Abstract
Our understanding of the molecular control of many disease pathologies requires the identification of direct substrates targeted by specific protein kinases. Here we describe an integrated proteomic strategy, termed kinase assay linked with phosphoproteomics, which combines a sensitive kinase reaction with endogenous kinase-dependent phosphoproteomics to identify direct substrates of protein kinases. The unique in vitro kinase reaction is carried out in a highly efficient manner using a pool of peptides derived directly from cellular kinase substrates and then dephosphorylated as substrate candidates. The resulting newly phosphorylated peptides are then isolated and identified by mass spectrometry. A further comparison of these in vitro phosphorylated peptides with phosphopeptides derived from endogenous proteins isolated from cells in which the kinase is either active or inhibited reveals new candidate protein substrates. The kinase assay linked with phosphoproteomics strategy was applied to identify unique substrates of spleen tyrosine kinase (Syk), a protein-tyrosine kinase with duel properties of an oncogene and a tumor suppressor in distinctive cell types. We identified 64 and 23 direct substrates of Syk specific to B cells and breast cancer cells, respectively. Both known and unique substrates, including multiple centrosomal substrates for Syk, were identified, supporting a unique mechanism that Syk negatively affects cell division through its centrosomal kinase activity.
Protein kinases and their substrates represent the largest signaling network that regulates protein–protein interactions, subcellular localization, and ultimately cellular functions (1, 2). Deregulation of the signaling network often leads to disease states such as human malignancies, diabetes, and immune disorders. Although many kinases are excellent therapeutic targets, the precise connection between protein kinases and their direct substrates has not been fully elucidated for a majority of protein kinases. Besides classical genetic and biochemical methods, there have been a number of high throughput approaches for the identification of potential kinase substrates. Common methods include in vitro kinase assays using libraries of synthetic peptides (3), phase expression libraries (4), protein/peptide arrays (5–7), or cell extracts (8, 9), but these methods can often be misleading and provide many false positive results. The discovery of physiological substrates for specific protein kinases has remained challenging, even with recent advances in mass spectrometry.
Mass spectrometry-based proteomics has become a powerful tool and been applied to map protein interaction networks, including kinase/phosphatase-substrate networks (10). Large-scale phosphoproteomics, however, does not typically reveal precise connections between protein kinases and their direct substrates (11, 12). In recent years, there have been increasing attempts to develop mass spectrometry-based proteomic strategies for the identification of elusive kinase substrates (7, 13, 14). These attempts commonly used purified, active kinases to phosphorylate cell lysate (or fractions of cell lysate) in vitro, followed by mass spectrometric analysis to identify phosphoproteins. The main challenge for these approaches is how to distinguish the kinase reaction from background phosphorylation events resulting from endogenous kinase activities. Radioisotope labeling using [γ-32P]ATP (8), the employment of a high concentration of purified kinase (8, 15), an additional heating step to inactivate endogenous kinase activities (9), and quantitative proteomics (10) are a few means to address these issues. An elegant chemical genetic approach was developed by Shokat and coworkers to use kinases engineered to accept bulky ATP analogs that can distinguish between wild-type and mutated or analog-sensitive kinases (16). Recently it has been coupled to quantitative proteomics, termed quantitative identification of kinase substrates (10), to identify substrate proteins of mitogen-activated protein kinase/Erk kinase. All these methods, however, have been limited to the identification of in vitro kinase substrates.
In this study, we have devised an integrated strategy termed kinase assay linked with phosphoproteomics (KALIP) for determining the substrate specificity and identifying direct substrates of protein kinase with high sensitivity and confidence. The strategy is based on a kinase reaction using formerly in vivo phosphorylated peptides as candidates. This step efficiently improves the sensitivity of the kinase reaction. The kinase reaction is further linked to endogenous phosphoproteomics modulated by the kinase of interest to detect bona fide substrates. To demonstrate the KALIP strategy, we used spleen tyrosine kinase (Syk) as our target kinase. Syk is a 72-kDa protein-tyrosine kinase known to have a crucial role in adaptive immune receptor signaling, in particular in B cells by coupling the B-cell receptor (BCR) for antigen to multiple intracellular signaling pathways and also in modulating cellular responses to inducers of oxidative stress in a receptor-independent fashion (17, 18). However, Syk also mediates diverse biological functions including cellular adhesion, innate immune recognition, osteoclast maturation, platelet activation, and vascular development (17). In addition, the expression of Syk is reduced or absent in many highly malignant carcinomas, suggesting that it can function as a tumor suppressor in some contexts (19). Although the role of Syk in signaling through antigen receptors is relatively well characterized, little is known about its direct substrates and the pathways that it regulates in epithelial cells. Therefore, in this study, we attempt to demonstrate the specificity and sensitivity of the KALIP approach by identifying Syk substrates in B cells and breast cancer cells to gain molecular insight of Syk’s distinctive roles in different cell types.
Results
The KALIP Strategy for the Identification of Direct Kinase Substrates.
