Abstract
Reactive oxygen species (ROS) are toxic oxygen-containing molecules that can damage multiple components of the cell and have been proposed to be the primary cause of aging. The antioxidant enzyme superoxide dismutase (SOD) is the only eukaryotic enzyme capable of detoxifying superoxide, one type of ROS. The fact that SOD is present in all aerobic organisms raises the question as to whether SOD is absolutely required for animal life and whether the loss of SOD activity will result in decreased lifespan. Here we use the genetic model organism Caenorhabditis elegans to generate an animal that completely lacks SOD activity (sod-12345 worms). We show that sod-12345 worms are viable and exhibit a normal lifespan, despite markedly increased sensitivity to multiple stresses. This is in stark contrast to what is observed in other genetic model organisms where the loss of a single sod gene can result in severely decreased survival. Investigating the mechanism underlying the normal lifespan of sod-12345 worms reveals that their longevity results from a balance between the prosurvival signaling and the toxicity of superoxide. Overall, our results demonstrate that SOD activity is dispensable for normal animal lifespan but is required to survive acute stresses. Moreover, our findings indicate that maintaining normal stress resistance is not crucial to the rate of aging.
Keywords: oxidative stress, reactive oxygen species-mediated signaling, free radical theory of aging, sod-2
The contribution of reactive oxygen species (ROS) to aging was first suggested by the free radical theory of aging, which postulates that aging results from damage caused by ROS that accumulate over time, leading to cellular dysfunction and an increased probability of death (1). In aerobic organisms, ROS are produced as a byproduct of normal metabolism when electrons that are being passed down the electron transport chain are leaked directly to oxygen to form superoxide. In addition to ROS generated by electron transport, a number of other enzymes are known to produce ROS, such as P450 oxidase and NADPH oxidase.
Although the role of ROS in aging is still controversial (2–4), it is clear that high levels of ROS are toxic. Accordingly, cells have evolved a number of both enzymatic and nonenzymatic antioxidant defenses that function to detoxify ROS. Superoxide dismutase (SOD) is the first line of antioxidant defense against ROS generated by respiration and, among eukaryotic organisms, SOD is the only enzyme that can detoxify superoxide (5). SOD acts by converting superoxide to hydrogen peroxide, which can subsequently be converted to water by catalase or peroxiredoxin. All aerobic organisms, and even some anaerobic organisms, express SOD. Anaerobes that do not express SOD, use other mechanisms to detoxify superoxide, such as superoxide reductase (6) or increasing intracellular levels of manganese (7), to allow them to survive brief encounters with oxygen. The fact that all known organisms, both aerobic and anaerobic, have some form of superoxide scavenging activity clearly indicates the importance of eliminating superoxide.
Whereas no naturally occurring organism has been identified without any form of superoxide scavenging ability, a number of groups have examined the consequences of eliminating the expression of individual sod genes in genetic model organisms. Consistent with the view that superoxide scavenging activity is important for survival, deletion of either cytoplasmic or mitochondrial sod genes in yeast (8–11), flies (12–14), and mice (15–17) results in decreased lifespan (Table 1). In contrast, deletion of individual sod genes has been found to have little or no detrimental effect on lifespan in the roundworm Caenorhabditis elegans (18–22) (Table 1).
Table 1.
Effect of decreasing expression of superoxide dismutase genes on lifespan
| Organism | No. of SOD genes | Effect of decreasing cytoplasmic SOD on lifespan, % | Effect of decreasing mitochondrial SOD on lifespan, % | Reference |
| Yeast | 2 | ↓ | ↓ | (9) |
| ↓63 | ↓63 | (10) | ||
| ↓40 | ↓70 | (11) | ||
| Worm | 5 | ↓1 (NS) | (21) | |
| ↓18 | ↓1 (NS) | (18) | ||
| ↓5 (NS) | ↑15 (NS) | (19) | ||
| ↓3 (NS) | ↑53 | (22) | ||
| ↓20 | (20) | |||
| Fly | 2 | ↓80 | (12) | |
| ↓81* | (13) | |||
| Lethality (<24 h) | (14) | |||
| Mouse | 3 | ↓30 | (15) | |
| Lethality (<10 d) | (16) | |||
| Lethality (<18 d) | (17) |
NS, Not significant.
*Sod2 levels were decreased by RNAi.
One explanation for the ability of C. elegans to accommodate for the loss of sod genes might be the number of sod genes present. Yeast and flies both have two SODs, one cytoplasmic and one mitochondrial. In addition to the cytoplasmic and mitochondrial SODs, mice also express an extracellular SOD. In contrast, C. elegans has five sod genes. sod-1, sod-2, and sod-4 are the primary cytoplasmic, mitochondrial, and extracellular sod genes, respectively, whereas sod-3 and sod-5 are inducible mitochondrial and cytoplasmic sod genes, respectively. Thus, it is possible that the loss of individual SODs is compensated for by the presence of these additional sod genes. In fact, up-regulation of other sod genes has been observed in individual sod deletion mutants (20, 22).
