Abstract
IRE1 couples endoplasmic reticulum unfolded protein load to RNA cleavage events that culminate in the sequence-specific splicing of the Xbp1 mRNA and in the regulated degradation of diverse membrane-bound mRNAs. We report on the identification of a small molecule inhibitor that attains its selectivity by forming an unusually stable Schiff base with lysine 907 in the IRE1 endonuclease domain, explained by solvent inaccessibility of the imine bond in the enzyme-inhibitor complex. The inhibitor (abbreviated 4μ8C) blocks substrate access to the active site of IRE1 and selectively inactivates both Xbp1 splicing and IRE1-mediated mRNA degradation. Surprisingly, inhibition of IRE1 endonuclease activity does not sensitize cells to the consequences of acute endoplasmic reticulum stress, but rather interferes with the expansion of secretory capacity. Thus, the chemical reactivity and sterics of a unique residue in the endonuclease active site of IRE1 can be exploited by selective inhibitors to interfere with protein secretion in pathological settings.
Keywords: 8-formyl-umbelliferone, unfolded protein response, high-throughput screening, reversible covalent inhibitor, aldehydes
Perturbation of the protein folding environment in the endoplasmic reticulum (ER) leads to rectifying changes in gene expression and protein synthesis. These are mediated by an unfolded protein response (UPR), whose most conserved arm is initiated by an ER localized transmembrane protein, IRE1 (1, 2). The lumenal domain of IRE1 senses the perturbation in the ER and transmits the ER stress signal across the ER membrane to the effector cytosolic domain of the protein. This effector domain is endowed with two linked enzymatic activities: a protein kinase and an RNase, and both are activated by ER stress.
The most conserved output of IRE1 signaling is the site-specific cleavage of an mRNA, the product of the yeast HAC1 gene (3) and its metazoan orthologue Xbp1 (4, 5). Cleavage occurs at two distinct sites and is followed by ligation of the 5′ and 3′ fragments to generate an ER stress-dependent spliced mRNA encoding a potent transcription factor. The target genes of spliced XBP1 (and Hac1p) enhance the ability of the ER to cope with unfolded proteins (6) and also act more broadly to upregulate secretory capacity (7). In addition, mammalian IRE1 contributes to the promiscuous degradation of membrane-associated mRNAs in a process known as regulated IRE1-dependent degradation (or RIDD), but whose mechanistic basis and functional consequences are incompletely understood (8, 9).
Fluctuating levels of ER stress accompany diverse physiological conditions. In metazoans, IRE1 signaling constitutes one arm of a three-pronged UPR. The other two arms are mediated by the translation initiation factor 2α (eIF2α) kinase PERK, which attenuates ER load by inhibiting protein synthesis in stressed cells, and by a parallel transcriptional pathway mediated by ATF6. Redundancy between the long-term transcriptional programs mediated by the three arms of the UPR has obscured the interpretation of genetic experiments to ascertain the unique role of IRE1 (10). Furthermore, the UPR is engaged in cancer cells as a survival pathway (11, 12), in effector cells of the immune response as a means of enhancing secretion of inflammatory mediators (13, 14), in virally infected cells to support viral replication (15), and negative feedback loops connect the UPR to autophagy (16, 17). These considerations have generated an interest in IRE1 inhibitors both as tools for fundamental research and as potential anticancer, antiinflammatory, and antiviral therapeutic agents (18).
IRE1 activation is initiated by homotypic interactions of the stress-sensing lumenal domain (19–21) favoring transautophosphorylation of the kinase-extension nuclease (KEN) domain on the cytoplasmic side of the ER membrane (22, 23). Phosphorylation promotes nucleotide binding in a pocket common to other protein kinases (24). Nucleotide binding affects a subtle conformational change in IRE1, stabilizing an unusual back-to-back dimer of the KEN domain (24, 25). This active IRE1 dimer, which may be further reinforced by higher-order oligomerization (26, 27), juxtaposes the RNase active site of two adjacent protomers, orienting the relevant residues for catalysis (25). Ligands have been proven to regulate (activate or inhibit) the essential dimerization step by engaging the nucleotide-binding site or by binding in a deep hydrophobic pocket at the dimer interface (28). Thus, our understanding of the molecular events leading to IRE1 activation suggest at least three sites that can be engaged by small molecule inhibitors: the nucleotide-binding pocket, the dimer-interface pocket, and the RNase active site.
Here, we describe the identification and characterization of a small molecule selective inhibitor of IRE1 RNase activity. By exploiting the unique properties of this covalent inhibitor we have been able to uncover the molecular basis for IRE1 inhibition by a class of aromatic aldehydes. Our observations identify and unify the mechanism of action of extant IRE1 inhibitors and provide insight into the participation of IRE1 in Xbp1 splicing and RIDD and their functional consequences.
Results
Identification of 8-formyl-7-hydroxy-4-methylcoumarin as a Potent Inhibitor of IRE1.
A previously described fluorescent-based in vitro FRET-derepression assay for IRE1 RNase activity (28) was adapted to high-throughput screening for small molecule inhibitors. The cytoplasmic domain of human IRE1α (HsIRE1α464–977), purified from insect SF9 cells, possesses potent sequence-specific endoribonuclease activity and proved well suited for this assay (Fig. S1 and SI Materials and Methods).
A collection of 238,287 pure compounds and partially purified natural extracts was screened for inhibitors of IRE1 RNase activity (Fig. S1). Inhibitors that satisfied our stringent in vitro test criteria were further assayed for their ability to inhibit IRE1-mediated Xbp1 splicing in ER stressed cultured cells. Members of only one class of inhibitors, umbelliferones (7-hyroxy-4-methylcoumarin derivatives), were active in both the in vitro and in vivo assays and were thus selected for further study.
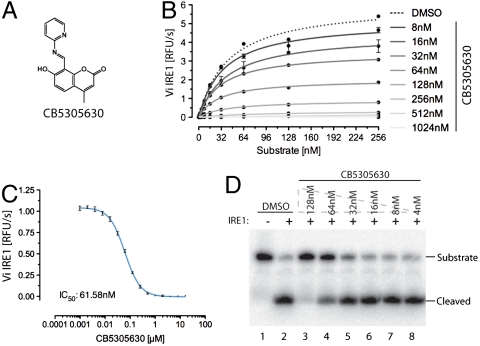
Kinetic analysis of the most potent compound, CB5305630 (Fig. 1A), displayed noncompetitive inhibition of IRE1 RNase activity (Fig. 1B) with an apparent IC50 of 60 nM in the FRET-derepression assay (Fig. 1C). CB5305630 also inhibited the cleavage of a radiolabeled Xbp1-derived RNA probe in vitro (Fig. 1D), confirming the block in IRE1 RNase activity.