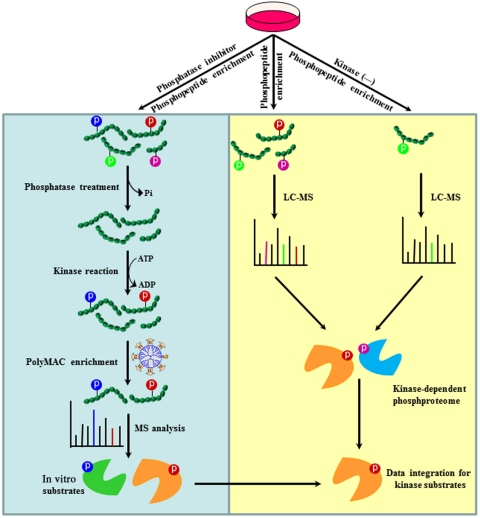
Current proteomics strategies using cell lysates for kinase assay can only identify potential in vitro kinase substrates. Furthermore, a majority of these strategies face interferences from endogenous kinase activities and enormous background phosphorylation in cell lysate. To address these issues, we devised the KALIP strategy that consists of two sets of experiments run in parallel: a newly designed sensitive kinase reaction carried out in vitro, and kinase-modulated phosphoproteomics in vivo (Fig. 1). To improve the sensitivity and specificity of the in vitro kinase reaction, we introduce a critical step to generate a pool of peptides derived from actual kinase substrates. The step includes first treating specific cell types with phosphatase inhibitors to increase the overall level of protein phosphorylation. Proteins are then extracted and digested with a protease. Phosphopeptides are isolated using affinity-based enrichment. A phosphatase is subsequently used to remove all phosphate groups from the phosphopeptides to generate a pool of highly relevant peptide substrate candidates for use in an in vitro kinase reaction. Note that the kinase reaction is performed at the peptide stage which efficiently eliminates any problem related to endogenous kinase contaminations (all endogenous kinases were digested before the kinase reaction). Phosphopeptides generated by in vitro phosphorylation with the kinase of interest are enriched using highly efficient polymer-based metal ion affinity capture (PolyMAC) (20), followed by mass spectrometric analyses to identify their sequences and sites of phosphorylation. This procedure generates a list of peptide substrates that can be compared to reveal the substrate specificity of the enzyme.
Fig. 1.
Workflow for the KALIP strategy to identify kinase candidates. It involves in vitro kinase reaction and in vivo phosphoproteome. In the in vitro kinase reaction, the critical step is the generation of substrate peptides, which are directly isolated from cell lysate through affinity purification and dephosphorylation. After the kinase reaction, phosphopeptides are further enriched and analyzed by mass spectrometry for sequencing and site identification. In in vivo phosphoproteome, phosphopeptides are enriched from two cell lines (+/- kinase) and analyzed by mass spectrometry. Kinase-modulated phosphorylation events are identified by comparing two phosphoproteomes (qualitatively or quantitatively). Phosphopeptides present within both datasets from in vitro kinase reaction and in vivo phosphoproteomics represent candidate proteins with the highest probability of being genuine Syk substrates. LC-MS, liquid chromatography-mass spectrometry.
Because in vitro kinase reactions typically display a degree of promiscuity (21), we propose here to link the in vitro kinase reaction to endogenous phosphoproteomic analyses using cells in which the kinase of interest is either active or inhibited. The kinase reaction generates in vitro phosphorylated peptides that include bona fide kinase substrates and a large number of artificial candidates due to the loss of its physiological regulatory mechanisms under in vitro condition. On the other hand, endogenous phosphoproteomic data include not only direct kinase substrates but also downstream proteins phosphorylated by other kinases activated by this particular kinase. We reason that the overlap between in vitro and endogenous kinase-modulated phosphoproteomic data represents genuine direct kinase substrates with highest possibility. The total number of direct kinase substrates depends on the specificity of the kinase in actual cell types.
Identification of Direct Syk Substrates in B Cells.
We applied the KALIP strategy to examine potential substrates for Syk in both B cells and breast cancer cells (the step-by-step scheme is illustrated in Fig. S1). Human DG75 B cells were first treated with pervanadate, an inducer of oxidative stress to activate Syk (18), and potent inhibitor of protein-tyrosine phosphatases, to elevate the overall level of phosphotyrosine-containing proteins in the cell and to increase the yield of phosphopeptides. Proteins were extracted from cell lysates, digested with trypsin, and peptides containing phosphotyrosine were isolated using a cocktail of immobilized antibodies against phosphotyrosine (4G10, PT66, and PY20). Phosphate groups were then removed from the phosphopeptides using an alkaline phosphatase, which was subsequently inactivated by pulse heating. Control experiments were carried out to examine the efficiency of dephosphorylation and the following deactivation of phosphatases from several commercial sources (Figs. S2–S4). Mass spectrometric analysis of former tyrosine phosphopeptides directly isolated from cell lysate revealed that virtually no phosphopeptides remained following phosphatase treatment.
The collection of candidate substrate peptides were evenly split and then incubated in a kinase reaction buffer containing ATP and Mg2+, with or without the addition of purified active Syk. The resulting phosphopeptides were enriched using PolyMAC, a highly efficient, titanium functionalized dendrimer support (20), and then analyzed by mass spectrometry. We identified 142 tyrosine phosphorylated peptides (out of a total of 156 peptide identifications; Dataset S1) in a sample that originated from 3 mg of DG75 whole cell extract, whereas virtually no phosphopeptides were detected in the sample without the addition of Syk. The experiment suggested that, although quantitative proteomics is highly desirable, tyrosine phosphorylation change with or without kinase was drastic and therefore we felt it was sufficient to make side-by-side comparison without actual use of stable isotope labeling. We also examined the exacted ion chromatograms to quantify the relative abundance of phosphopeptides in both samples. The majority of phosphopeptides that were not identified as being present in one sample were hardly detectable in the corresponding raw MS data. Only a small fraction of these phosphopeptides were detected with extremely low intensity and poor MS/MS spectra.