To determine whether the presence of additional sod genes in C. elegans masks a detrimental effect of sod gene deletion on lifespan, we generated a sod quintuple mutant that lacks all five sod genes (sod-12345 worms). sod-12345 worms were found to be viable and fertile but exhibited multiple alterations of physiologic rates. Despite having markedly increased sensitivity to multiple stresses, the lifespan of sod-12345 worms was not different from wild-type worms. Overall, this suggests that SOD function is important in reacting to environmental stresses but appears to be dispensable with respect to normal lifespan.
Results and Discussion
Generation of a sod Quintuple Mutant with No SOD Activity.
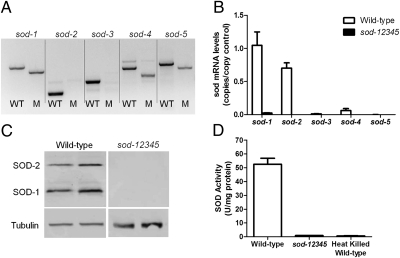
To determine whether the total loss of SOD activity would result in decreased lifespan, we generated mutants lacking all five sod genes that are normally present in C. elegans. The sod quintuple mutants were made by progressively crossing pairs of sod single mutants, double mutants, triple mutants, and quadruple mutants to obtain a strain with deletions in all five sod genes (Fig. 1A). To confirm that we had generated a bona fide sod quintuple mutant, we examined sod gene expression at the mRNA and protein levels. Quantitative real-time RT-PCR showed that sod-12345 worms express little or no sod mRNA (Fig. 1B). Similarly, Western blotting using polyclonal antibodies generated against SOD-1 and SOD-2, the primary cytoplasmic and mitochondrial SODs respectively, revealed no detectable SOD-1 or SOD-2 protein in sod-12345 worms (Fig. 1C). Importantly, examination of SOD activity revealed no detectable SOD activity in sod-12345 worms (Fig. 1D). This result confirms that we had successfully generated a sod quintuple mutant and that worms are able to survive and reproduce in the absence of SOD.
Fig. 1.
sod quintuple mutant worms have no SOD activity. (A) PCR genotyping reveals that sod-12345 worms have deletions in all five sod genes. (B) Quantitative real-time RT-PCR shows that sod-12345 worms have little or no sod mRNA expression. (C) Western blotting using polyclonal antibodies against SOD-1 and SOD-2 reveals that sod-12345 worms have no SOD-1 or SOD-2 protein expression. (D) sod-12345 worms have no detectable SOD activity. The generation of a bona fide sod quintuple mutant was thus confirmed by DNA, mRNA, protein, and activity. Error bars indicate SEM. M, sod-12345 worms.
sod Quintuple Mutants Exhibit Abnormal Physiologic Rates.
Having shown that sod quintuple mutants are viable and fertile, we sought to determine whether the absence of SOD activity would affect worm development and physiologic rates. As with unicellular mutants lacking SOD activity (23, 24), sod-12345 worms were found to exhibit slow development and reduced fertility (Fig. S1 A and B). These worms were also found to have other physiologic abnormalities such as a slower defecation cycle and decreased movement (thrashing in liquid) (Fig. S1 C and D). These deficits appeared to result primarily from the loss of SOD-1 and/or SOD-2 expression, which is consistent with the fact that these are the primary cytoplasmic and mitochondrial SODs, respectively. Similarly, the absence of detectable phenotypic abnormalities in sod-3, sod-4, and sod-5 mutant worms likely stems from the fact that these sod genes are normally expressed at very low levels (18). Because ROS-mediated signaling has previously been shown to affect physiology in C. elegans (25), the absence of SOD activity may influence physiologic rates through an increase in superoxide-mediated signaling or a decrease in H2O2-mediated signaling.
sod Quintuple Mutants Are Sensitive to Multiple Stresses.
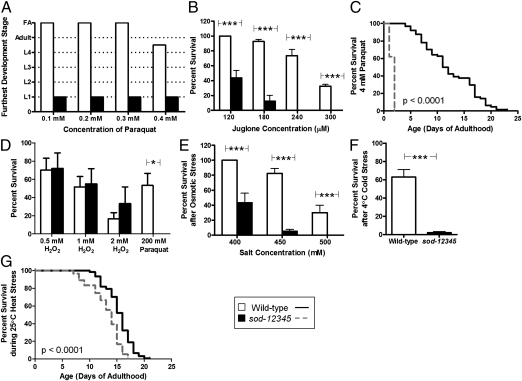
As SOD functions to detoxify superoxide, we sought to determine whether sod-12345 mutants would exhibit increased sensitivity to superoxide-mediated oxidative stress. In three different paradigms involving exposure to the superoxide-generating compounds, paraquat and juglone, sod-12345 worms were found to have markedly increased sensitivity to oxidative stress both during development and adulthood. This included exposure to paraquat during development (Fig. 2A), acute exposure to juglone at day 1 of adulthood (Fig. 2B), and chronic exposure to paraquat from day 1 of adulthood until death (Fig. 2C). In each of these paradigms, sod-12345 worms exhibited markedly increased sensitivity to oxidative stress, which appeared to result primarily from the absence of SOD-1, SOD-2, or both (Fig. S2).