Fig. 1.
Coumarin-based inhibition of IRE1. (A) Structure of CB5305630 [7-hydroxy-4-methyl-8-((pyridine-2-ylimino)methyl)-2H-chromenone], the most potent IRE1 inhibitor identified in a screen of 238,287 compounds. Note the imine bond between the umbelliferone and the pyridine. (B) A plot of IRE1’s RNase initial velocity (Vi) as a function of RNA substrate concentration in the presence of the indicated concentration of CB5305630 (mean ± SD, n = 2). Km for the RNA substrate is approximately 37 nM and is unaffected by the inhibitor, whereas the Vmax is inhibited in a concentration-dependent manner with a Ki of approximately 60 nM. (C) A plot of the concentration-dependent inhibition of IRE1’s RNase activity by CB5305630 (mean ± SD, n = 4). (D) Autoradiograph of 32P-labeled RNA substrate following cleavage by IRE1 in the absence or presence of the indicated concentration of CB5305630.
CB5305630 is 8-formyl-7-hydroxy-4-methylcoumarin conjugated to 2-aminopyridine via an aldimine. Structure activity relationship analysis of the primary data from the screen suggested the importance of this linkage to the activity, as the corresponding reduced amines were inactive (Fig. S2). Imines of the structural form of CB5305630 will be subject to rapid hydrolytic cleavage in aqueous media. Indeed, time-resolved NMR analysis of CB5305630 demonstrated facile hydrolysis to its constituent fragments (aldehyde and 2-aminopyridine) with a half-life of approximately 10 min (Fig. S3). Of the product of this hydrolytic event, 2-aminopyridine was inactive while 8-formyl-7-hydroxy-4-methylcoumarin had an indistinguishable inhibitory profile to CB5305630 when tested in pure form (Fig. S4) and therefore likely represents the active component in CB5305630. It was given the lab name 4μ8C (4-methyl umbelliferone 8-carbaldehyde) and characterized further.
Identification of 4μ8C Binding Sites in IRE1.
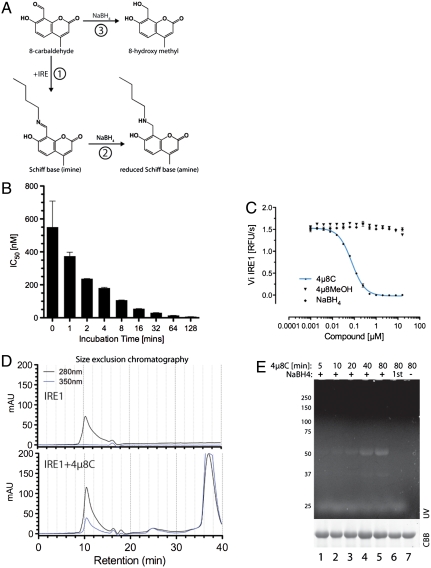
Aldehydes, such as 4μ8C, can react covalently with diverse nucleophiles, forming a Schiff base with the epsilon amino group of lysine side chains (Fig. 2A). A possible covalent mode of action for 4μ8C was suggested by the observation that the inhibitor showed noncompetitive inhibition (Vmax reduction) (Fig. 1B). Furthermore, 4μ8C displayed a strong inverse relationship between its apparent IC50 and the time it was allowed to react with the enzyme before substrate addition (Fig. 2B). The importance of the aldehyde functionality to IRE1 inhibition was supported by the observation that following 4μ8C reduction to a primary alcohol by treatment with sodium borohydride the product was inactive (see reaction 3 in Fig. 2 A and C).
Fig. 2.
Stable Schiff base formation between 4μ8C and IRE1. (A) Predicted reaction schemes for Schiff base formation of 4μ8C to form a 4μ8C-lysyl imine (in reaction 1) that can be reduced to the stable lysyl amine by borohydride (NaBH4, reaction 2). Reductive inactivation of 4μ8C to the 8-hydroxymethyl is depicted in reaction 3. (B) Plot of the relationship between the incubation time of IRE1 with 4μ8C and the apparent IC50 in the in vitro RNase assay (mean ± SEM, n = 2). (C) In vitro RNase activity of IRE1 following incubation with 4μ8C and 4μ8C that had been previously reduced with borohydride (NaBH4; mean ± SEM, n = 2). (D) Absorbance trace of material eluting from a size exclusion chromatography column loaded with IRE1 or IRE1 and 200 μM 4μ8C. Note the emergence of a novel absorbance peak at 350 nm that comigrates with IRE1 in the 4μ8C-treated sample. (E) Fluorescent micrograph (excitation 340 nm, emission 450 nm) of an SDS-PAGE loaded with IRE1, following exposure 120 μM 4μ8C for the indicated times with or without subsequent reduction by borohydride. The sample in lane 6 was reacted with 4μ8C that had been previously reductively inactivated by borohydride.
Coumarins absorb light strongly between 310 and 350 nm. When resolved by size exclusion chromatography, IRE1 gave rise to a single peak of absorbance at 280 nm. However, following incubation with 4μ8C, a new comigrating absorbance peak at 350 nm was observed, as well as a late-eluting 350-nm peak, consistent with free 4μ8C (Fig. 2D). These findings suggest a stable chemical bond forms between a residue of 4μ8C and IRE1. To explore this further, we exploited the fluorescent properties of a borohydride-reduced apparent covalent conjugate between a residue of 4u8C and IRE1 (Fig. S5). IRE1 that had been exposed to 4μ8C without subsequent reduction by borohydride produced no fluorescent signal on denaturing SDS-PAGE, consistent with the lability of the putative imine bond between the inhibitor and denatured IRE1. However, reduction to a stable amine (with borohydride), led to time-dependent fluorescence of the IRE1–4μ8C complex (Fig. 2E).
