulatory aspects of its chromatin structure are now relatively well understood at the molecular level, including the decisive influence of DNA methylation on transcriptional activity (12) and on defining different chromatin types (13, 14). In plants, DNA methylation is found at cytosines in CG and non-CG sequence contexts, depending on the bases neighboring methylated C (mC). Faithful maintenance of mCGs through DNA replication cycles is conferred by the maintenance methyltransferase MET1. MET1 acts at replication forks, recognizing newly synthesized, hemimethylated DNA and copying methylation patterns from the template strand. MET1-mediated propagation of mCG patterns occurs during somatic plant development and gametogenesis, promoting accurate transgenerational epigenetic inheritance (15). Non-CG methylation can be divided further into CHG and CHH methylation (H equals A, T, or C), which are propagated by the plant-specific chromo methyltransferase 3 (CMT3) and domains rearranged methyltransferase 2 (DRM2), respectively (16). Deficiencies in propagating mCG patterns due to mutations in the MET1 gene, or in the DECREASE IN DNA METHYLATION1 (DDM1) gene encoding a chromatin remodeling factor, lead to heritable alterations of mCG distribution that cannot be reversed simply by reintroduction of MET1 or DDM1 function (15, 17, 18). It was shown that such changes can be inherited over several generations, even in the presence of a functional MET1 or DDM1 (18–21). Moreover, deficiencies in mCGs observed in met1 and ddm1 mutants trigger the redistribution of other chromatin marks and lead to heritable alterations in chromatin properties (12, 14, 22).
We sought to examine whether loss of DNA methylation can alter recombination rates by analyzing mapping populations derived from parents with variable methylation levels and distribution. For this approach, we first crossed a wild-type (WT) plant with a hypomethylated mutant (met1) and analyzed MRRs in the F2 segregating population, which reflects CO distribution in F1 meiosis. We observed significant changes in CO distribution in comparison with the control cross between two WT parents. Surprisingly, we found an additional increase in COs in distal chromatin and CO suppression in pericentromeric regions. To examine whether MRR gradients were altered due to the met1 mutation itself or due to DNA methylation changes in cis, we examined MRRs in two additional mapping populations derived from two different epigenetic recombinant inbred lines (epiRILs). The epiRILs are themselves derived from a cross between met1 mutant and WT, but were inbred for eight generations in the presence of the WT homozygous MET1 gene (20). In other words, epiRILs share the same WT genetic background but differ in their DNA methylation and, thus, epigenetic landscape. Mosaic methylation patterns were shown directly by genome-wide DNA methylation profiling that assigns the parental origin of chromosomal segments based on the inheritance of DNA methylation patterns (20). Thus, an epiRIL epigenome is composed of a particular mosaic of WT- and met1-derived chromosomal regions. However, certain changes in DNA methylation during epiRIL inbreeding have been observed (19, 20). Using this unique material, we analyzed MRRs along this epigenetic mélange of chromosomes. In this experimental setup, very drastic changes were also observed, notably on chromosome 2. Thus, we concluded that such MRR changes were independent of the met1 mutation. To determine whether the mechanisms of MRR alteration can be linked in cis to the DNA methylation levels or whether they act in trans by transcriptional activation of genes involved in the recombination process, we examined genome-wide transcript levels for one epiRIL used for MRR determination. We found no transcriptional misregulation of genes encoding known factors involved in meiotic recombination. Therefore, we propose that changes in the level of DNA methylation can affect CO distribution in cis and that hypomethylated pericentromeric regions on some chromosomes can retain their hyporecombinogenic properties, even after several generations of inbreeding.
Results
Distinct Effects of DNA Methylation on Recombination in Euchromatin vs. Heterochromatin.