Fig. 2.
Loss of SOD activity results in increased sensitivity to multiple stresses. sod-12345 worms have markedly increased sensitivity to superoxide-mediated oxidative stress induced by exposure to low concentrations of paraquat during development (A), by acute exposure to juglone during adulthood (B), and by chronic exposure to 4 mM paraquat beginning on day 1 of adulthood (C). In contrast, sod-12345 worms exhibit normal sensitivity to H2O2-mediated oxidative stress (D). sod-12345 worms are also sensitive to osmotic stress (E), cold stress (F), and heat stress (G). Results from each stress assay represent the average of at least three independent trials with 20 worms or 40 eggs per trial. Whereas there was a trend toward increased resistance to H2O2-mediated oxidative stress at 2 mM H2O2, the difference was not significant. Error bars indicate SEM. *P < 0.05, **P < 0.01, ***P < 0.001. FA, fertile adult.
To determine whether the sensitivity of sod-12345 worms to oxidative stress was specific to superoxide or whether these worms were sensitive to other forms of oxidative stress, we exposed sod-12345 and wild-type worms to H2O2. Using three different concentrations of H2O2, we found that sod-12345 worms survived at least as well as wild-type worms (Fig. 2D and Fig. S3 A–C). Although there was a trend toward increased survival in the sod quintuple mutant at the highest concentration of H2O2, this difference did not reach significance. Because this assay was performed in liquid, we assessed the sensitivity of sod-12345 worms to paraquat in liquid as a control and found that these worms were still very sensitive to paraquat in liquid (Fig. 2D and Fig. S3D). This indicates that sod-12345 worms exhibit a specific sensitivity to superoxide-mediated oxidative stress, which is consistent with the function of SOD in detoxifying superoxide. Having shown that sod-12345 worms are sensitive to specific forms of oxidative stress, we next sought to determine whether the absence of SOD activity would make sod-12345 worms susceptible to other forms of stress. We found that sod-12345 worms also exhibited increased sensitivity to osmotic stress, cold stress, and heat stress (Fig. 2 E–G).
sod Quintuple Mutants Have a Normal Lifespan.
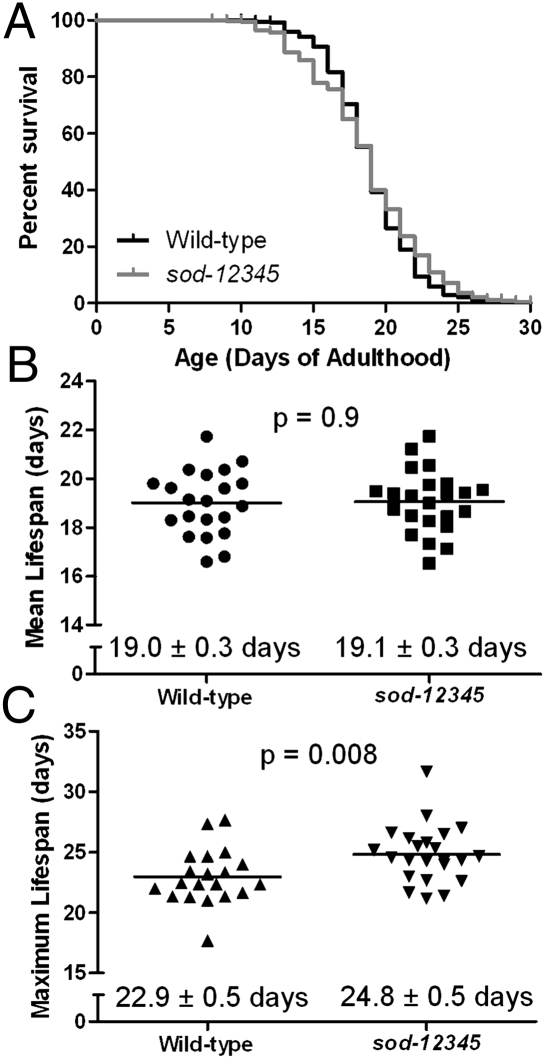
We next sought to determine whether the markedly increased sensitivity to multiple stresses in sod-12345 worms would result in decreased lifespan. As the weight of evidence suggests that increased resistance to multiple forms of stress may be the key to longevity (multiplex stress resistance theory of aging) (26) and oxidative stress is widely believed to be the primary cause of aging (free radical theory of aging) (1), sod-12345 worms would be predicted to have a decreased lifespan. Surprisingly, despite their marked increase in stress sensitivity, sod quintuple mutant worms live as long as wild-type worms (Fig. 3A). In 17 independent trials, the average lifespan of wild-type worms was 19.0 d, whereas the average lifespan for sod-12345 worms was 19.1 d (Fig. 3B). Interestingly, the maximum lifespan of sod-12345 worms was significantly increased compared with wild-type worms (Fig. 3C). Thus, whereas a proportion of sod-12345 worms died earlier than wild-type worms, the longest lived sod-12345 worms lived longer than wild type. This is in contrast to sod double mutants in yeast that exhibit a 90% decrease in lifespan (10) and individual sod deletion mutants in yeast, flies, and mice, which all show decreased lifespan (8–17).