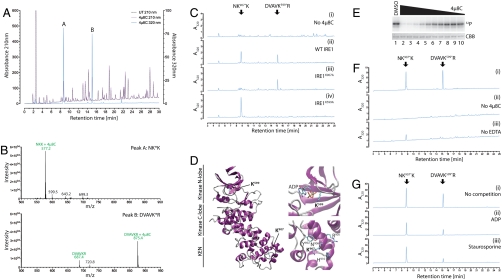
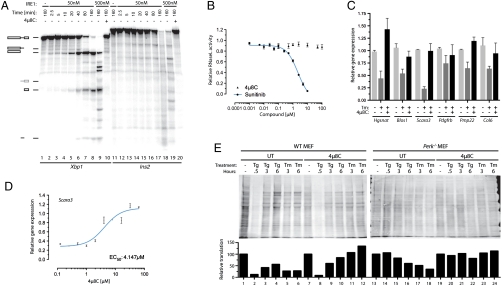
The covalent linkage between the reduced IRE1 and 4μ8C upon borohydride reduction suggested a method for mapping the reactive sites in IRE1. Following denaturation, the borohydride-reduced complex of IRE1 and 4μ8C was subjected to tryptic digestion and the fragments were resolved by reverse-phase HPLC. Multiple peaks absorbing at 210 nm were observed, consistent with the generation of many tryptic peptides. However, only two prominent species absorbing at 320 nm (the absorbance peak of reduced 4μ8C, Fig. S5) were observed, and these were unique to traces from the 4μ8C-treated samples (Fig. 3A, peaks A and B).
Fig. 3.
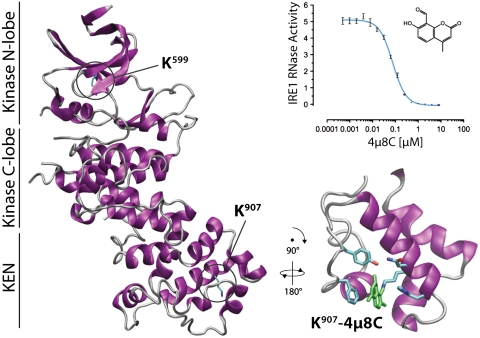
Two critical lysines in IRE1 are selectively targeted by 4μ8C. (A) Absorption traces of peptides eluting from a C18 reverse-phase HPLC. The column was loaded with a tryptic digest of baculovirus expressed IRE1 that had or had not been exposed to 120 μm 4μ8C followed by reduction with borohydride. Note the appearance of two peaks at 320 nm (labeled “A” and “B”) in the sample exposed to 4μ8C. (B) MALDI-TOF spectra of the material recovered in peaks “A” and “B” from A above. Peaks, corresponding in mass to predicted singly charged 4μ8C-modified peptides NH2-Asn-Lys-Lys-COOH (labeled NKK + 4μ8C in upper trace) and NH2-Asp-Val-Ala-Val-Lys-Arg-COOH (labeled DVAVKR + 4μ8C in lower trace) are singled out for attention. The unmodifed DVAVKR peptide is also detectable, likely resulting from in-source loss of the modification. (C) Absorption traces at 320 nm of peptides derived as in A from samples of immunopurified FLAG-tagged IRE1WT that had not been modified (−4μ8C, panel i) or IRE1WT, IREK907A, and IRE1K599A that had been modified with 120 μm 4μ8C (panels ii–iv). Note the absence of peak “A” in the IRE1K907A sample and the absence of peak “B” in the IRE1K599A sample. (D) Position of K599 and K907 in human IRE1α (from PDB ID code 3P23 A chain). (E) (Upper) Autoradiograph of SDS-PAGE of IRE1 following incubation with γ32P-ATP in the presence of decreasing concentrations of 4μ8C in twofold dilutions from 32 μM. (Lower) Coomassie Brilliant Blue (CBB) staining of the same gel. (F) Absorption traces (320 nm) of peptides derived from wildtype baculovirus expressed IRE1 or immunopurified Flag-IRE1 purified in the absence of either 4μ8C (ii) or EDTA (iii). (G) As in C and F. Where indicated, the nucleotide-binding pocket ligands ADP (ii) and staurosporine (iii) were preincubated prior to addition of 4μ8C.
The HPLC fractions containing the labeled peptides were recovered and subjected to MALDI-TOF mass spectrometry. This analysis indicated the presence of species with masses predicted for the partially cleaved (skipped) tryptic peptides NH2-N-K907-K-COOH and NH2-D-V-A-V-K599-R-COOH, each decorated with a single 4μ8C residue via an amine bond, in fractions A and B, respectively (Fig. 3B). NanoLC-ESI-tandem mass spectrometry of doubly charged precursor ions of the above peptides confirmed their identity and established that 4μ8C modified IRE1 residues K599 and K907 (Fig. S6). These assignments were confirmed by mutational analysis: The IREK907A mutation selectively abolished peak A, while the IREK599A mutation selectively abolished peak B (Fig. 3C).
K599 is a phosphate-coordinating residue common to all kinases, whereas the side chain of K907 protrudes into the RNase active site (22, 24) (Fig. 3D). Mutation of either residue interferes with IRE1 function in vivo (29, 30), thus the ability of 4μ8C to target these residues explains its ability to inhibit both the RNase (Fig. 1B) and kinase activity of IRE1 in vitro (Fig. 3E).
IRE1 binds nucleotide with high affinity (24), but modeling of 4μ8C in contact with K599 predicts that the two ligands would compete for the same site (Fig. S7). Mutually exclusive binding of nucleotide and inhibitors is consistent with the observation that modification of K599 requires that IRE1 be purified away from the cell lysate in the presence of EDTA (compare Fig. 3C, ii and Fig. 3F, iii) and with the ability of staurosporine and ADP to selectively interfere with modification of K599 by 4μ8C in vitro (Fig. 3G).
A Unifying Mechanism for Targeted Inhibition of IRE1.
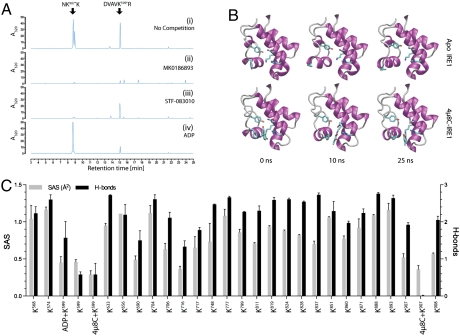
Two other classes of IRE1 inhibitor have recently been identified. The compounds used by Volkman et al. (31), including MK0186893, are all aldehydes, whereas STF083010 (32) decomposes to an aldehyde in water (Fig. S8). To determine if the chemical commonality among extant IRE1 inhibitors is also reflected in a shared mechanism of action, MK0186893 and STF083010 were allowed to compete with 4μ8C for IRE1 binding, and the outcome was measured by analysis of the HPLC spectra. Preincubation with MK0186893 inhibited both peaks A and B (Fig. 4A, ii), pointing to its reactivity with both active sites of IRE1. STF083010, by contrast, selectively competed with 4μ8C for K907, whereas ADP, used as a control, only competed for K599, as expected (Fig. 4A, iii and iv). These observations are consistent with the reported selectivity of STF083010 for the IRE1 RNase activity (32) and with the ability of the compounds described by Volkman et al. to inhibit both functions of IRE1 (31). Moreover, IRE1 K907R a mutant with residual RNase activity in vitro, is refractory to inhibition by any of the three inhibitors (Fig. S9). Thus, Schiff base formation at K907 represents a consensus mechanism by which all three inhibitors inactivate the IRE1 endoribonuclease.