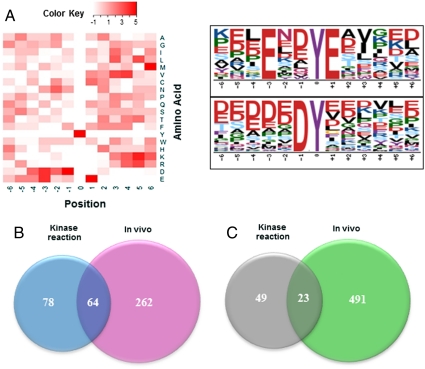
The sites on substrates that are phosphorylated by many protein kinases are dependent on the surrounding sequence of amino acids. The primary specificity determinants for a particular kinase are often identified through the use of extensive libraries consisting of multiple peptides of random sequences. The approach described here provides an alternative method for determining the substrate specificity of a kinase by generating an extensive collection of kinase reaction products derived from substrates that were actually phosphorylated in an intact cell. It also provides an extensive collection of peptides with sequences that actually exist on cellular proteins. An analysis of the consensus sequences (Fig. 2A) for substrates of Syk indicates an abundance of acidic residues surrounding the phosphorylated tyrosine, which was also identified using Motif-X (22). This consensus is consistent with those determined by screening a phage-display library or by a comparison of known substrates for Syk (23).
Fig. 2.
(A) Motif analysis of in vitro kinase phosphorylation for Syk substrate specificity; the distribution of amino acid residues is depicted by heat map (Left) and by Motif-X (22) (Right). (B) Venn diagram illustrating the overlap in the sets of tyrosine phosphopeptides identified using in vitro kinase reaction and in vivo Syk-modulated tyrosine phosphoproteome in DG-75 cells; and (C) Venn diagram illustrating the overlap in the sets of tyrosine phosphopeptides identified using in vitro kinase reaction and in vivo Syk-modulated tyrosine phosphoproteome in MDA-MB-231 cells. The overlaps between the data from in vitro kinase reaction and from kinase-dependent phosphoproteome represent genuine Syk substrates in B cells (64) and breast cancer cells (23), respectively.
Tyrosine kinases vary in the degree of their selectivity for specific primary sequences surrounding their sites of phosphorylation. To demonstrate the sensitivity and specificity of this approach, we compared the repertoire of peptides phosphorylated by Syk to those phosphorylated by a different protein-tyrosine kinase with less well-defined substrate specificity. We carried out in vitro kinase reactions using cellular-Src kinase (Csk) with either a mixture of dephosphorylated peptides generated from cellular proteins present in a DG75 cell lysate or a mixture of formerly tyrosine phosphorylated and then dephosphorylated peptides prepared from DG75 cells. Our proteomic analysis of dephosphorylated peptides generated directly from whole cell extracts identified over 200 peptides phosphorylated by Csk in vitro, but none were known Csk substrates. The analysis of formerly tyrosine phosphorylated peptides did identify one known Csk substrate (Lyn) and the false positive identification was significantly lower (Dataset S1). In both cases certain selectivity for substrates of Csk was determined (Fig. S5), which is consistent with biochemical experiments based on in vitro kinase assays using a synthetic peptide library (24). The comparison revealed higher sensitivity and specificity of kinase reactions when formerly phosphorylated peptides were used as potential substrates. It is also important to note that the list of peptides phosphorylated by Csk was distinct from those phosphorylated by Syk, illustrating how different kinases can be distinguished from one another using this approach and that the appearance in the analysis of the highly acidic Syk phosphorylated peptides was not an artifact of the recovery procedure.
The list of substrates phosphorylated in the in vitro kinase assay using Syk provides important clues to the identity of actual protein substrates. However, removal of the kinase from the cell often results in a loss of physiological regulatory mechanisms and kinases can be particularly promiscuous when peptide substrates are employed in vitro. The use of high concentration of purified kinase in vitro is partially responsible for lower specificity as well. To enhance our ability to identify true substrates, we compared endogenous phosphotyrosine-containing peptides derived from phosphoproteins from DG75 cells that were treated with or without the Syk inhibitor, piceatannol. Isolation of phosphopeptides by a tandem purification scheme using antiphosphotyrosine antibody and PolyMAC (20) and analysis by mass spectrometry resulted in the identification of over 400 phosphotyrosine-containing peptides. A larger number were identified in samples from untreated as compared to inhibitor treated cells. This observation was consistent with a comparison of levels of phosphotyrosine-containing proteins detected by Western blotting with antibodies against phosphotyrosine. Selectivity for the recovery of phosphopeptides using the PolyMAC reagent was excellent (only 24 nonphosphorylated peptides were identified among a total of 472 peptides, corresponding to > 95% selectivity). Again, to complement identification, we measured the relative abundance of tyrosine phosphopeptides from DG75 cells expressing or inhibiting Syk with normalized exacted ion chromatograms. Overall, we identified 326 unique sites of tyrosine phosphorylation representing 235 phosphoproteins that were present dominantly in samples in which Syk was not inhibited (Dataset S1).