Fig. 3.
Worms lacking SOD activity live as long as wild-type worms. (A) Examination of worm lifespan revealed no difference in overall survival between sod-12345 and wild-type worms. This survival plot is the average of 17 independent trials with a total of 1,571 deaths recorded. (B) Scatterplot of the mean lifespan from 17 independent lifespan assays shows no difference between sod-12345 and wild-type lifespan. (C) Maximum lifespan of sod-12345 is significantly greater than wild-type worms.
As sod-2 deletion mutants have increased lifespan, the loss of the other four sod genes abolishes their extended longevity. To determine which combinations of the remaining four sod genes are required for the long life of sod-2 mutants, we examined the lifespan of sod triple and quadruple mutants. Overall, only strains with a deletion in sod-2 exhibited increased lifespan and this effect was eliminated by the loss of sod-1 and sod-3 together (Fig. S4). As SOD-1 has been observed in the mitochondria (20, 27), this suggests the possibility that it is the complete absence of any SOD in the mitochondria that abolishes the positive effect of sod-2 deletion on lifespan.
sod-12345 Worms Respire Aerobically and Have Normal Levels of Oxidative Damage.
One possible explanation for the worm's unique ability to survive without SOD activity is its capacity to generate energy by fermentative pathways. By switching to fermentation, worms could generate less superoxide and have less need for superoxide detoxification. To determine whether sod-12345 worms continue to generate energy by oxidative phosphorylation, we measured oxygen consumption and found that sod quintuple mutants were still carrying out aerobic respiration but that the rate of oxygen consumption was significantly reduced compared with wild type (Fig. S5A). Assuming that an equal percentage of electrons that are passed down the electron transport chain are leaked to form superoxide, this decreased level of respiration would result in decreased production of ROS. However, despite their decreased levels of oxygen consumption, sod-12345 worms were found to have normal levels of ATP (Fig. S5B). This suggests that to maintain normal ATP levels, sod-12345 worms decrease their energy utilization resulting in the observed slow physiologic rates.
To gain further insight into the mechanism by which sod-12345 worms survive as well as wild-type worms, we examined oxidative damage by measuring protein carbonyl levels. Despite their markedly increased sensitivity to oxidative stress, we found that sod-12345 worms have normal levels of protein carbonyls (Fig. S5 C and D). In addition to decreased ROS production, increased expression of other antioxidants, increased damage repair, and/or increased protein turnover could contribute to the normal level of oxidative damage in sod-12345 worms. Investigation of these different mechanisms revealed that sod-12345 worms exhibit up-regulation of genes coding for other antioxidant enzymes, such as catalase (Fig. S5E) as well as for glutathione S-transferase (gst) repair genes (Fig. S5F) but no change in proteasome activity (Fig. S5G). Thus, there are multiple mechanisms that contribute to the normal levels of oxidative damage in sod-12345 worms including decreased rate of oxidative phosphorylation, increased expression of other antioxidant genes, and increased expression of repair genes.
The fact that sod-12345 worms have increased sensitivity to a variety of stresses indicates that the compensatory mechanisms that allow for normal levels of oxidative damage are not sufficient to allow sod-12345 worms to survive acute stresses as well as wild-type worms. Importantly, we have previously shown that oxidative damage does not cause worm aging because increasing levels of oxidative damage were shown not to impact lifespan (28) and increased oxidative damage was shown to be compatible with long life (22). In future studies, it would be interesting to examine other forms of molecular damage to determine whether these are affected in sod-12345 worms. In addition, a comparison of mitochondrial and cytoplasmic oxidative damage in long-lived strains with a deletion in sod-2 (e.g., sod-2, sod-1; sod-2) and normal-lived strains with a deletion in sod-2 (e.g., sod-1; sod-2; sod-3) may provide insight into the role of subcellular compartment-specific oxidative damage in the increased longevity of sod-2 mutants.
sod-12345 Lifespan Results from a Balance Between the Prosurvival Signaling and the Toxicity of Superoxide.
Recent work from our laboratory and others has shown that both decreasing mitochondrial superoxide detoxification through the deletion of sod-2, or increasing superoxide levels through the addition of low concentrations of paraquat, results in increased lifespan (22, 29, 30). These results suggest that increased levels of mitochondrial superoxide can trigger a prosurvival signal that leads to increased longevity (29–31). Thus, it is possible that increased levels of superoxide resulting from the absence of SOD activity in sod-12345 worms engages similar prosurvival mechanisms that contribute to the normal longevity of sod-12345 worms by compensating for deleterious effects that result from lacking a crucial mechanism of ROS detoxification. To investigate this possibility, we examined the effect of increasing and decreasing superoxide levels on the lifespan of wild-type and sod-12345 worms.