Fig. 4.
Stable binding of 4μ8C to IRE1 lysine 907. (A) Absorption traces of peptides eluting from a C18 reverse-phase HPLC (as in Fig. 3). Where indicated, the recently described IRE1 inhibitors MK0186893 (31) and STF083010 (32) were allowed to compete with 4μ8C for IRE1 binding. (B) Snapshots of the IRE1 RNase domain at the indicated time points from a molecular dynamics simulation of the apo state and with 4μ8C attached to K907 via an imine. Note the relative flexibility of the apo state and stable conformation in the presence of 4μ8C. Residues highlighted are Y892, F889, N906, K907, and H910. Five additional repeat simulations with different starting velocities were carried out for both protomers in PDB ID code 3P23, and yielded comparable results. (C) Predicted solvent accessibility (SAS) and hydrogen bonding to water of lysine residues in IRE1 (based on PDB ID code 3P23) in the apo state and in presence of the indicated bound ligands derived from the MD simulations.
The selectivity of 4μ8C for K907 is striking given the multitude of other seemingly accessible nucleophiles on IRE1. To gain insight into the basis of the apparent selectivity of 4μ8C, we performed molecular dynamics simulations using the apo state of human IRE1 [Protein Data Bank (PDB) ID code 3P23, A-chain] as a point of departure (Fig. S7). The RNase active site proved quite dynamic, with rapid conformational rearrangements over the 25 ns of simulation (Fig. 4B, Upper). This flexibility is almost entirely eliminated by the docking of 4μ8C (Fig. 4B, Lower, and Movies S1 and S2). Schiff base formation between K907 and the 8-carbaldehyde appears to constrain the RNase site by immersion of the ligand in a hydrophobic pocket formed by the nonpolar residues of this domain, in particular F889 that stacks with the coumarin ring structure. In this stable configuration, 4μ8C is interposed between side chains of RNase catalytic residues Y892 and H910, disrupting catalysis.
Denaturation led to dissociation of 4μ8C from IRE1 (Fig. 2E), attesting to the inherent susceptibility of the Schiff base to hydrolysis. However, in the native state both K907 and K599 are relatively unexposed: their epsilon amino group having relatively low solvent accessibility (SAS) and having weak hydrogen bonding potential with water (Fig. 4C). Furthermore, docking of 4μ8C diminishes predicted accessibility of these two residues to solvent (Fig. 4C). Thus, while transient Schiff base formation may occur with other nucleophiles on IRE1 (and indeed on other proteins), this analysis suggests that a low off rate due to diminished hydrolysis accounts for the selectivity of 4μ8C toward IRE1 K907. Indeed, in vitro 4μ8C modification of IRE1 has a half-life of approximately 1.6 h (Fig. S10 A and B), attesting to the stability of the adduct with IRE1.
In Vivo 4μ8C Iinhibits both Xbp1 Splicing and RIDD but not IRE1 Kinase Activity.
When applied to the media of ER stressed cultured cells, 4μ8C inhibited Xbp1 splicing in a concentration-dependent manner (Fig. 5A). Inhibition was observed with diverse agents that induce ER stress; the glycosylation inhibitor tunicamycin and thapsigargin, an inhibitor of the SERCA Ca2+ pump (Fig. 5). By contrast, IRE1 autophosphorylation, reflected in reactivity to an antisera specific for phospho-serine 724 and in reduced mobility on Phos-tag™ SDS-PAGE, was unaffected by exposure to the inhibitor (Fig. 5B). The latter is consistent with the effective competition by endogenous nucleotides (and other ligands of the nucleotide-binding pocket) for access to K599 (Fig. 3F). Interestingly, and in contrast to the stability of the IRE1–4μ8C complex in vitro, Xbp1 splicing recovered rapidly from 4μ8C-mediated inhibition following exchanges of the culture medium (Fig. S10C).
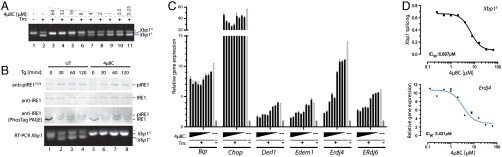
Fig. 5.
Selective targeting of the IRE1 RNase by 4μ8C in vivo. (A) Fluorescent stained agarose gel of an RT-PCR assay of the Xbp1 mRNA from MEF cells treated simultaneously with increasing concentrations of 4μ8C and either the ER stress causing agent tunicamycin (Tm 2.5 μg/mL) or a vehicle control for 6 h. The migration of the spliced (Xbp1S) and unspliced (Xbp1U) forms are indicated. (B) Immunoblot of endogenous IRE1α immunopurified from MEF cells treated simultaneously with thapsigargin (Tg 0.5 μM) and 4μ8C (32 μM) or a vehicle control for the indicated times analyzed by SDS-PAGE or PhosTag™-PAGE and probed either with phosphospecific antsera to IRE1S724 or total IRE1 as indicated. The lower panel is an RT-PCR of endogenous (mouse) Xbp1 mRNA from the same cells. (C) Quantitative PCR (qPCR) analysis of mRNAs expressed in MEF cells following 6-h treatment with vehicle control (dark gray bar); tunicamycin (2.5 μg/mL) alone (light gray) or with cotreatment of twofold increasing concentrations of 4μ8C (black bars), from 0.125 to 64 μM (mean ± SEM, n = 2). (D) Plots of tunicamycin-induced Xbp1 splicing and Erdj4 expression in cells cotreated with the indicated concentration of 4μ8C for 6 h. Note the similarity in the IC50 for both in vivo read outs of 4μ8C activity (mean ± SEM, n = 2 for Erdj4).
Spliced Xbp1 serves as an important effector of IRE1, activating its target genes. Accordingly, the ER stress-induced up-regulation of known XBP1 target genes: Derl1, Edem1, Erdj4, and Erdj6 was inhibited in cells exposed to 4μ8C with a similar concentration dependence as that observed for inhibition of Xbp1 splicing (Fig. 5C). In contrast, stress-induced expression of Bip was unaffected by the inhibitor (Fig. 5C), an observation consistent with redundancy of the XBP1 and ATF6 arms of the UPR in regulating the activity of this key ER chaperone (33). Similarly, expression of the PERK and ATF4-controlled proapoptotic gene Chop was unaffected by 4μ8C (Fig. 5C), indicating selectivity of 4μ8C for the IRE1 arm of the UPR and resistance of the PERK kinase to the compound.