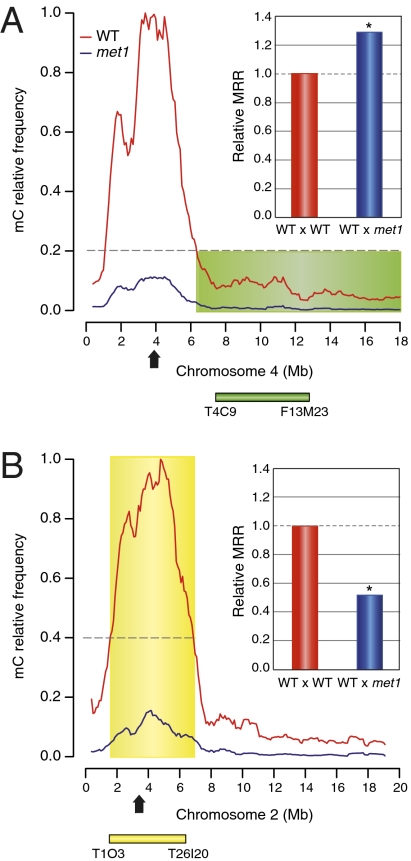
We crossed a WT plant and a met1 mutant (met1-3 allele), both of the Columbia accession, to a WT plant of the Landsberg accession and obtained two F2 populations, hereafter named WTxWT and WTxmet1, respectively. The F2 plants were analyzed for the segregation of genetic markers specific for each parental accession. Initially, two intervals were analyzed for meiotic recombination frequencies by using insertion/deletion (INDEL) PCR markers. One 5.5 megabases (Mb) interval is located on the long arm of chromosome 4, and the other is a 4.8-Mb interval spanning the centromere on chromosome 2 (Fig. 1). These two intervals were selected for their contrasting levels of DNA methylation in WT plants, one being hypermethylated on chromosome 2 and the other having a low methylation level on chromosome 4 (Fig. 1). The recombination rates were calculated at both intervals, and the results obtained for the WTxmet1 population were normalized to the MRR of the WTxWT population (set at 1.0). For the euchromatic interval, we observed that the WTxmet1 MRR increased to 1.29 (P < 0.02; Fig. 1A). Conversely, the WTxmet1 MRR decreased to 0.50 for the heterochromatic interval (P < 0.05; Fig. 1B). These initial observations suggested that meiotic CO distribution on WT chromosome pairs differs when one chromosome is hypomethylated. Moreover, the loss of DNA methylation could have either a positive or a negative impact on MRRs within euchromatin or heterochromatin, respectively.
Fig. 1.
The met1 mutation effect on MRRs depends on chromatin states. The MRR in plants derived from the cross between WT (Landsberg accession) and met1 mutant (Columbia accession) was analyzed at two intervals with contrasted DNA methylation levels. The methylcytosine (mC) density plots were obtained from bisulfite sequencing data (3, 4) by using a 100,000-base sliding window with 10-base steps. The WT Col-0 (red curve) and met1-3 (blue curve, met1) mC densities are relative to the highest methylation level detected in WT Col-0 plants. Based on the relative DNA methylation in WT plants, we defined euchromatic and heterochromatic intervals as chromosomal regions having <20% (green shading) or >40% (yellow shading) relative mC levels in WT, respectively. F2 plants originating from the cross between WT and WT (WTxWT) and WT and met1 (WTxmet1) were genotyped for INDEL markers at two intervals as shown below the mC density graphs (green box, euchromatic interval; yellow box, heterochromatic interval). MRR was calculated from 192 F2 plants originating from two different F1 plants for both WTxWT and WTxmet1 populations. WTxmet1 MRR is compared with WTxWT MRR set at 1 (red bar, WTxWT; blue bar, WTxmet1). Arrow, centromere. (A) MRR is higher in WTxmet1 at the euchromatic interval located on the long arm of chromosome 4 (green bar). *P < 0.02, t test. (B) MRR is lower in WTxmet1 at the heterochromatic interval spanning the centromeric region on chromosome 2 (yellow bar). *P < 0.05, t test.
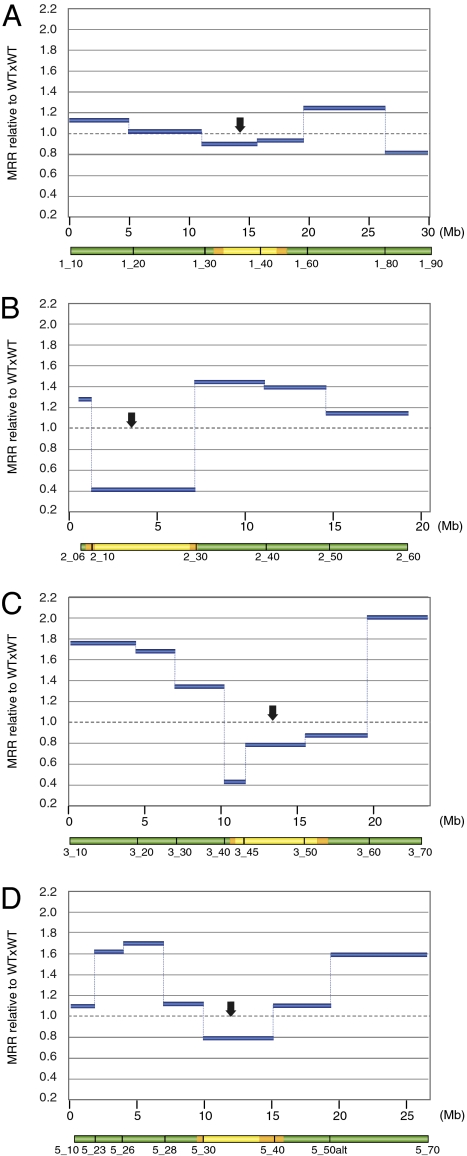
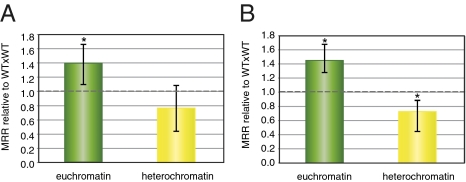
To better understand whether the contrasting effects of DNA methylation observed within these two intervals reflect a general trend, we analyzed genome-wide MRRs further in the same populations using SNP markers distributed across chromosomes 1, 2, 3, and 5 (Table S1 and Fig. S1). Because chromosome 4 has a heterochromatic knob region in the Columbia accession that is absent in Landsberg, we excluded this chromosome from these analyses to avoid a possible additional variable influencing the MRR of this chromosome pair. We used 29 KASP genotyping markers (KBioscience: Materials and Methods) along the four remaining chromosomes