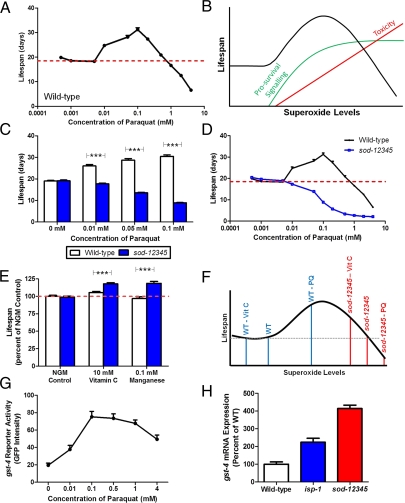
To study the effect of superoxide levels on wild-type lifespan, we measured the lifespan of wild-type worms on plates containing increasing concentrations of the superoxide generator paraquat. We observed a biphasic pattern in which increasing concentrations of paraquat initially resulted in a dose-dependent increase in lifespan until an optimum superoxide concentration at about 0.1 mM paraquat, after which further increases in paraquat resulted in decreased lifespan (Fig. 4A). To explain this pattern, we propose a model in which superoxide has two opposing effects on lifespan. First, mitochondrial superoxide triggers a prosurvival signal that results in a dose-dependent increase in lifespan until this mechanism is maximally engaged (Fig. 4B). At the same time, increasing levels of superoxide are also toxic, resulting in a dose-dependent decrease in lifespan. Thus, at levels of superoxide where lifespan is increased, the effect of prosurvival signaling is greater than the toxic effect, whereas at superoxide concentrations where lifespan is decreased the toxicity of superoxide overwhelms its prosurvival signaling effect. The maximum lifespan occurs at the optimum superoxide concentration.
Fig. 4.
Genotype determines the effect of superoxide on lifespan. (A) Increasing superoxide levels through addition of paraquat has a biphasic effect on the lifespan of wild-type worms. (B) This pattern can be explained by a model for the effect of superoxide on lifespan in which superoxide has two opposing effects on lifespan: prosurvival signaling and toxicity. Initially, the effect of the prosurvival signaling is greater than the toxic effect, resulting in a dose-dependent increase in lifespan. At higher superoxide levels, the toxic effect of superoxide overwhelms the prosurvival signaling effect leading to decreased lifespan. (C) Increasing superoxide levels with paraquat resulted in a dose-dependent decrease in sod-12345 worm lifespan but a dose-dependent increase in wild-type lifespan. (D) Paraquat did not increase the lifespan of sod-12345 worms at any concentration. (E) Decreasing ROS with 10 mM vitamin C or 0.1 mM manganese increased the lifespan of sod-12345 worms with little or no effect on wild-type lifespan. (F) Superoxide levels in sod-12345 worms are past their optimum superoxide concentration. (G) gst-4 promoter shows increased expression levels with increasing concentrations of paraquat. (H) isp-1 and sod-12345 worms show increased gst-4 expression, suggesting elevated levels of superoxide. Results from the paraquat and antioxidant survival studies are the average of at least three independent trials of at least 20 worms per strain per trial. Error bars indicate SEM. ***P < 0.001.
On the basis of this model, the normal longevity of sod-12345 worms could result from a balance between the prosurvival signaling and toxicity of superoxide if the superoxide levels in sod-12345 worms are above their optimum superoxide concentration (as would be predicted from their decreased ability to detoxify superoxide). If the superoxide levels in these worms are already above their optimum superoxide concentration, then further increases in superoxide levels should only result in decreased lifespan. To test this possibility, we measured the lifespan of sod-12345 worms on plates containing paraquat at concentrations that resulted in an increase in wild-type lifespan. In contrast to the increase in lifespan observed in wild-type worms, sod-12345 worms exhibit a dose-dependent decrease in lifespan with increasing concentrations of paraquat (Fig. 4C). In fact, even at very low concentrations of paraquat, we never observed a positive effect of paraquat on sod-12345 lifespan (Fig. 4D and Fig. S6). This finding suggests that superoxide-mediated prosurvival signaling is maximally engaged in sod-12345 worms, such that further increases in superoxide are toxic and result in decreased lifespan.
Next, we examined the effect of decreasing superoxide levels on wild-type and sod-12345 lifespan through the addition of the antioxidant vitamin C, or manganese, which has superoxide scavenging ability. In support of our model, we found that both treatments increased the lifespan of sod-12345 worms by 15–20% but had little or no impact of wild-type lifespan (Fig. 4E). Thus, the levels of superoxide in wild-type worms are below their optimum superoxide concentration such that reducing superoxide levels with antioxidants has a minimal effect on lifespan, whereas increasing superoxide levels with paraquat brings superoxide levels closer to their optimum concentration, resulting in increased lifespan (Fig. 4F). In contrast, our results suggest that the superoxide levels in sod-12345 worms are beyond their optimum concentration. In these worms the level of superoxide is brought closer to their optimum concentration by decreasing superoxide with vitamin C, resulting in increased lifespan, whereas increasing superoxide with paraquat pushes them away from their optimum concentration, resulting in decreased lifespan (Fig. 4F). This finding suggests that the normal longevity of sod-12345 worms represents a balance between the prolongevity signaling and the toxicity of superoxide.