In addition to site-specific endonucleolytic activity (leading to Xbp1 mRNA splicing), metazoan IRE1 has also been implicated in the promiscuous degradation of diverse mRNAs encoding secreted proteins; a process referred to as RIDD (8, 9). We were able to recapitulate this phenomenon in vitro: highly purified mammalian IRE1 degraded the insulin mRNA. This reaction, which proceeds more slowly than the site-specific cleavage of Xbp1, was nonetheless blocked by 4μ8C (Fig. 6A). By contrast, the RNase activity of the IRE1-related endoribonuclease, RNase L, was unaffected by 4μ8C (Fig. 6B), attesting to the specificity of 4μ8C for the IRE1 RNase domain. In vivo, 4μ8C reversed the ER stress-dependent loss of several known RIDD targets (Fig. 6C), with an EC50 of approximately 4 μM, approximating that of inhibition of XBP1 target gene activation (Fig. 6D). Together, these observations indicate that 4μ8C inhibits IRE1-mediated gene expression and mRNA degradation by targeting the RNase domain.
Fig. 6.
Inhibition of RIDD by 4μ8C. (A) Autoradiograph of in vitro transcribed 32P-labeled mouse Xbp1 or Insulin2 (Ins2) mRNA separated by electrophoresis after incubation with the indicated concentration of IRE1 in the absence or presence of 4μ8C (10 μM). (B) RNase L activity measured with a FRET assay in the presence of 2–5A cofactor and in the presence or absence of the indicated concentrations of 4μ8C or Sunitinib (45). (C) Expression levels determined by qPCR of known IRE1 RIDD target genes following 6 h of treatment with tunicamycin (2.5 μg/mL) in the presence or absence of coapplied 32 μM 4μ8C (mean ± SEM, n = 2). (D) Plot of mRNA level of the RIDD target gene Scara3 in tunicamycin-treated cells exposed simultaneously to the indicated concentrations of 4μ8C (mean ± SEM, n = 2) for 6 h. (E) Immunoblot of puromycinylated proteins recovered from the membrane fraction of semipermeabilized wild-type and Perk-/- mutant MEFs that had been exposed to thapsigargin (500 nM) or tunicamycin (2.5 μg/mL) in the absence and presence of 32 μM 4μ8C for the indicated times. The intensity of the signal, integrated over the surface of each lane, is plotted under the image of the immunoblot.
In vivo, RIDD selectively targets membrane-associated mRNAs for degradation (8, 9). Therefore to gauge the contribution of RIDD to homeostatic control of ER biosynthetic load, we compared the rate of membrane-associated protein synthesis in the ER of stressed cells with and without 4μ8C in both wild type and Perk-/- mutant cells. Following a brief (10 min) pulse of puromycin, mouse embryonic fibroblast (MEF) cells were semipermeabilized with low concentrations of digitonin, centrifuged to wash off cytosolic proteins, then triton extracted to recover the membrane fraction. The puromycinylated proteins in the membrane fraction were quantified by immunoblot. In wild-type MEF cells, ER stress led to a steep transient decline in rates of protein synthesis that was more conspicuous in response to the rapidly acting drug thapsigargin than the slower acting tunicamycin (Fig. 6E, lanes 1–6). The compound had no effect on the intensity of the dip in membrane protein synthesis (Fig. 6, lanes 7 and 8) but accelerated the recovery of protein synthesis in thapsigargin treated cells (Fig. 6, lanes 9 and 10) and attenuated the tunicamycin-mediated repression of protein synthesis (Fig. 6, lanes 11 and 12). The steep transient phase of translation repression is entirely missing in PERK mutant cells (Fig. 6, lanes 13 and 14), as expected (34), however the PERK-independent decline in membrane protein synthesis observed at later times of the stress response was conspicuously alleviated by 4μ8C (Fig. 6, compare lanes 15–18 with lanes 21–24), arguing that it was a consequence of RIDD. Thus, ER stress-induced mRNA decay by IRE1 likely impacts the biosynthetic output of secretory cells.
IRE1 Inhibition Interferes with Programmed Expansion of Secretory Capacity but Has no Measurable Role in Defending Cells Against Acute ER Stress.
IRE1 loss-of-function mutations markedly sensitize yeast and worms to the acute toxicity of ER stress. However, mammalian cells lacking IRE1 are not hypersensitive to the consequences of ER stress, unlike cells lacking PERK or ATF6. Yet, unlike PERK or ATF6, which are not essential genes, mammalian embryos lacking IRE1 (or its XBP1 effector) succumb early in embryonic development (35, 36). The availability of a potent and rapidly acting inhibitor, might enable, for the first time, determination of whether the aforementioned observations reflect the gradual emergence of redundant mechanisms for coping with acute ER stress in cultured IRE1-deficient mammalian cells or if IRE1 might have roles that are played out selectively in a developmental context.
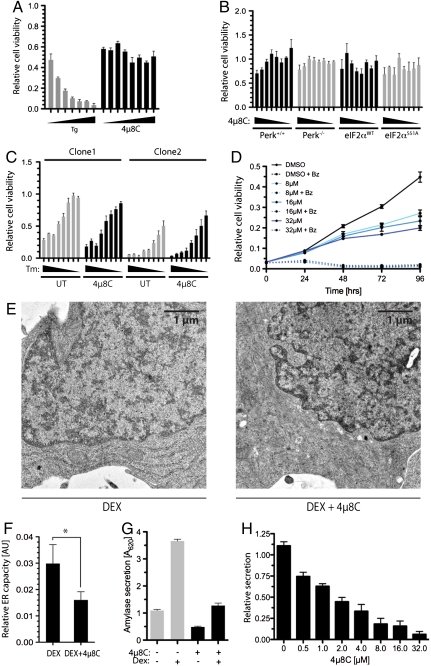
Exposure of wild-type MEFs to 4μ8C at concentrations of up to 128 μM had no measureable toxicity, while the ER stress-inducing agent thapsigargin markedly diminished cell viability in a dose dependent manner (Fig. 7A). The IRE1 inhibitor was also devoid of apparent toxicity in cells lacking PERK or cells with a mutation in its substrate, rendering the latter nonphosphorylatable (Fig. 7B). This finding suggests that the conspicuous complementation between the PERK-mediated arm that regulates the flux of proteins into the ER (see Fig. 6E) and IRE1 signaling in simple eukaryotes (37) is not a feature of cultured mammalian cells. Furthermore, there was no apparent synergism of 4μ8C with the toxicity of the ER stress-inducing agents tunicamycin or thapsigargin (Fig. 7C).