in both WTxWT and WTxmet1 populations, analyzing 302 chromosomes from WTxWT F2 and 364 chromosomes from WTxmet1 F2 plants for all 29 markers. We detected 792 and 1,050 COs, respectively. Comparing the WTxmet1 population to WTxWT, we again observed a decrease in MRRs in the heterochromatic chromosomal regions spanning the centromeric and pericentromeric DNA (Fig. 2 and Fig. S1); this decrease reached 10% on chromosome 1 (Fig. 2A), and 21% on chromosome 5 (Fig. 2D). The effect was more pronounced for chromosomes 2 and 3, where we observed 59% and 57% reductions in MRRs, respectively (Fig. 2 B and C). In euchromatic regions, elevated levels of MRR were observed relative to the WTxWT population for all four chromosomes of the WTxmet1 population. The increase was lower on chromosomes 1 and 2, where it reached 124% and 144%, respectively, than on chromosomes 3 and 5, where it reached 200% and 170% (Fig. 2). The only exception was a terminal region of chromosome 1 (Fig. 2A). To calculate the overall effect of the methylation losses due to the met1 mutation on MRR, we used MRR data obtained for the four chromosomes, dividing them into euchromatic and heterochromatic regions depending on their DNA methylation levels (Fig. S1 and Table S1). In euchromatin, the relative MRR in WTxmet1 increased to 140% (P < 0.01), whereas in heterochromatin it decreased to 80% (Fig. 3A). However, the decrease in heterochromatin was only significant if we considered only chromosomes 2 and 3, where the MRR decreased to 73% (P < 0.01; Fig. 3B), indicating that the impact of DNA methylation loss differed between different chromosomes. In conclusion, the genome-wide analyses of MRRs further supported our initial observation that loss of DNA methylation has a stimulatory effect on MRRs in euchromatic chromosomal regions, whereas it has a suppressive effect within heterochromatic areas. These altered rates were especially pronounced for chromosomes 2 and 3.
Fig. 2.
Contrasted effect of the met1 mutation on MRR along Arabidopsis chromosomes. F2 plants originating from the cross between WT and met1 (WTxmet1 population) were genotyped by using KASP markers. Intervals defined by these markers are represented below the graphs and color-coded to indicate their relative mC level in WT (green box, 0–20% mC; orange box, 20–40% mC; yellow box, >40% mC; see Fig. S1 for details). Calculated MRR was normalized to the data obtained with the WTxWT control cross. Relative MRR (WTxmet1 relative to WTxWT) is represented along Arabidopsis chromosomes (arrow, centromere; Mb, coordinates in Mb; dotted lines, WTxWT MRR set at 1). (A) Chromosome 1. (B) Chromosome 2. (C) Chromosome 3. (D) Chromosome 5.
Fig. 3.
MRR in euchromatic vs. heterochromatic intervals. MRR for the WTxmet1 population is shown relative to the data obtained with the control cross (WTxWT). (A) MRR for intervals located on chromosomes 1, 2, 3, and 5 according to their euchromatic (green bar) or heterochromatic (yellow bar) content. (B) MRR for intervals located on chromosomes 2 and 3. *P = 0.01, χ2.
We examined whether the observed changes of MRRs induced by the met1 mutation could be correlated with the alteration in transcription caused by this mutation by superimposing transcription (3) and recombination changes (this study) along all four chromosomes (Fig. S2). In euchromatin regions, we could not find any correlation between the density of activated or repressed transcription units and changes in recombination rates. In contrast, there was a massive up-regulation of transcription in met1-derived heterochromatin, which did not prevent suppression of recombination (Fig. S2).
Effects of DNA Methylation on MRRs Are Independent of the met1 Mutation.