Many kinases do not have specific inhibitors available and, to further demonstrate its generality of the strategy present here, we carried out experiments with Syk-deficient DT40 B cells in parallel with cells in which the expression of Syk was restored by transfection with plasmids coding for the wild-type kinase. A similar number of Syk-dependent phosphoproteins was identified in DT-40 cells, including a number of known Syk kinases reported previously (Dataset S1), showing that multiple strategies can be used to inhibit the kinase activity.
Phosphopeptides present within both datasets (i.e., from proteins phosphorylated in cells expressing Syk and those phosphorylated by Syk by the in vitro kinase reaction) should represent sites on candidate proteins with the highest probability of being genuine Syk substrates (Fig. 2B) (See Table S1 for the full list.) Among the candidate proteins, nine are known substrates for Syk, representing over two-thirds of known substrates in this cell type and more than 50% of Syk substrates reported previously in the literature (Table 1). The sites that were phosphorylated were also consistent with previous literature reports, although not all sites of phosphorylation were previously characterized for some substrates. At the same time, many potential substrates have not been described previously. To further validate the methodology for the identification of unique Syk substrates, we selected a limited number of candidates to explore further. These included the microtubule-associated protein, MAPRE1; the nuclear, casein kinase and cyclin-dependent kinase substrate, NUCKS; hepatocellular carcinoma-associated antigen 59 (HCA59); methylosome subunit pICln, CLNS1A; Pit-Oct-Unc (POU) domain class 2-associating factor 1, BOB1; and NIMA (never in mitosis gene a)-related kinase 9, Nek9 (Table 1). Each protein was immunoprecipitated from lysates of DG75 cells and then incubated with Syk in a reaction buffer containing ATP. Analyses of the reaction products by Western blotting with antibodies against phosphotyrosine confirmed that all could serve directly as substrates for Syk (Fig. 3A). Because Syk is activated in B cells following aggregation of the BCR, we treated DG75 cells with antibodies against surface IgM (25). We detected the receptor-stimulated phosphorylation of HCA59, CLNS1A, Nek9, and BOB1, indicating that these substrates may be involved in signaling pathways downstream of the BCR. We were unable to demonstrate the phosphorylation of either MAPRE1 or NUCKS in this assay (Fig. 3A).
Table 1.
Known substrates and confirmed other substrates of Syk in DG75
| Substrate | Site of phosphorylation | Reference | |
| Known substrates identified in B cells | |||
| HPK1 | LSESSDDDyDDVDIP | Y381 | (33) |
| BLNK | LLEDEADyVVPVEDN | Y178 | (34) |
| BTK | RYVLDDEyTSSVGSK | Y550 | (35) |
| GCET2 | GNSAEEYyENVPCKA | Y107 | (36) |
| HS1 | EPEPENDyEDVEEMD | Y378 | (37) |
| LAT | EDEESEDyQNSASIH | Y193 | (38) |
| PLCG1 | IGTAEPDyGALYEGR | Y771 | (39, 40) |
| TUBA | MAALEKDyEEVGVDS | Y432 | (41) |
| Syk | LPMDTEVyESPYADPTEVYESPyADPEEIR | Y348Y352 | (42) |
|
Confirmed new Syk substrates in B cells | |||
| MAPRE1 | FFDANYDGKDyDPVAAR | Y124 | |
| NUCKS | SQFQESDDADEDyGR | Y13 | |
| BOB1 | LLLEEEDSDAyALNHTLS | Y245 | |
| CLNS1A | TEDSIRDyEDGMEVDT | Y214 | |
| HCA59 | NAEDcLyELPENIR | Y147 | |
| Nek9 | LGLDSEEDyYTPQK | Y520 | |
HPK1, mitogen-activated protein kinase kinase kinase kinase 1; BLNK, B-cell linker protein; BTK, bruton tyrosine kinase; GCET2, germinal center B-cell-expressed transcript 2 protein; HS1, hematopoietic lineage cell-specific protein; LAT, linker for activation of T-cells family member 2; PLCG1, 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-1; TUBA, tubulin alpha; Syk, spleen tyrosine kinase; MAPRE1, microtubule-associated protein RP/EB family member 1; NUCKS, nuclear ubiquitous casein and cyclin-dependent kinases substrate; BOB1, Pit-Oct-Unc (POU) domain class 2-associating factor 1; CLNS1A, methylosome subunit pICln; HCA59, hepatocellular carcinoma-associated antigen 59; Nek9, NIMA-related kinase 9.
Fig. 3.
(A) Confirmation of Syk kinase substrates through in vitro kinase assay and endogenous phosphorylation changes in response to IgM stimulation in DG-75 cells. For in vitro kinase assay, individual proteins were isolated by immunoaffinity purification from cell lysate and kinase reaction was carried out directly on the beads with ATP and Syk. The reaction mixture was separated by SDS-PAGE and analyzed by Western blotting (WB) with the indicated antibodies and antiphosphotyrosine antibody (4G10). For IgM stimulation, individual proteins were isolated from cell lysate, directly separated by SDS-PAGE, and analyzed by WB with the indicated antibodies and antiphosphotyrosine antibody (4G10); (B) in vitro kinase assays of GST-Nek9 full length, truncated, and mutant (Y520I/Y521D) with or without the presence of Syk.