Fig. 7.
The expansion of secretory capacity is selectively inhibited by 4μ8C. (A) Viable cell mass of MEF following 24-h exposure to escalating (2-fold) concentrations of 4μ8C (from 1 to 128 μM) or thapsigargin (from 16 to 1,024 nM; mean ± SEM, n = 3). (B) Viable cell mass of MEF of the indicated genotypes following 24-h exposure to escalating (2-fold) concentrations of 4μ8C (from 1 to 128 μM; mean ± SEM, n = 3). (C) Viable cell mass of three different clones of wild-type MEFs following 24-h exposure to escalating (2-fold) concentrations of tunicamycin (from 16 to 2,048 ng/mL), and in the absence or presence of 4μ8C (32 μM; mean ± SEM, n = 3). (D) Time-dependent growth of multiple myeloma MM.1R cells exposed to the indicated concentrations of 4μ8C in the absence or presence of bortezomib (Bz, 6 nM) as assessed by WST1 assay (mean ± SEM, n = 3). (E) Representative electron micrographs of AR42J cells treated for 48 h with dexamethansone (100 nM, an inducer of differentiation to the secretory state) in the absence or presence of 4μ8C (32 μM) (F) Quantitation showing the ratio of ER cross-section area to cytosol cross-section area (AU) in the samples treated as in E (mean ± SEM, n = 5; * indicates p < 0.05 by T test). (G) Amylase secretion detected in the media of AR42J cells cultured for 48 h in the absence or presence of dexamethasome (DEX, 100 nM) with or without 4μ8C (32 μM; mean ± SEM, n = 3). (H) Amylase secretion as in G, with dexamethasome (DEX, 100 nM) and a range of concentrations of 4μ8C (mean ± SEM, n = 3).
Loss of function mutations in Xbp1 and Ire1 selectively compromise the development and integrity of specialized secretory tissues (7, 36, 38–40). Consistent with this theme, the inhibitor attenuated the growth of multiple myeloma cell lines that are characterized by a robust secretory apparatus (Fig. 7D). However, this interference with growth was not associated with evidence of acute toxicity in these cells, nor was there evidence for synergism with the robust antimyeloma effects of the proteasome inhibitor Bortezomib. Furthermore, in a cell culture model of exocrine pancreatic development induced in AR42J cells by dexamethasone, 4μ8C interfered with ER expansion (Fig. 7 E and F) and amylase secretion (Fig. 7 G and H). Together, these observations point to a role for IRE1 in building ER capacity but not in preserving ER integrity of stressed cells.
Discussion
The structure of IRE1 suggests three alternative sites that could be targeted by small molecules: the nucleotide-binding pocket, the dimer-interface hydrophobic pocket, and the RNase active site. Unbiased screens for inhibitors have all yielded compounds whose activity converges on the RNase domain, and this study provides a unifying molecular mechanism for this surprising observation. All extant inhibitors are aldehydes, but the spectral properties of 4μ8C have enabled us to track its reactivity with IRE1 and revealed a highly selective and stable modification of lysine 907 in the RNase active site. IRE1 has 24 other lysines in its cytosolic domain, and most are surface exposed and flexible. By contrast, K907 is buried in the enzyme active site. The aldehyde moiety, which is required for inhibition, forms a Schiff base with lysyl side chains that is rapidly reversed by hydrolysis. However, the imine bond with K907 is shielded from the solvent by the protein and by the inhibitor that is held in place by hydrophobic and stacking interactions with side chains in the active site. Thus, steric effects specify an unusually long off-rate for inhibitor interactions with a specific lysine.
In vitro, 4μ8C shows affinity for lysine K599, which, like K907, is shielded from the surface, being partially buried in the nucleotide-binding pocket. However, endogenous nucleotides compete effectively with the inhibitor for this site, and in vivo inhibition is limited to the RNase of IRE1. Similar considerations likely apply to the compounds described by Volkman et al. (31), whereas STF083010 is restricted in its reactivity to K907. In vitro 4μ8C dissociates slowly from IRE1, but washout of inhibitor leads to rapid recovery of Xbp1 splicing in cells. The molecular dynamics simulation suggests that the observed slow off-rates in vitro are due to rigidity of the enzyme–inhibitor complex shielding the imine bond. The higher apparent off-rates in cells is consistent with a more flexible enzyme active site in vivo and reveals that despite its covalent mechanism of action 4μ8C is a reversible inhibitor of the enzyme. These observations highlight the interplay between steric and chemical effects in the function of IRE1 inhibitors, an interplay that features in the growing class of drugs that are covalent inhibitors (41).
K907 is not believed to play a role in hydrolysis of the phosphodiester bond of RNA substrates of IRE1 (24, 25). Indeed, the corresponding residue in the related enzyme, RNaseL, is a leucine. Consistent with this observation, RNaseL is refractory to the inhibitory effect of 4μ8C. Surprisingly, however, the lysine side chain is critical for IRE1 activity, as its replacement with alanine, leucine and arginine inactivates or substantially retards IRE1 activity (29) (Fig. S9). Thus, the side-chain amino group of K907 is likely to be pKa perturbed and engage in charge interactions with substrate or other residues in the active site that affect its protonation state, and thereby also influence the rate of Schiff base formation via acid catalysis.
Despite its potential pleiotropic reactivity with other nucleophiles, in cultured cells 4μ8C has remarkable selectivity for IRE1. Furthermore, at concentrations that block all measurable IRE1 RNase activity, the compound exhibits no measureable acute toxicity. And, while the theoretical possibility of reactivity with the counterpart of K599—found in nearly all protein kinases—conjures the possibility of inhibiting other kinases, no evidence for that was observed. For example, the PERK kinase remains active in 4μ8C-treated cells (Fig. 6E, compare lanes 1 and 2 with lanes 7 and 8).
Unfavorable pharmacokinetics limit the utility of 4μ8C (and likely that of the other aldehyde IRE1 inhibitors) for animal studies. However, in cultured cells inhibition of IRE1 by 4μ8C has provided valuable insights into the normal role of the enzyme in physiology. The strong suppression of ER stress mediated degradation of membrane-bound mRNA in compound-treated cells supports the notion that the IRE1 RNase is directly implicated in RIDD (8, 29). The impact of RIDD on membrane-associated ER protein synthesis has been hitherto difficult to assess, because of the dominance of PERK-mediated translational suppression in stressed cells. However, comparing the rates of membrane-associated protein synthesis in ER stressed PERK deficient cells in the absence and presence of 4μ8C has enabled us to estimate the quantitative significance of RIDD to ER associated protein synthesis. While its physiological significance remains to be explored in further detail, our observations suggest that RIDD is likely to exert a powerful influence on the protein composition of the ER in severely stressed cells.