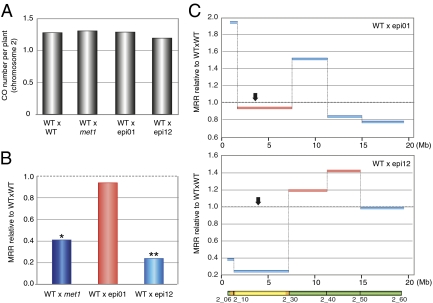
To determine whether the observed changes in MRRs are due to the met1 mutation itself or whether they are more directly associated with changes in DNA methylation, we examined MRRs using epiRILs (20). Based on their DNA methylation profiles, we selected two contrasting lines (epi01 and epi12) displaying different arrangements of met1 or WT Columbia chromosomal segments and, thus, different DNA methylation patterns (20). We crossed each of the two selected epiRILs (of the Columbia accession) to WT plants of the Landsberg accession to obtain two F2 populations, hereafter named WTxepi01 and WTxepi12. We performed the recombination analyses using KASP markers as described above and analyzed 340 WTxepi01 F2 chromosomes and 336 WTxmet1 F2 chromosomes, detecting 888 and 900 COs, respectively.
To better understand whether CO distribution was affected by the loss of DNA methylation, we analyzed the total number of COs for all F2 populations analyzed. The total number of COs per chromosome was not statistically different from the WTxWT control (Fig. 4A and Fig. S3), indicating that loss of DNA methylation affects the distribution of COs but not their number. We analyzed recombination at centromeric intervals on chromosome 2, where the most extreme suppression in the WTxmet1 population was previously observed (Fig. 2B). Noticeably, the chromosome 2 pericentromeric regions in epi01 and epi12 were derived from WT and met1, respectively, thereby providing a direct contrast of the inherited mosaic epigenomes (Fig. S4). The MRR was not significantly affected in WTxepi01 (MRR reduced to 90%), whereas it was strongly suppressed in WTxepi12 compared with WTxWT (MRR reduced to 22%; P < 0.0001) (Fig. 4B). Thus, on chromosome 2, the epiRIL heterochromatic regions retained the distinct parental properties, which continued to impinge on meiotic recombination efficiency despite eight generations of inbreeding. A similar trend was observed for the heterochromatic region of chromosome 5 (Fig. S5). However, only slightly decreased recombination rates were observed for chromosomes 1 and 3 in both epiRIL populations, irrespective of the parental origin of their heterochromatic regions (Fig. S5). Together, these data suggest that the suppressive effect of the met1 mutation on MRRs in heterochromatin can also be observed and inherited independently of the met1 mutation itself; however, the degree of this suppression may vary between different chromosomes.
Fig. 4.
The combination of DNA hypomethylation and inbreeding affects MRR in epiRILs. F2 plants originating from the cross between WT and epi01 (WTxepi01) and WT and epi12 (WTxepi12) were genotyped for SNPs by using KASP markers. (A) Total number of COs per plant for chromosome 2 for the four F2 populations. (B) MRR at the centromeric region of chromosome 2. MRR obtained from F2 plants originating from two different F1 plants is presented relative to WTxWT MRR and is compared with the data obtained from the cross between WT and met1 mutant (WTxmet1, blue bar). The bars are color-coded to indicate the global methylation status of the parental epiRIL at the analyzed heterochromatic intervals (light blue, met1-like methylation; light red, WT-like methylation). Dotted lines mark the WTxWT MRR set at 1. *P < 0.001; **P < 0.0001 (ANOVA). (C) Relative MRR (WTxepi01 relative to WTxWT, Upper; WTxepi12 relative to WTxWT, Lower) is represented along chromosome 2 (arrow, centromere; Mb, coordinates in Mb; gray dotted lines, WTxWT MRR set at 1).
Next, we investigated whether MRRs in euchromatic chromosomal areas are also influenced by their parental origin. Although an increase in MRR was detected in the chromosome 2 euchromatic regions of the WTxmet1 population (7- to 19-Mb interval; Fig. 2B), the MRR ranged from 0.75 to 1.5 across the met1-derived segment in the WTxepi01 population (Fig. 4C). In the WTxepi12 population, this interval is derived from the WT Columbia epigenome, but the MRR was unexpectedly increased relative to the WTxWT MRR (1.35–1.6; Fig. 4C). On chromosome 5, we detected higher MRRs in WTxmet1 than in WTxWT—for example, on the right arm (20- to 26-Mb interval, 1.58-fold compared with WTxWT; Fig. 2D). This interval spans a Columbia-derived region in both epi01 and in epi12, with the exception of a short met1 segment in epi12 (Fig. S4). We observed a modest increase in the MRR in WTxepi12 but not in WTxepi01 (Fig. S4).