Identification of Direct Syk Substrates in Breast Cancer Cells.
Because Syk plays the unusual role of tumor suppressor in breast cancer cells but few direct substrates and interacting proteins have been reported (26), we applied the KALIP strategy to explore Syk substrates that might be specific to these cells. To facilitate this analysis, we used a line of highly invasive MDA-MB-231 breast cancer cells that normally lack Syk, but were engineered such that the expression of an EGFP-tagged form of the kinase could be induced by treatment with tetracycline (27). We carried out in vitro kinase reaction and cellular tyrosine phosphoproteome analyses in both Syk+ and Syk− breast cancer cells using the same strategy described above. A total of 72 substrate peptides for Syk were identified following the in vitro kinase reaction (Fig. 2C and Dataset S1). In the analysis of the phosphotyrosine proteome (20), a much larger number of phosphotyrosine-containing peptides was identified in samples from cells induced to express Syk-EGFP as compared to those lacking Syk (820 versus 377 unique phosphopeptides). Selectivity for the recovery of phosphopeptides using the PolyMAC reagent in this experiment was excellent (only 51 nonphosphopeptides were identified among a total of 871 peptides identified, corresponding to > 94% selectivity in the Syk-EGFP-induced sample). Overall, we identified 514 unique sites of tyrosine phosphorylation on 458 different proteins that were present only in samples from Syk-EGFP-expressing cells. The overlap of the two datasets allowed us to identify 23 potential substrates for Syk in breast cancer cells (see Fig. 2C and Table S2 for the full list). The list includes the only known substrate for Syk in breast cancer cells, cortactin (27).
Active Syk Phosphorylates Multiple Centrosomal Substrates.
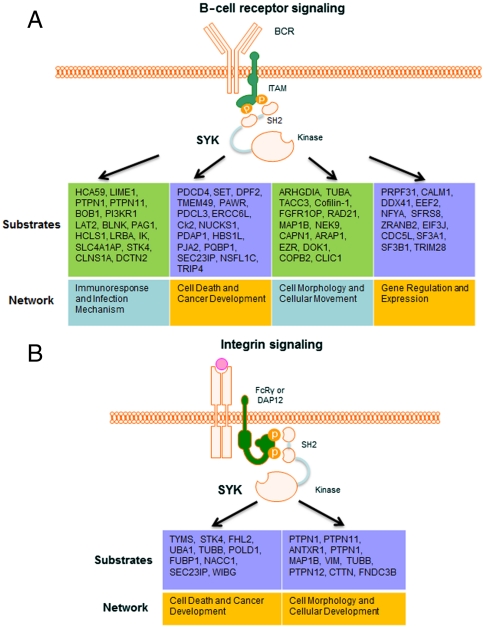
To globally compare functional roles of identified Syk substrates in B cells and breast cancer cells, we performed bioinformatic analyses to identify the signaling pathways and networks to which these substrates belong (Fig. 4). The identified Syk substrates in B cells are mainly involved in four signaling pathways, one related to immune cell responses and three related to cellular movement and cancer development. Proteins involved in immune cell signaling include multiple adaptor proteins [B-cell linker protein (BLNK), linker for activation of T-cells family member 2 (LAT2), Lck-interacting transmembrane adapter 1 (LIME1), and phosphoprotein associated with glycosphingolipid-enriched microdomains 1 (PAG1)] as well as BOB1, a transcriptional coactivator required for transcription of the B29 gene that codes for the CD79b component of the B-cell antigen receptor and a protein not known previously to be phosphorylated on tyrosine (28). The identified substrates in breast cancer cells are mainly involved in two processes, cellular organization and cancer development.
Fig. 4.
Identified Syk substrates and their functional networks in B cells (A) and epithelial cells (B).
The observation that multiple substrates for Syk involved in cellular organization are centrosomal proteins [e.g., MAPRE1, Microtubule-associated protein 1B (MAP1B), Cofilin-1 (CFL), Dynactin subunit 2 (DCTN2), Fibroblast growth factor receptor 1 oncogene partner (FGFR1OP), NIMA-related kinase 9 (Nek9), and Transforming acidic coiled-coil-containing protein 3 (TACC3)] is in particular intriguing, and it provides the direct evidence for previous reports of Syk’s centrosomal localization and kinase activity and its role in the negative regulation of mitosis in multiple cell types (29–31). In our experiment, we confirmed that Syk is indeed localized to the centrosome in MDA-MB-231. In addition, we treated MDA-MB-231 cells expressing Syk-EGFP with H2O2 to induce oxidative stress and activate Syk. In these cells, treatment with H2O2 resulted in the localization of a substantial portion of Syk-EGFP to the centrosome (Fig. S6). To confirm the discovery by mass spectrometry, we selected one centrosomal protein, Nek9, for further studies. Nek9 interacts with other NIMA-family kinases to form a mitotically activated module with key roles during mitotic progression, but the module activation mechanism is not totally clear (32). The KALIP method identified Nek9 as a potential Syk substrate at Y520 and Y521 sites (Y520 and Y521 were phosphorylated in in vitro kinase reaction, whereas only Y520 was phosphorylated endogenously). GST-Nek9 fusion proteins in full length and truncated with Y520I/Y521D mutants were cloned and purified from Escherichia coli. Using an in vitro kinase assay and probing for phosphotyrosine using anti-pY antibody, we confirmed the loss of tyrosine phosphorylation in the Y520I/Y521D mutant (Fig. 3B and Fig. S7). Thus, our study has established strong connections between centrosomal substrates and the kinase and provides important clues in terms of the regulatory role of Syk in cell apoptosis and survival.