Much of what we know about IRE1 has come from genetic loss-of-function experiments. The potential for adaptation by the feedback loops in the two other arms of the UPR has obscured the interpretation of such experiments. The availability of a tool to rapidly block IRE1 activity has revealed that in mammalian cells, unlike yeast and worms, IRE1 has no measureable role in protecting cells against acute ER stress. Moreover, the complementation between IRE1 and PERK, conspicuous in worms, is also not apparent in cultured mammalian cells. Instead, our observations suggest that in mammals the IRE1 branch of the UPR has specialized in adapting cells to a heavy secretory load and is not important to the day-to-day ER protein folding homeostasis.
These observations may bear on the clinical application of IRE1 inhibitors. The lack of a major role for IRE1 in maintaining protein folding homeostasis of differentiated secretory cells in mammals suggests that IRE1 inhibitors may have limited toxicity in these species. Perhaps IRE1 inhibitors will find their greatest utility in situations with ongoing differentiation of precursors into pathogenic secretory cells. This might be the case in diseases caused by the secretory products of plasma cells or their malignant counterpart in multiple myeloma. Inhibitors of IRE1 might also have utility for local administration; for example, to block the development of mucin-producing goblet cells in the respiratory tract, which continuously differentiate from precursors in the mucosa and contribute to pathogenesis of asthma and chronic obstructive pulmonary disease. Thus, compounds like 4μ8C, which are not suited for systemic administration, may nonetheless provide a platform for developing new locally acting drugs.
Materials and Methods
Cell Culture and Analysis.
MEFs, AR42J, and HEK-293T cells were maintained in DMEM supplemented with 10% FCS, 2 mM glutamine, nonessential amino acids, and 50 μM 2-mercaptoethanol. MM1.R cells were maintained in RPMI containing 10% FCS, Pen/strep, and glutamine. Xbp1 splicing was determined by RT-PCR as previously described (4, 28), and the resulting amplified products were stained with SYTO-60 dye (Invitrogen) and visualized using the Odyssey scanner (LI-COR). Amylase secretion was measured as described in SI Materials and Methods.
Kinase Analysis.
In vitro kinase assays were performed as described previously (28). For in vivo analysis, IRE1 was collected from AR42J or MEF cells as previously described (19) and probed with anti-pIRE1S724 (Cell Signalling), before the membranes were stripped and reprobed with anti-IRE1 (Cell Signalling). Alternatively, cell lysates were run on SDS-PAGE incorporating 50 μM Phos-tag reagent (NARD institute) and 50 μM MnCl2. Gels were then incubated in 1 mM EDTA before transfer and probing with anti-IRE1. Membranes were probed with secondary antibodies conjugated to near-infrared dyes and visualized with the LI-COR Odyssey scanner.
Quantitative RT-PCR.
RNA from treated cells was harvested as described previously (28), and used in duplicate for quantitative real-time PCR analysis using the SYBR Green reagent system (Applied Biosystems) and an ABI 7900 (PerkinElmer Biosystems). Relative quantities of amplified cDNAs were then determined using SDS software (Applied Biosystems) and normalized to cyclophilin A mRNA.
In Vitro IRE1 RNase and RIDD Assays.
Analysis of radiolabeled Xbp1 substrate cleavage was performed as previously (28) except that mammalian IRE1 reaction buffer (see HTS screen, SI Materials and Methods) was used. In vitro RIDD substrates were synthesized by in vitro transcription using the T7-MAXIscript Kit (Ambion) in the presence of 32P ATP or Cy5-UTP on templates isolated by RT-PCR from mouse Min6 cells (Ins2) or PCR from cloned XBP1 cDNA using the primers listed in Table S1. The resulting products were gel purified to obtain full-length substrate. Reactions were then separated by 15% UREA-PAGE for analysis by phosphorimaging or by near-infrared imaging using the LI-COR Odyssey scanner.
HPLC.
Recombinant HsIRE1[464–977] was incubated for 3 h with 120 μM 4μ8C in 50 mM Tris pH 7.4 100 mM NaCl prior to addition of 6 mM NaBH4. The reaction was buffer exchanged on equilibrated PD-10, digested with trypsin for 16 h and collected on C18 hypersep columns (Thermo). Eluted peptides were dried and separated by reverse-phase HPLC on a poroshell 120 C18 column using a multistep acetonitrile gradient with 0.1% TFA on an Agilent LC1290 system with a diode array detection of absorbance from 180–400 nm. Two-drop (approximately 13 μL) fractions were collected, dried, and analyzed by MALDI-TOF mass spectrometry and nanoLC-ESI-MS/MS (see SI Materials and Methods). Alternatively, HEK-293T cells were transfected with expression vectors encoding preprotrypsin-FLAG-HsIRE1[19-977] wild type, K907A, or K599A mutants and treated with 100 μM 4μ8C for 1 h prior to lysis in buffer (50 mM Tris pH 7.5, 150 mM NaCl, 10% glycerol, and 1% Triton X-100) supplemented with 5 mM NaBH4 where indicated and protease inhibitors. Postlabeling analysis was by lysis in the above buffer with the addition of 50 mM EDTA, followed by incubation with 100 μM 4μ8C and reduction with NaBH4. Cleared lysates were then immunopurified with anti-FLAG-M2-aggarose beads, washed, and analyzed by trypsin digest and HPLC as above.
Modeling and Molecular Dynamics.
Docking and molecular dynamics (MD) simulations of 4μ8C–IRE1 are described in detail in SI Materials and Methods. Briefly, MD systems contained dimeric IRE1 (PDB ID code 3P23), surrounded by a 0.1 M NaCl solution in a truncated octahedral box; system sizes were approximately 250,000 atoms. Systems included either the unmodified protein, or the K907–4μ8C/K599–4μ8C states, all in the presence of ADP.Mg2, along with an additional ADP-free K599–4μ8C complex. The 4μ8C-bound states were obtained via a tailored geometry optimization protocol carried out within CHARMM (42). Each system under went multiple stages of energy minimization and protein-restrained equilibration, prior to production MD. The CHARMM22/CMAP all-atom force field was used to represent the protein, and parameters for 4μ8C were adapted from the CHARMM CGenFF general force field for small drug-like molecules. All MD simulations were performed with GROMACS 4.5.3 (43).