The met1 mutation might also affect MRRs in trans through misregulation of genes encoding factors influencing MRRs. To address this possibility, we analyzed transcription profiling data of met1-3 (3) for transcript levels of genes encoding proteins implicated in meiotic recombination (Table S2). In addition, to determine whether any of these genes were misregulated in epiRILs, we performed a genome-wide transcription analysis of epi12. We found no significant change in expression of these genes in either dataset of transcription profiling (Table S2) (3). Therefore, the formation and persistence of a met1-triggered epiallele encoding recombination factor acting in trans seems to be doubtful. As a consequence, the observed effects of DNA methylation loss on MRRs are likely to be due to cis effects of chromosomal DNA hypomethylation, which is known to influence the distribution of other epigenetic marks and structural chromatin properties (12, 14).
Discussion
By analyzing recombination in an F2 population originating from a met1 mutant in which the properties and stability of the epigenome are strongly affected (12, 14), we have demonstrated that the met1 mutation affects the CO landscape in Arabidopsis. In general, the mutation tends to yield stimulatory and suppressive effects on MRRs within euchromatin and heterochromatin, respectively. These general effects are clearest on chromosome 2; however, this trend is much less pronounced on chromosomes 1 and 5, indicating chromosome-specific variations. In euchromatin, we also observed variations in the level of MRR enhancement particularly on the long arm of chromosome 1. Because these variations could not be explained by variations in transcription in the met1 mutant, they imply that other local chromosomal features influence recombination. In experiments using epigenetic recombinant lines derived from WT and met1 (epiRILs epi01 and epi12), for which we have previously characterized mosaic methylation maps, we observed that the met1-specific effects on recombination were again clearest for chromosome 2 and also observable for chromosome 5. However, for chromosomes 1 and 3, the trend in recombination changes was not consistent with parental chromosomal origin. We believe that the observed variations could be attributed to the stochastic methylation changes well-documented for met1 chromatin (22), which are exaggerated in met1-derived epiRILs (20). Such methylation dynamics results from the activity of the supplementary methylation pathway directed by small RNAs (19) and from suppression of DNA demethylation activities (22). In addition, MRRs in particular regions of epiRIL chromosomes could have been influenced by the epigenetic status of the surrounding areas. Hence, although our epiRIL data support the general notion that loss of DNA methylation affects the global recombination landscape, the met1-derived chromosomal segments inherited through eight generations of epiRIL inbreeding did not affect MRRs as consistently as the met1 mutant directly (Fig. 4C and Fig. S4).
Notably, the met1 mutation did not significantly alter the total number of COs but, rather, led to their redistribution along the chromosomes. This redistribution could either be due to indirect effects of the met1 mutation influencing gene expression (15) or, more directly, to altered DNA methylation and chromatin properties. To distinguish between these two possibilities, we examined MRRs in F2 populations originating from epiRIL parents and transcriptional changes in genes encoding proteins implicated in meiotic recombination. Because the epiRILs maintained the MET1 function yet possessed chromosomal regions with mosaic epigenetic patterns and the transcription of recombination-related genes was unchanged, it can be concluded that the observed CO redistribution in two different epiRILs was caused by altered chromatin properties and not by changes in expression of genes directly involved in the recombination pathway. Further cytogenetic studies would help address the altered chromatin properties in epiRILs.
The finding that hypomethylated euchromatic DNA stimulated MRR points to the possibility that further relaxation of these already decondensed chromosomal parts promotes accessibility for the recombination machinery. This observation is supported by comparable results obtained in another hypomethylated mutant (ddm1) by Melamed-Bessudo and Levy (23). One implication of these findings is that gene body methylation, which is an ancestral property of eukaryotic genomes (24, 25), could protect genome integrity by inhibiting supernumerary COs within exons. On a similar note, it has been shown that the controlled addition of DNA methylation leads to a decrease in meiotic recombination for a particular locus in Ascobulus (26, 27).