Discussion
Despite increasing efforts in the analysis of phosphoproteomes, a majority of mass spectrometry-based proteomics studies cannot pinpoint kinase-substrate pairs under physiological environment. The KALIP strategy distinguishes itself from others by virtue of linking comparative phosphoproteomics to kinase reactions for comprehensive identification of bona fide kinase substrates. Further, kinase reaction with peptides derived directly from potential kinase substrates improves the sensitivity and specificity of in vitro kinase assay. In our search for direct target proteins of Syk in both B cells and breast cancer cells, more than two-thirds of known Syk substrates were among the short lists of potential Syk substrates in two cell types. We further verified a few other substrates identified by the strategy through immunoprecipitation and in vitro kinase assay. Some of these substrates are involved in the BCR pathway, whereas others are independent of BCR activation, according to their phosphorylation status changes in response to anti-IgM stimulation. The identification of multiple centrosomal proteins as potential Syk substrates supports previous reports of Syk’s centrosomal kinase activity and offers possible mechanism in terms of a unique role of Syk in cell apoptosis and survival. The discovery of distinct Syk substrates in B cells and breast cancer cells has also confirmed much recent evidence that, as a group, tyrosine kinases such as Syk play complex roles in multiple signaling pathways that are essential to both the positive and negative regulation of cell growth and proliferation. In addition, most chemotherapies, even those directed against tyrosine kinases, face the inevitable problem of relapse, often due to the activation of alternative signaling pathways. There is great need, therefore, for highly effective systems approaches to dissect these multiple signaling pathways at the molecular level. The proteomics-based KALIP strategy can be a powerful tool to decipher complex signaling cascades one kinase and one cell type at a time.
We have demonstrated the successful identification of direct substrates of a tyrosine kinase and it is conceivable that the KALIP can also be applied to serine/threonine kinases as well. Serine and threonine phosphopeptides can be enriched and dephosphorylated to provide a pool of candidates for sensitive kinase reaction in vitro. However, compared to tyrosine phosphorylation, serine/threonine phosphorylation is much more extensive. As a result, the preenrichment of serine/threonine phosphopeptides may be not as inclusive as that of tyrosine phosphopeptides. Furthermore, large amount of serine and threonine phosphorylation also makes it difficult to achieve subsequent dephosphorylation completely. In contrast to this present study, quantitative proteomics, based on either stable isotope labeling or label-free experiments, is unquestionably necessary to distinguish phosphorylation events as a result of kinase reaction from background phosphorylation.
The kinase reaction is an important component of the KALIP method. Although it is performed at the peptide stage, efficiently eliminating any problem related to endogenous kinase contaminations, the KALIP method may not be effective for kinases that require a priming phosphorylation event, additional interacting surfaces, or a docking site on the protein (e.g., in the case of Csk). In addition, the KALIP approach cannot eliminate certain false-positives in cases where substrates of other downstream kinases have similar motifs as the kinase of interest due to the loss of localization information when the cell is lysed. Furthermore, tryptic digestion will abolish certain motifs containing multiple basic residues that are required by basophilic kinases, although this issue can be partially resolved by employing a different protease to generate peptide substrates. On another note, because peptides typically have relatively high Km values for phosphorylation by tyrosine kinases, it requires higher concentrations of peptides in a solution assay. The design of KALIP strategy addresses this issue. Peptides are derived from those formerly tyrosine phosphorylated, effectively removing highly abundant background peptides from cell lysate that may interfere with the kinase reaction and result in high false-positives. In order to further increase the sensitivity, an effective phosphatase inhibitor (pervandate in our study) is applied to elevate the phosphorylation level, resulting in the isolation of large amount of phosphopeptides for the kinase reaction.
The KALIP strategy also underscores the need for highly specific inhibitors for the kinase of interest. The use of kinase knockout such as siRNA can be an alternative solution to generate kinase-dependent phosphoproteomics. Overall, by manipulating the types of phosphatase inhibitors or kinase activator that are used, methods of phosphopeptide enrichment, the nature of protease used to generate peptide substrates, and types of cells or model organisms, the KALIP approach should be generally applicable to the analysis of the substrate specificity and to the identification of unique substrates for virtually any protein kinase.
Materials and Methods
Details on cell culture, phosphopeptide enrichment, in vitro kinase assay, mass spectrometric data acquisition and data analysis, immunoprecipitation, cloning, and protein purification are provided in SI Materials and Methods. All data files after mass spectrometric database searches are provided as Tables S1 and S2 and Dataset S1. Tandem mass spectra showing the sequence and phosphorylation sites of identified Syk substrates are available at the public domain https://proteomecommons.org/tranche/data-downloader.jsp?fileName=NJIzQonHf%2FtJl7of9zJDWVFtiYWPAPfJh22qiicZH%2BN4DQfXPHFhlfgmF9khR6UnNNp9tr7zzReiTBKRZCtwS6RnQSsAAAAAAAABtw%3D%3D.