According to this model, it should also be possible to decrease the lifespan of worms close to their optimum superoxide concentration by treatment with antioxidants. As we have previously shown that the mitochondrial mutant isp-1 worms have increased mitochondrial superoxide and an extended longevity that can be suppressed by antioxidant treatment (29, 32), we examined whether treatment of isp-1 worms with vitamin C under the exact same conditions that increased sod-12345 worm lifespan would result in decreased lifespan. In support of our model, we found that the treatment with vitamin C markedly decreased lifespan in isp-1 worms (Fig. S7). Thus, increasing superoxide levels with paraquat can either increase lifespan, as in the case of wild-type worms, or decrease lifespan, as in the case of sod-12345 worms. Similarly, decreasing superoxide levels with antioxidants can either increase lifespan, as in the case of sod-12345 worms, or decrease lifespan, as in the case of isp-1 worms. Overall this demonstrates that the effect of superoxide on lifespan is entirely dependent on the genotype of the strain examined and their baseline levels of superoxide.
Increased Superoxide Reporter Expression in sod-12345 Worms.
To provide additional support for our model, we indirectly measured superoxide levels using a gst-4 reporter construct, because it is currently not possible to accurately measure mitochondrial superoxide levels in vivo in worms. The GST gene, gst-4, has been previously shown to be highly induced by superoxide levels (33) and on the basis of this property, the promoter from this gene was used to generate a reporter construct expressing GFP (34). To validate the use of this reporter construct to measure superoxide, we examined the level of reporter activity at increasing concentrations of paraquat. We observed a dose-dependent increase in gst-4 reporter activity with increasing concentrations of paraquat up to 0.1 mM (Fig. 4G and Fig. S8). At higher concentrations, there was no further increase in expression from the gst-4 promoter.
Having shown that the gst-4 reporter can respond to increasing levels of superoxide, we generated isp-1; Pgst-4::gfp and sod-1235; Pgst-4::gfp worms to examine gst-4 reporter activity on an isp-1 and sod mutant background (the sod-12345; Pgst-4::gfp mutant could not be constructed because of linkage between sod-4 and the insertion of the Pgst-4::gfp transgene; however, sod-1235 worms have deletions in all four intracellular sod genes, have a similar lifespan to that of sod-12345 worms, and would be predicted to have elevated superoxide levels less than or equal to sod-12345 worms). Our model would predict increased levels of superoxide in both strains with the levels in sod-1235 worms being equal to or greater than those in isp-1 worms. Quantification of gst-4 reporter activity showed significantly increased GFP expression in both strains compared with control with greater activity in the sod mutant worms compared with isp-1 worms (Fig. S9 A and B). To ensure that gst-4 promoter activity is also increased in sod quintuple mutant worms, we examined gst-4 expression by quantitative real-time RT-PCR. We observed increased gst-4 mRNA expression in both isp-1 and sod-12345 worms, thereby confirming the results obtained using the Pgst-4::gfp reporter construct (Fig. 4H). The fact that the gst-4 promoter responds to increasing concentrations of superoxide, and its activity was shown to be increased in isp-1 and sod-12435 worms, suggests that these strains have increased levels of superoxide and provides further support for our model.
Prosurvival Superoxide Signaling Contributes to the Longevity of sod-2 Mutant Worms.
Because sod-2 deletion mutants are long lived we sought to determine whether prosurvival superoxide signaling contributes to their long lifespan. If this were true, we would predict that sod-2 mutants (i) would exhibit elevated levels of superoxide, (ii) would exhibit decreased lifespan at a lower concentration of paraquat than wild-type worms, and (iii) would exhibit decreased lifespan when treated with antioxidants. To test the first prediction, we examined Pgst-4::gfp reporter activity in sod-2 worms. We found increased reporter activity, suggesting elevated levels of superoxide (Fig. S10A). To test the second prediction, we examined the lifespan of sod-2 worms treated with 0.2 mM paraquat, a concentration that is sufficiently mild to increase the lifespan of wild-type worms. We found that 0.2 mM paraquat decreases the lifespan of sod-2 mutant worms, thereby confirming that the lifespan of sod-2 worms begins to decline at lower concentrations of paraquat than wild-type worms (Fig. S10B). To test the third prediction, we treated sod-2 worms with 10 mM vitamin C and observed a small, yet significant, decrease in lifespan (Fig. S10C). Together these observations suggest that mitochondrial superoxide levels are increased in sod-2 worms and contribute to their extended longevity. Moreover it suggests that sod-2 worms are closer to their optimum superoxide concentration than wild-type worms. The fact that sod-2 worms are nearer to their optimum superoxide concentration than wild-type worms suggests that the mechanism by which the deletion of sod-1 and sod-3 abolishes the increased longevity of sod-2 worms (Fig. S4) is by pushing sod-2 worms past their optimum superoxide concentration. This conclusion is supported by the fact that deletion of sod-1 makes sod-2 worms more sensitive to paraquat, although not as sensitive as sod-12345 worms (Fig. S10D).