The suppressive effect of the loss of DNA methylation on MRR in heterochromatin is less intuitive. Methylation of heterochromatic DNA is responsible for transcriptional silencing of centromeric and pericentromeric repeats and TEs (reviewed in refs. 16 and 28). Alleviation of this transcriptional suppression could lead to transposon mobility and an increasing load of DSBs that interfere with meiotically programmed DSBs and, thus, have an impact on the control of the recombination frequencies (29). Alternatively, a particular degree of heterochromatin compaction in pericentromeres, known to be stabilized by DNA hypermethylation (12, 14, 30), could facilitate chromosome pairing and the formation of synapses in WT meiosis, as well as impinge on the particular timing of this process. In many eukaryotes, including plants, chromosome pairing is tightly linked to the progression of the early steps of meiotic recombination by the process of homology search, which involves the single-end invasion step of meiotic recombination (31). Opening chromosome structure in pericentromeric regions could, perhaps, still allow for more recombination initiation events in these regions. At the same time, however, these early events may lead to undesirable ectopic pairing between repetitive genome regions. Indeed, loss of DNA methylation at tandem repeats in centromeric heterochromatin resulted in incomplete chromosome synapsis in mice (32). Moreover, depletion in methylation of lysine 9 on histone H3 (H3K9me) in spermatocytes with a mutated gene encoding the histone methyltransferase Suv39h delayed synapses and provoked higher frequencies of nonhomologous chromosome interactions in mice (33, 34). In Arabidopsis, chromosome pairing is telomere-led (35), and centromeric regions pair late. Therefore, if hypomethylation would lead to further delay in centromeric regions, pairing could happen after CO designation has already occurred in the arms, thus decreasing CO number in centromeric regions. Notably, in our experimental system, the meiotic recombination was monitored between one hypomethylated chromosome and its normally methylated homolog; thus, the effect of hypomethylation seems to be dominant. Because of the lack of a met1 null allele in another accession than Columbia, we could not examine whether this effect would persist when both homologs are hypomethylated. However, Melamed-Bessudo and Levy (23) could show, using another hypomethylated mutant, that the effect of the ddm1 mutation was similar in homozygous and heterozygous ddm1 meioses.
The influence of DNA methylation on MRR could also be exerted at steps subsequent to early recombination events. For example, CO interference documented in Arabidopsis (36, 37)—possibly together with the CO homeostasis observed so far in yeast and in worms—ensures that one or only a few DSBs will be repaired as COs (38–40). It has been suggested that the limited number of COs and their positions could be regulated by physical properties of chromosomes determined by their chromatin structure (41). If this hypothesis was true, it could be envisaged that met1- or epiRIL-derived chromosomes acquire distinct chromatin states affecting the redistribution of COs to varying degrees on different chromosomes.
Epigenetic diversity, as well as genetic diversity, contributes to plant and animal phenotypic variation (42, 43). Identifying factors influencing meiotic CO frequency are undoubtedly important for refining breeding strategies because hyperrecombinant lines could be selected to facilitate breeding programs (44). Increased MRRs in euchromatic chromosome arms could contribute to accelerated selection for novel haplotypes without the need for very large populations. However, our epiRIL results indicate that the magnitude of the effect of hypomethylation on MRRs may decline over generations, likely because of the accumulation of compensatory epigenetic changes in physical properties of the epiRIL chromosomes. Indeed, compensation mechanisms are established shortly after the genome undergoes hypomethylation stress (14, 19–22).
Our results demonstrate that the role of DNA methylation in regulating meiotic recombination is more complex than previously anticipated. First, the impact of DNA methylation on MRR depends on the epigenetic chromosomal context. Furthermore, after the confrontation of two different parental epigenomes, alteration of the MRR can persist for several generations, independently of the met1 mutation. By combining genome-wide DNA methylation and other epigenomic maps with high-throughput genotyping for determination of meiotic recombination frequencies, it is now possible to address with unprecedented accuracy the remaining questions linked to the influence of chromatin structure on meiotic recombination. Furthermore, detailed analysis of the impact of recently defined, multiple chromatin types (13, 45) will help in the precise dissection of chromatin impact on meiotic recombination. Interestingly, in an identical WT genetic background, the MRR was found to be significantly higher in Arabidopsis male than female meiosis (46–48), suggesting that epigenetic regulation could also be responsible for these differences. Finally, because DNA methylation is subjected to natural variation (48, 49), we propose that it could influence the evolution of plant genomes through the control of meiotic recombination.
Materials and Methods
Plant Materials and Growing Conditions.
The met1-3 mutant (15) and two epiRILs (20) (all in the Columbia background) were crossed with WT plants in the Landsberg accession. As a control, we crossed WT Columbia to Landsberg. All plants were grown in a controlled environment (Percival chamber, 21 °C, 16-h light, 8-h dark). F1 progeny obtained from crosses were grown on soil simultaneously, and seeds were harvested after 4, 5, and 6 wk. To eliminate bias due to age effects on recombination (50), only seeds collected 6 wk after germination were used for genotyping, thus ensuring that the same developmental stage was used for all experiments. From each cross, 192 F2 plants originating from at least 2 F1 plants were used per cross for the INDEL analyses. For the KASP (KBioscience) analyses, at least 144 plants from two different F1 progenies were analyzed (Tables S3 and S4). See SI Materials and Methods and Table S5 for INDEL and KASP genotyping details.
MRR Calculations.