Supplementary Material
Acknowledgments.
This project has been funded in part by an National Science Foundation CAREER Award, Che-0645020 (to W.A.T.) and by National Institutes of Health Grants 1R01GM088317 and 5R21RR025802 (to W.A.T.), CA115465-03 (to R.L.G. and W.A.T.), and CA037372 (to R.L.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The tandem mass spectrum gallery is available free of charge at https://proteomecommons.org/tranche/data-downloader.jsp?fileName=NJIzQonHf%2FtJl7of9zJDWVFtiYWPAPfJh22qiicZH%2BN4DQfXPHFhlfgmF9khR6UnNNp9tr7zzReiTBKRZCtwS6RnQSsAAAAAAAABtw%3D%3D.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119418109/-/DCSupplemental.
References
- 1.Hunter T. Signaling—2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 2.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 3.Yaffe MB, et al. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat Biotechnol. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- 4.Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mok J, Im H, Snyder M. Global identification of protein kinase substrates by protein microarray analysis. Nat Protoc. 2009;4:1820–1827. doi: 10.1038/nprot.2009.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam KS, Liu R, Miyamoto S, Lehman AL, Tuscano JM. Applications of one-bead one-compound combinatorial libraries and chemical microarrays in signal transduction research. Acc Chem Res. 2003;36:370–377. doi: 10.1021/ar0201299. [DOI] [PubMed] [Google Scholar]
- 7.Amanchy R, et al. Identification of c-Src tyrosine kinase substrates using mass spectrometry and peptide microarrays. J Proteome Res. 2008;7:3900–3910. doi: 10.1021/pr800198w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knebel A, Morrice N, Cohen P. A novel method to identify protein kinase substrates: eEF2 kinase is phosphorylated and inhibited by SAPK4/p38delta. EMBO J. 2001;20:4360–4369. doi: 10.1093/emboj/20.16.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Troiani S, et al. Searching for biomarkers of Aurora-A kinase activity: Identification of in vitro substrates through a modified KESTREL approach. J Proteome Res. 2005;4:1296–1303. doi: 10.1021/pr050018e. [DOI] [PubMed] [Google Scholar]
- 10.Morandell S, et al. QIKS—quantitative identification of kinase substrates. Proteomics. 2010;10:2015–2025. doi: 10.1002/pmic.200900749. [DOI] [PubMed] [Google Scholar]
- 11.Bodenmiller B, Aebersold R. Quantitative analysis of protein phosphorylation on a system-wide scale by mass spectrometry-based proteomics. Methods Enzymol. 2010;470:317–334. doi: 10.1016/S0076-6879(10)70013-6. [DOI] [PubMed] [Google Scholar]
- 12.Kubota K, et al. Sensitive multiplexed analysis of kinase activities and activity-based kinase identification. Nat Biotechnol. 2009;27:933–940. doi: 10.1038/nbt.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang SY, Tsai ML, Chen GY, Wu CJ, Chen SH. A systematic MS-based approach for identifying in vitro substrates of PKA and PKG in rat uteri. J Proteome Res. 2007;6:2674–2684. doi: 10.1021/pr070134c. [DOI] [PubMed] [Google Scholar]
- 14.Coba MP, et al. Neurotransmitters drive combinatorial multistate postsynaptic density networks. Sci Signal. 2009;2:ra19. doi: 10.1126/scisignal.2000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen P, Knebel A. KESTREL: A powerful method for identifying the physiological substrates of protein kinases. Biochem J. 2006;393:1–6. doi: 10.1042/BJ20051545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah K, Liu Y, Deirmengian C, Shokat KM. Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc Natl Acad Sci USA. 1997;94:3565–3570. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mocsai A, Ruland J, Tybulewicz VL. The SYK tyrosine kinase: A crucial player in diverse biological functions. Nat Rev Immunol. 2010;10:387–402. doi: 10.1038/nri2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geahlen RL. Syk and pTyr’d: Signaling through the B cell antigen receptor. Biochim Biophys Acta. 2009;1793:1115–1127. doi: 10.1016/j.bbamcr.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coopman PJ, et al. The Syk tyrosine kinase suppresses malignant growth of human breast cancer cells. Nature. 2000;406:742–747. doi: 10.1038/35021086. [DOI] [PubMed] [Google Scholar]
- 20.Iliuk AB, Martin VA, Alicie BM, Geahlen RL, Tao WA. In-depth analyses of kinase-dependent tyrosine phosphoproteomes based on metal ion functionalized soluble nanopolymers. Mol Cell Proteomics. 2010;9:2162–2172. doi: 10.1074/mcp.M110.000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manning BD, Cantley LC. Hitting the target: Emerging technologies in the search for kinase substrates. Sci STKE. 2002;2002:49pe. doi: 10.1126/stke.2002.162.pe49. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 23.Schmitz R, Baumann G, Gram H. Catalytic specificity of phosphotyrosine kinases Blk, Lyn, c-Src and Syk as assessed by phage display. J Mol Biol. 1996;260:664–677. doi: 10.1006/jmbi.1996.0429. [DOI] [PubMed] [Google Scholar]