Conclusions
Overall, the normal longevity of sod quintuple mutant worms clearly demonstrates that SOD activity is not required for normal lifespan in C. elegans. Although sod-12345 worms use multiple compensatory mechanisms to maintain low levels of oxidative damage, these worms are highly sensitive to superoxide levels and exhibit increased sensitivity to a variety of stresses. This suggests that SOD activity is necessary to respond to acute stresses and indicates that increased sensitivity to multiple stresses, including oxidative stress, does not result in decreased lifespan. Our results also indicate that superoxide is not simply a toxic byproduct of metabolism but is involved in a type of ROS-mediated signaling that can result in increased longevity. Thus, whether a particular concentration of superoxide will increase or decrease lifespan depends on the genotype of the strains examined and their initial levels of superoxide. This work casts doubt on the notion that oxidative stress is the primary cause of aging.
Materials and Methods
Strains.
C. elegans strains were cultured as described on nematode growth medium (NGM) agar plates seeded with OP50 bacteria at 20 °C (35). Wild-type animals were N2 Bristol strain. sod-12345 worms were generated by sequentially crossing single sod deletion mutants, double sod deletion mutants, triple sod deletion mutants, and finally quadruple sod deletion mutants. At each stage, the presence of sod deletion was confirmed by PCR genotyping.
SOD Levels and Activity.
sod mRNA and SOD protein levels were measured by quantitative real-time RT-PCR and Western blotting as described previously (22, 36). SOD activity was measured using the Superoxide Dismutase Assay kit from Cayman Chemical according to the manufacturer's instructions.
Lifespan Studies.
Lifespan studies were completed at 20 °C on plates containing 100 μM 5-fluoro-2′-deoxyuridine (FUdR) (Sigma). A total of 17 independent trials were completed for sod-12345 worms. Lifespan on plates containing paraquat, vitamin C, or manganese were begun on day 1 of adulthood.
Physiologic Rates.
Postembryonic development time, self-brood size, defecation cycle length, and thrashing rate were measured as described previously (22, 36).
Stress Assays.
Four paradigms were used to measure sensitivity to oxidative stress: (i) exposure to 0.1–0.4 mM paraquat during development, (ii) exposure to 120–240 μM juglone on day 1 of adulthood, (iii) exposure to 4 mM paraquat from day 1 of adulthood until death, and (iv) exposure to 0.5–2 mM H2O2 on day 1 of adulthood. Sensitivity to osmotic stress was assessed by placing day 1 adult worms on plates containing 400–500 mM NaCl and examining survival after 3 d. Sensitivity to cold stress was assessed by transferring worms to 4 °C for 3 d and examining survival after recovery at room temperature. Sensitivity to chronic heat stress was examined by measuring lifespan at 25 °C.
See SI Materials and Methods for full experimental details.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis elegans Knockout Consortium, the National BioResource Project of Japan (Mitani laboratory), and the Caenorhabditis Genetics Center for providing strains used in this research. J.M.V.R. is supported by the Canadian Institutes of Health Research, Parkinson Society Canada, the Hereditary Disease Foundation, and McGill Tomlinson Fellowships. S.H. is supported by Canadian Institutes of Health Research MOP-89761 and McGill University. S.H. is Campbell Chair of Developmental Biology and Strathcona Chair of Zoology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116158109/-/DCSupplemental.
References
- 1.Harman D. Aging: A theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 2.Muller FL, Lustgarten MS, Jang Y, Richardson A, Van Remmen H. Trends in oxidative aging theories. Free Radic Biol Med. 2007;43:477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 3.Van Raamsdonk JM, Hekimi S. Reactive oxygen species and aging in Caenorhabditis elegans: Causal or casual relationship? Antioxid Redox Signal. 2010;13:1911–1953. doi: 10.1089/ars.2010.3215. [DOI] [PubMed] [Google Scholar]
- 4.Hekimi S, Lapointe J, Wen Y. Taking a “good” look at free radicals in the aging process. Trends Cell Biol. 2011;21:569–576. doi: 10.1016/j.tcb.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fridovich I. The biology of oxygen radicals. Science. 1978;201:875–880. doi: 10.1126/science.210504. [DOI] [PubMed] [Google Scholar]
- 6.Jenney FE, Jr, Verhagen MF, Cui X, Adams MW. Anaerobic microbes: Oxygen detoxification without superoxide dismutase. Science. 1999;286:306–309. doi: 10.1126/science.286.5438.306. [DOI] [PubMed] [Google Scholar]
- 7.Archibald FS, Fridovich I. Manganese and defenses against oxygen toxicity in Lactobacillus plantarum. J Bacteriol. 1981;145:442–451. doi: 10.1128/jb.145.1.442-451.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Longo VD, Liou LL, Valentine JS, Gralla EB. Mitochondrial superoxide decreases yeast survival in stationary phase. Arch Biochem Biophys. 1999;365:131–142. doi: 10.1006/abbi.1999.1158. [DOI] [PubMed] [Google Scholar]
- 9.Longo VD, Gralla EB, Valentine JS. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J Biol Chem. 1996;271:12275–12280. doi: 10.1074/jbc.271.21.12275. [DOI] [PubMed] [Google Scholar]
- 10.Wawryn J, Krzepiłko A, Myszka A, Biliński T. Deficiency in superoxide dismutases shortens life span of yeast cells. Acta Biochim Pol. 1999;46:249–253. [PubMed] [Google Scholar]