Most markers followed Mendelian expectations; however, we observed rare segregation distortion as reported (51) at similar levels (52). Genotyping data were uploaded into MapDisto (http://mapdisto.free.fr/) for the MRR calculation (Table S1). The markers used for pericentromeric regions analysis were 1–30/1–40, 2–10/2–30, 3–40/3–50, and 5–30/5–40 for chromosomes 1, 2, 3, and 5, respectively.
Transcription Profiling.
RNA was extracted from rosette leaves and analyzed as described (53) by using the Affymetrix GeneChip Arabidopsis Tiling 1.0R array. Data have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE34173).
Supplementary Material
Acknowledgments
We thank all current and past members of the J.P. laboratory for helpful discussions; Cathy Melamed-Bessudo and Avi Levy for sharing unpublished data; Claude Becker, Jörg Hagmann, and Detlef Weigel for kindly providing mC density plots; Wojtek Pawlowski for helpful comments on an earlier version of the manuscript; Taisuke Nishimura for INDEL markers; Mathias Lorieux for advice on MapDisto; Pat King for English editing; and Larissa Broger, Maryline Freyre, and Christian Mégies for technical assistance. The KASP genotyping data were generated at the Genetic Diversity Centre at Eidgenössiche Technische Hochschule Zurich, with the help of Tania Torossi and support from the Swiss Plant Science Web. This paper received the contribution number 261 by the Department of Soil, Plant, Environmental and Animal Production Sciences. This work was supported by Swiss National Science Foundation Grant 31003A-125005 and by the European Commission through “Acquired Environmental Epigenetics Advances: From Arabidopsis to Maize” Project Grant FP7-226477 and RECBREED Consortium Grant FP7-227190.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The expression data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE34173).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120841109/-/DCSupplemental.
References
- 1.Yanowitz J. Meiosis: Making a break for it. Curr Opin Cell Biol. 2010;22:744–751. doi: 10.1016/j.ceb.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ronceret A, Sheehan M, Pawlowski W. Chromosome dynamics in meiosis. In: Verma DPS, Hong Z, editors. Cell Division Control in Plants. Heidelberg: Springer; 2007. pp. 103–124. [Google Scholar]
- 3.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eichten SR, et al. Heritable epigenetic variation among maize inbreds. PLoS Genet. 2011;7:e1002372. doi: 10.1371/journal.pgen.1002372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaut BS, Wright SI, Rizzon C, Dvorak J, Anderson LK. Recombination: An underappreciated factor in the evolution of plant genomes. Nat Rev Genet. 2007;8:77–84. doi: 10.1038/nrg1970. [DOI] [PubMed] [Google Scholar]
- 7.Liu S, et al. Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet. 2009;5:e1000733. doi: 10.1371/journal.pgen.1000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lichten M. Meiotic chromatin: The substrate for recombination initiation. In: Egel R, Lankenau D-H, editors. Genome Dynamics and Stability. Vol 3. Berlin: Springer; 2008. pp. 165–193. [Google Scholar]
- 9.Buard J, Barthès P, Grey C, de Massy B. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 2009;28:2616–2624. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perrella G, et al. Histone hyperacetylation affects meiotic recombination and chromosome segregation in Arabidopsis. Plant J. 2010;62:796–806. doi: 10.1111/j.1365-313X.2010.04191.x. [DOI] [PubMed] [Google Scholar]
- 12.Soppe WJJ, et al. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;21:6549–6559. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roudier F, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tariq M, et al. Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc Natl Acad Sci USA. 2003;100:8823–8827. doi: 10.1073/pnas.1432939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saze H, Mittelsten Scheid O, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet. 2003;34:65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- 16.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kakutani T, Munakata K, Richards EJ, Hirochika H. Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics. 1999;151:831–838. doi: 10.1093/genetics/151.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kakutani T, Jeddeloh JA, Flowers SK, Munakata K, Richards EJ. Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci USA. 1996;93:12406–12411. doi: 10.1073/pnas.93.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teixeira FK, et al. A role for RNAi in the selective correction of DNA methylation defects. Science. 2009;323:1600–1604. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- 20.Reinders J, et al. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 2009;23:939–950. doi: 10.1101/gad.524609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johannes F, et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009;5:e1000530. doi: 10.1371/journal.pgen.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Melamed-Bessudo C, Levy A. Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1120742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 25.Feng S, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colot V, Maloisel L, Rossignol JL. Interchromosomal transfer of epigenetic states in Ascobolus: Transfer of DNA methylation is mechanistically related to homologous recombination. Cell. 1996;86:855–864. doi: 10.1016/s0092-8674(00)80161-0. [DOI] [PubMed] [Google Scholar]