Cell Viability Assays.
Cells were seeded in phenol red-free cell culture medium in 96 or 24 well dishes at a density of 5 × 103 or 5 × 104 cells per well, respectively. Cultures were incubated for 16 h before treatment with 4μ8C, bortezomib, tunicamycin, or thapsigargin for 24 to 96 h. Cultures were then analyzed by the addition of 200 μM WST1 (Dojindo) and 10 μM phenazine metho-sulfate (Sigma). After development of the reagent for 2 h at 37 °C, the hydrolyzed dye was detected by absorbance at 450 nm, after subtracting background and absorbance at 595 nm. Alternatively, cell viability was determined by staining of the adherent culture with crystal violet. Quantitation of the dye uptake was analyzed by extensive washing of the stained cells with water and solublization of the crystal violet in methanol followed by absorbance measurements at 595 nm.
Translational Regulation/Puromycin Labeling Assays.
Treated wild-type or Perk-/- cells were labeled for 10 min with 10 μg/mL puromycin followed by addition of 100 μg/mL cycloheximide. Collected cells were then washed twice in ice-cold HMN buffer (50 mM HEPES pH 72, 2 mM MgCl2, 150 mM NaCl) before semipermeabilization of 2 × 106 cells/100 μL with 0.2% digitonin for 10 min. Pelleted cells were then resuspended in HMN buffer supplemented with 0.5% Triton X-100 and protease inhibitors, cleared by centrifugation, and analyzed by probing with antipuromycin antibody (44).
Statistics.
Curve fitting and statistical values were calculated using Prism 4.0 for MacOSX. The Ki for CB5305630 was determined using a noncompetitive model for inhibition: Vmaxinh = Vmax/(1 + I/Ki), Y = Vmaxinh ∗ X/(Km + X), where I is the concentration of CB5305630. Ki and Km for substrate are shared for each concentration, and R2 for the model (global) was 0.9967.
Supplementary Material
Acknowledgments.
We thank Patrick Liu, Matt Shoulders, and Jeff Kelly (Scripps Research Institute) for fruitful discussions, the staff of the Harvard–Institute of Chemistry and Cell Biology screening center for their help with the inhibitor screen, and Alice Liang from the imaging core facility of New York University School of Medicine for processing and acquiring the EM images. All simulations were executed using the Darwin Supercomputer of the University of Cambridge High Performance Computing Service (www.hpc.cam.ac.uk), provided by Dell Inc. using Strategic Research Infrastructure Funding from the Higher Education Funding Council for England. Supported by a Wellcome Trust Principal Research Fellowship to D.R. and by National Institutes of Health Grants NS050276 and CA016087, a grant from the 100 Women In Hedge Funds Foundation to T.A.N., and The Royal Society to I.R.B.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 5559 (volume 109, number 15).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115623109/-/DCSupplemental.
References
- 1.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 2.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 3.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 4.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing theXBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida H, et al. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell. 2003;4:265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 7.Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrineglands. EMBO J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hollien J, et al. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 10.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 11.Feldman DE, Chauhan V, Koong AC. The unfolded protein response: A novel component of the hypoxic stress response in tumors. Mol Cancer Res. 2005;3:597–605. doi: 10.1158/1541-7786.MCR-05-0221. [DOI] [PubMed] [Google Scholar]
- 12.Romero-Ramirez L, et al. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 2004;64:5943–5947. doi: 10.1158/0008-5472.CAN-04-1606. [DOI] [PubMed] [Google Scholar]
- 13.Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Todd DJ, et al. XBP1 governs late events in plasma cell differentiation and is not required for antigen-specific memory B cell development. J Exp Med. 2009;206:2151–2159. doi: 10.1084/jem.20090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu CY, Hsu YW, Liao CL, Lin YL. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J Virol. 2006;80:11868–11880. doi: 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding W-X, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cellviability. Am J Pathol. 2007;171:513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogata M, et al. Autophagy is activated for cell survival after ER stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahoney DJ, et al. Virus-tumor interactome screen reveals ER stress response can reprogram resistant cancers for oncolytic virus-triggered caspase-2 cell death. Cancer Cell. 2011;20:443–456. doi: 10.1016/j.ccr.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Bertolotti A, Zhang Y, Hendershot L, Harding H, Ron D. Dynamic interaction of BiP and the ER stress transducers in the unfolded protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 20.Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimata Y, et al. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J Cell Biol. 2007;179:75–86. doi: 10.1083/jcb.200704166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ali MM, et al. Structure of the Ire1 autophosphorylation complex and implications for the unfolded proteinresponse. EMBO J. 2011;30:894–905. doi: 10.1038/emboj.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 24.Lee KP, et al. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell. 2008;132:89–100. doi: 10.1016/j.cell.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korennykh AV, et al. Structural and functional basis for RNA cleavage by Ire1. BMC Biol. 2011;9:47. doi: 10.1186/1741-7007-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korennykh AV, et al. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2099;457:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Korennykh AV, Behrman SL, Walter P. Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc Natl Acad Sci USA. 2010;107:16113–16118. doi: 10.1073/pnas.1010580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiseman RL, et al. Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Mol Cell. 2010;38:291–304. doi: 10.1016/j.molcel.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han D, et al. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 2000;14:2725–2736. doi: 10.1101/gad.839400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Volkmann K, et al. Potent and selective inhibitors of the inositol-requiring enzyme 1 endoribonuclease. J Biol Chem. 2011;286:12743–12755. doi: 10.1074/jbc.M110.199737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papandreou I, et al. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117:1311–1314. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 35.Iwawaki T, Akai R, Yamanaka S, Kohno K. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc Natl Acad Sci USA. 2009;106:16657–16662. doi: 10.1073/pnas.0903775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reimold AM, et al. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- 37.Shen X, Ellis RE, Sakaki K, Kaufman RJ. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet. 2005;1:e37. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reimold AM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 39.Zhang K, et al. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hess DA, et al. Extensive pancreas regeneration following acinar-specific disruption of Xbp1 in mice. Gastroenterology. 2011;141:1463–1472. doi: 10.1053/j.gastro.2011.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh J, Petter RC, Baillie TA, Whitty A. The resurgence of covalent drugs. Nat Rev Drug Discov. 2011;10:307–317. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- 42.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30:1545–1615. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 45.Jha BK, et al. Inhibition of RNase l and PKR by sunitinib impairs antiviral innate immunity. J Biol Chem. 2011;286:26319–26326. doi: 10.1074/jbc.M111.253443. [DOI] [PMC free article] [PubMed] [Google Scholar]