- 11.Unlu ES, Koc A. Effects of deleting mitochondrial antioxidant genes on life span. Ann.N.Y. Acad.Sci. 2007;1100:505–509. doi: 10.1196/annals.1395.055. [DOI] [PubMed] [Google Scholar]
- 12.Phillips JP, Campbell SD, Michaud D, Charbonneau M, Hilliker AJ. Null mutation of copper/zinc superoxide dismutase in Drosophila confers hypersensitivity to paraquat and reduced longevity. Proc Natl Acad Sci USA. 1989;86:2761–2765. doi: 10.1073/pnas.86.8.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirby K, Hu J, Hilliker AJ, Phillips JP. RNA interference-mediated silencing of Sod2 in Drosophila leads to early adult-onset mortality and elevated endogenous oxidative stress. Proc Natl Acad Sci USA. 2002;99:16162–16167. doi: 10.1073/pnas.252342899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duttaroy A, Paul A, Kundu M, Belton A. A Sod2 null mutation confers severely reduced adult life span in Drosophila. Genetics. 2003;165:2295–2299. doi: 10.1093/genetics/165.4.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elchuri S, et al. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene. 2005;24:367–380. doi: 10.1038/sj.onc.1208207. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 17.Lebovitz RM, et al. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proc Natl Acad Sci USA. 1996;93:9782–9787. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doonan R, et al. Against the oxidative damage theory of aging: Superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev. 2008;22:3236–3241. doi: 10.1101/gad.504808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yen K, Patel HB, Lublin AL, Mobbs CV. SOD isoforms play no role in lifespan in ad lib or dietary restricted conditions, but mutational inactivation of SOD-1 reduces life extension by cold. Mech Ageing Dev. 2009;130:173–178. doi: 10.1016/j.mad.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Yanase S, Onodera A, Tedesco P, Johnson TE, Ishii N. SOD-1 deletions in Caenorhabditis elegans alter the localization of intracellular reactive oxygen species and show molecular compensation. J Gerontol A Biol Sci Med Sci. 2009;64:530–539. doi: 10.1093/gerona/glp020. [DOI] [PubMed] [Google Scholar]
- 21.Honda Y, Tanaka M, Honda S. Modulation of longevity and diapause by redox regulation mechanisms under the insulin-like signaling control in Caenorhabditis elegans. Exp Gerontol. 2008;43:520–529. doi: 10.1016/j.exger.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Van Raamsdonk JM, Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000361. doi: 10.1371/journal.pgen.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassett DJ, Schweizer HP, Ohman DE. Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J Bacteriol. 1995;177:6330–6337. doi: 10.1128/jb.177.22.6330-6337.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: Is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shibata Y, Branicky R, Landaverde IO, Hekimi S. Redox regulation of germline and vulval development in Caenorhabditis elegans. Science. 2003;302:1779–1782. doi: 10.1126/science.1087167. [DOI] [PubMed] [Google Scholar]
- 26.Lithgow GJ, Miller RA. The determination of aging rate by coordinated resistance to multiple forms of stress. In: Guarente L, Partridge L, Wallace DC, editors. The Molecular Biology of Aging. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2008. [Google Scholar]
- 27.Sturtz LA, Diekert K, Jensen LT, Lill R, Culotta VC. A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J Biol Chem. 2001;276:38084–38089. doi: 10.1074/jbc.M105296200. [DOI] [PubMed] [Google Scholar]
- 28.Yang W, Li J, Hekimi S. A measurable increase in oxidative damage due to reduction in superoxide detoxification fails to shorten the life span of long-lived mitochondrial mutants of Caenorhabditis elegans. Genetics. 2007;177:2063–2074. doi: 10.1534/genetics.107.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang W, Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 2010;8:e1000556. doi: 10.1371/journal.pbio.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SJ, Hwang AB, Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr Biol. 2010;20(23):2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walter L, Baruah A, Chang HW, Pace HM, Lee SS. The homeobox protein CEH-23 mediates prolonged longevity in response to impaired mitochondrial electron transport chain in C. elegans. PLoS Biol. 2011;9:e1001084. doi: 10.1371/journal.pbio.1001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng J, Bussière F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- 33.Tawe WN, Eschbach ML, Walter RD, Henkle-Dührsen K. Identification of stress-responsive genes in Caenorhabditis elegans using RT-PCR differential display. Nucleic Acids Res. 1998;26:1621–1627. doi: 10.1093/nar/26.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Link CD, Johnson CJ. Reporter transgenes for study of oxidant stress in Caenorhabditis elegans. Methods Enzymol. 2002;353:497–505. doi: 10.1016/s0076-6879(02)53072-x. [DOI] [PubMed] [Google Scholar]
- 35.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Raamsdonk JM, et al. Decreased energy metabolism extends life span in Caenorhabditis elegans without reducing oxidative damage. Genetics. 2010;185:559–571. doi: 10.1534/genetics.110.115378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.