- 27.Maloisel L, Rossignol JL. Suppression of crossing-over by DNA methylation in Ascobolus. Genes Dev. 1998;12:1381–1389. doi: 10.1101/gad.12.9.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reinders J, Paszkowski J. Unlocking the Arabidopsis epigenome. Epigenetics. 2009;4:557–563. doi: 10.4161/epi.4.8.10347. [DOI] [PubMed] [Google Scholar]
- 29.Grelon M, Gendrot G, Vezon D, Pelletier G. The Arabidopsis MEI1 gene encodes a protein with five BRCT domains that is involved in meiosis-specific DNA repair events independent of SPO11-induced DSBs. Plant J. 2003;35:465–475. doi: 10.1046/j.1365-313x.2003.01820.x. [DOI] [PubMed] [Google Scholar]
- 30.Probst AV, Fransz PF, Paszkowski J, Mittelsten Scheid O. Two means of transcriptional reactivation within heterochromatin. Plant J. 2003;33:743–749. doi: 10.1046/j.1365-313x.2003.01667.x. [DOI] [PubMed] [Google Scholar]
- 31.Bozza CG, Pawlowski WP. The cytogenetics of homologous chromosome pairing in meiosis in plants. Cytogenet Genome Res. 2008;120:313–319. doi: 10.1159/000121080. [DOI] [PubMed] [Google Scholar]
- 32.De La Fuente R, et al. Lsh is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells. Nat Cell Biol. 2006;8:1448–1454. doi: 10.1038/ncb1513. [DOI] [PubMed] [Google Scholar]
- 33.Peters AH, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 34.Tachibana M, Nozaki M, Takeda N, Shinkai Y. Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J. 2007;26:3346–3359. doi: 10.1038/sj.emboj.7601767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Armstrong SJ, Franklin FC, Jones GH. Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J Cell Sci. 2001;114:4207–4217. doi: 10.1242/jcs.114.23.4207. [DOI] [PubMed] [Google Scholar]
- 36.Copenhaver GP, Housworth EA, Stahl FW. Crossover interference in Arabidopsis. Genetics. 2002;160:1631–1639. doi: 10.1093/genetics/160.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mézard C, Vignard J, Drouaud J, Mercier R. The road to crossovers: Plants have their say. Trends Genet. 2007;23:91–99. doi: 10.1016/j.tig.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 38.Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126:285–295. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edlinger B, Schlögelhofer P. Have a break: Determinants of meiotic DNA double strand break (DSB) formation and processing in plants. J Exp Bot. 2011;62:1545–1563. doi: 10.1093/jxb/erq421. [DOI] [PubMed] [Google Scholar]
- 40.Rosu S, Libuda DE, Villeneuve AM. Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science. 2011;334:1286–1289. doi: 10.1126/science.1212424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleckner N, et al. A mechanical basis for chromosome function. Proc Natl Acad Sci USA. 2004;101:12592–12597. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mirouze M, Paszkowski J. Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol. 2011;14:267–274. doi: 10.1016/j.pbi.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 43.Johannes F, Colot V, Jansen RC. Epigenome dynamics: A quantitative genetics perspective. Nat Rev Genet. 2008;9:883–890. doi: 10.1038/nrg2467. [DOI] [PubMed] [Google Scholar]
- 44.Wijnker E, de Jong H. Managing meiotic recombination in plant breeding. Trends Plant Sci. 2008;13:640–646. doi: 10.1016/j.tplants.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 45.Filion GJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toyota M, Matsuda K, Kakutani T, Terao Morita M, Tasaka M. Developmental changes in crossover frequency in Arabidopsis. Plant J. 2011;65:589–599. doi: 10.1111/j.1365-313X.2010.04440.x. [DOI] [PubMed] [Google Scholar]
- 47.Giraut L, et al. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 2011;7:e1002354. doi: 10.1371/journal.pgen.1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X, Shiu SH, Cal A, Borevitz JO. Global analysis of genetic, epigenetic and transcriptional polymorphisms in Arabidopsis thaliana using whole genome tiling arrays. PLoS Genet. 2008;4:e1000032. doi: 10.1371/journal.pgen.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaughn MW, et al. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 2007;5:e174. doi: 10.1371/journal.pbio.0050174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Francis KE, et al. Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:3913–3918. doi: 10.1073/pnas.0608936104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Werner JD, et al. Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc Natl Acad Sci USA. 2005;102:2460–2465. doi: 10.1073/pnas.0409474102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singer T, et al. A high-resolution map of Arabidopsis recombinant inbred lines by whole-genome exon array hybridization. PLoS Genet. 2006;2:e144. doi: 10.1371/journal.pgen.0020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tittel-Elmer M, et al. Stress-induced activation of heterochromatic transcription. PLoS Genet. 2010;6:e1001175. doi: 10.1371/journal.pgen.1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.