- 24.Sondhi D, Xu W, Songyang Z, Eck MJ, Cole PA. Peptide and protein phosphorylation by protein tyrosine kinase Csk: Insights into specificity and mechanism. Biochemistry. 1998;37:165–172. doi: 10.1021/bi9722960. [DOI] [PubMed] [Google Scholar]
- 25.Harwood NE, Batista FD. Early events in B cell activation. Annu Rev Immunol. 2010;28:185–210. doi: 10.1146/annurev-immunol-030409-101216. [DOI] [PubMed] [Google Scholar]
- 26.Larive RM, et al. Phosphoproteomic analysis of Syk kinase signaling in human cancer cells reveals its role in cell-cell adhesion. Oncogene. 2009;28:2337–2347. doi: 10.1038/onc.2009.99. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Shrikhande U, Alicie BM, Zhou Q, Geahlen RL. Role of the protein tyrosine kinase Syk in regulating cell-cell adhesion and motility in breast cancer cells. Mol Cancer Res. 2009;7:634–644. doi: 10.1158/1541-7786.MCR-08-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schubart DB, Rolink A, Kosco-Vilbois MH, Botteri F, Matthias P. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature. 1996;383:538–542. doi: 10.1038/383538a0. [DOI] [PubMed] [Google Scholar]
- 29.Faruki S, Geahlen RL, Asai DJ. Syk-dependent phosphorylation of microtubules in activated B-lymphocytes. J Cell Sci. 2000;113:2557–2565. doi: 10.1242/jcs.113.14.2557. [DOI] [PubMed] [Google Scholar]
- 30.Zyss D, et al. The Syk tyrosine kinase localizes to the centrosomes and negatively affects mitotic progression. Cancer Res. 2005;65:10872–10880. doi: 10.1158/0008-5472.CAN-05-1270. [DOI] [PubMed] [Google Scholar]
- 31.Uckun FM, Ozer Z, Qazi S, Tuel-Ahlgren L, Mao C. Polo-like-kinase 1 (PLK1) as a molecular target to overcome SYK-mediated resistance of B-lineage acute lymphoblastic leukaemia cells to oxidative stress. Br J Haematol. 2010;148:714–725. doi: 10.1111/j.1365-2141.2009.07983.x. [DOI] [PubMed] [Google Scholar]
- 32.Bertran MT, et al. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO J. 2011;30:2634–2647. doi: 10.1038/emboj.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuji S, et al. B cell adaptor containing src homology 2 domain (BASH) links B cell receptor signaling to the activation of hematopoietic progenitor kinase 1. J Exp Med. 2001;194:529–539. doi: 10.1084/jem.194.4.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu C, Turck CW, Kurosaki T, Chan AC. BLNK: a central linker protein in B cell activation. Immunity. 1998;9:93–103. doi: 10.1016/s1074-7613(00)80591-9. [DOI] [PubMed] [Google Scholar]
- 35.Baba Y, et al. BLNK mediates Syk-dependent Btk activation. Proc Natl Acad Sci USA. 2001;98:2582–2586. doi: 10.1073/pnas.051626198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan Z, et al. Studies of a germinal centre B-cell expressed gene, GCET2, suggest its role as a membrane associated adapter protein. Br J Haematol. 2007;137:578–590. doi: 10.1111/j.1365-2141.2007.06597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brunati AM, et al. Thrombin-induced tyrosine phosphorylation of HS1 in human platelets is sequentially catalyzed by Syk and Lyn tyrosine kinases and associated with the cellular migration of the protein. J Biol Chem. 2005;280:21029–21035. doi: 10.1074/jbc.M412634200. [DOI] [PubMed] [Google Scholar]
- 38.Zhang W, Sloan-Lancaster J, Kitchen J, Trible RP, Samelson LE. LAT: The ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell. 1998;92:83–92. doi: 10.1016/s0092-8674(00)80901-0. [DOI] [PubMed] [Google Scholar]
- 39.Law CL, Chandran KA, Sidorenko SP, Clark EA. Phospholipase C-gamma1 interacts with conserved phosphotyrosyl residues in the linker region of Syk and is a substrate for Syk. Mol Cell Biol. 1996;16:1305–1315. doi: 10.1128/mcb.16.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulanova E, et al. The IL-15R alpha chain signals through association with Syk in human B cells. J Immunol. 2001;167:6292–6302. doi: 10.4049/jimmunol.167.11.6292. [DOI] [PubMed] [Google Scholar]
- 41.Peters JD, Furlong MT, Asai DJ, Harrison ML, Geahlen RL. Syk, activated by cross-linking the B-cell antigen receptor, localizes to the cytosol where it interacts with and phosphorylates alpha-tubulin on tyrosine. J Biol Chem. 1996;271:4755–4762. doi: 10.1074/jbc.271.9.4755. [DOI] [PubMed] [Google Scholar]
- 42.Keshvara LM, et al. Syk- and Lyn-dependent phosphorylation of Syk on multiple tyrosines following B cell activation includes a site that negatively regulates signaling. J Immunol. 1998;161:5276–5283. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.