Abstract
The cell envelope of Gram-negative bacteria protects the organism from environmental stresses, components of the innate immune response, and the actions of other antagonistic molecules. However, the complexity of the cell envelope dictated by these protective roles creates a significant challenge for assembly of the outer membrane. Extensive research has focused on the export and assembly of outer membrane proteins and there is continuing progress in this area. By contrast, knowledge of the export and assembly of complex glycoconjugates in the outer membrane has been limited until recently. New structural and biochemical information identifies an envelope-spanning molecular scaffold for the export of group 1 capsular polysaccharides and provides insight into a complex molecular machine.
Introduction
The Gram-negative cell envelope is a complex structure. The cytoplasmic (inner) membrane is protected from osmotic stress and turgor pressure by the rigid stress-bearing peptidoglycan layer. The critical role for peptidoglycan in cell viability is best exemplified in its status as the target for β-lactam antibiotics. The peptidoglycan layer resides in the periplasm, a region delineated by the cytoplasmic membrane and the characteristic outer membrane. The outer membrane is an atypical lipid bilayer with phospholipids in the inner leaflet and the unique glycolipid called lipopolysaccharide (LPS) in the outer leaflet (reviewed in [1]). Proteins are associated with (and embedded within) the bilayer. Many of these proteins create channels and receptors to facilitate import and export of substrates across the outer membrane. Until recently, outer membrane proteins were thought to contain only β-sheet structures in their transmembrane segments [2], whereas proteins in cytoplasmic membranes typically have α-helices. The presence of LPS makes the outer membrane resistant to many enzymes, detergents and other antagonistic factors that would compromise and breach a typical cytoplasmic membrane.
The outer membranes of many Gram-negative bacteria are covered with a capsule structure composed of capsular polysaccharides (CPSs). These are high molecular weight polymers (105–106 Da or greater) that are firmly associated with the cell surface, although the precise linkage is not always known. CPSs are essential virulence determinants in pathogens of plants, livestock and humans. Despite a remarkable diversity of CPS structures in Gram-negative bacteria, currently only two mechanisms of biosynthesis and assembly have been described and both are found in isolates of Escherichia coli. These processes differ fundamentally in membrane topology and characteristic components. These have been reviewed in detail elsewhere [3•].
The protective role of the envelope must be balanced against the need to assemble the outer membrane. The key question is: how does the cell accomplish trafficking of complex outer membrane components from their site(s) of synthesis in the cytoplasmic membrane to the outer membrane without compromising these essential barrier components? This hurdle to export (and import) has made assembly of outer membrane components interesting their own right but they may also have relevance to organellar assembly in eukaryotes. Two basic strategies can be envisaged for outer membrane assembly (reviewed in [ [4] and [5•]]). One involves the use of chaperone proteins to traffic material to insertion points in the outer membrane; outer membrane proteins provide an excellent example. The alternative is to use a molecular scaffold to overcome the barriers in a contiguous process and this is used for CPS assembly. Here, we examine the relationships between the various outer membrane assembly systems.
State of the art: outer membrane protein folding and assembly
Outer membrane proteins are typically synthesized as precursors with an N-terminal signal sequence and are exported across the cytoplasmic membrane by the Sec protein complex in the general secretory pathway [6]. After removal of the signal peptide, the protein is released into a periplasmic trafficking system where it can interact with various chaperone and folding activities [7]. Finally, it is transferred to a multi-component complex in the outer membrane that has at its core the YaeT (formerly Omp85) protein [8••]. SurA is a chaperone with a four-domain structure containing two internal peptidyl–prolyl isomerase domains (PPIase P1 and P2). Structures of a PPIase P1 fragment bound to two peptide substrates have revealed the importance of exposed aromatic residues in the substrates recognized by SurA [9]. Surprisingly, SurA adjusts its tertiary and quaternary structure dependent on the precise presentation of the aromatic residues. SurA interacts with YaeT [10•]. An additional folding pathway involves Skp and DegP and their role in protein export becomes evident under conditions of stress or in cells lacking SurA [10•]. Skp exists as a trimer with long α-helices extending from a β-barrel ‘association body’ and it has been proposed that these helices act as ‘tongs’ grasping the unfolded protein as it emerges from the Sec translocon [ [11] and [12]]. Cargo-bound Skp also binds LPS and undergoes a change in topology and is thought that this interaction facilitates delivery of its cargo to the outer membrane [ [13] and [14]].
At the outer membrane, the newly delivered protein interacts with a complex including YaeT and the lipoproteins YfgL, YfiO, NlpB and Smp [ [8••] and [15]]. YaeT and YfiO are essential for viability. YaeT has a C-terminal channel-forming β-barrel [16] preceded by five polypeptide transport-associated (POTRA) domains arranged in a periplasmic extension [17••]. POTRA domains are found in some other export-machinery proteins including Sam50 and Toc75 from mitochondria and chloroplasts, respectively. The POTRA domains are thought to provide a scaffold that facilitates the interaction of the various lipoproteins but the presence of all five is not essential [ [17••] and [18]]. YaeT binds the C-terminal region of most outer membrane proteins being translocated [19••]. Outer membrane proteins OmpA [19••] and TolC [20] (the drug efflux channel [21]) provide variations; these proteins possess an N-terminal β-barrel structure but their C-terminal domains are α-helical. It appears that an internal segment anneals to YaeT in such cases. The structural data support the hypothesis that the POTRA domains recognize hydrophobic periodicity in the exported substrate and they interact via a β-strand augmentation approach. This templating may initiate the process of assembly into a β-barrel directly adjacent to the membrane. However, the spatial interactions, and the means by which the new β-barrel escapes into the outer membrane are still subjects of debate [ [7] and [22]], as is the exact role played by the various accessory lipoproteins.
Variation on a theme: assembly of lipopoproteins
A superficially similar strategy applies to the export and assembly of outer membrane lipoproteins but the machinery is different. Mature lipoproteins have an N-terminal cysteine that is modified by a thioether-linked diacylglycerol and N-linked fattyacyl chains. Most of the 90 lipoproteins in E. coli reside in the outer membrane. Their location is determined by the presence (or absence) of an Asp inner membrane-retention signal at position 2 (reviewed in [23]) and is influenced by phospholipid composition in the membrane [24]. After export across the inner membrane by the Sec system and maturation, nascent lipoproteins are released from the inner membrane by the action of a protein complex comprised of LolCDE. Sequence motifs in LolD place it in the ABC (ATP-binding cassette) superfamily. While the complex requires ATP hydrolysis for lipoprotein release (reviewed in [25]), it is not an ABC ‘transporter’ in the conventional sense. The nascent lipoprotein is released by the LolCDE pump to a periplasmic chaperone (LolA), creating a soluble complex. At the outer membrane, LolA delivers its cargo to a receptor (LolB) and the new lipoprotein is incorporated into the outer membrane. LolA and LolB share structural features; both have a β-barrel with a hydrophobic cavity capped by an α-helical lid (reviewed in [23]). Unidirectional energy-independent transfer of lipoproteins from LolA to LolB is ensured by their relative binding affinities for their substrates [26].
Stay tuned: export and assembly of lipopolysaccharides
Despite the importance of LPS in almost all Gram-negative bacteria, surprisingly little is known about the export across the periplasm and outer membrane and there is debate over the export process. Attempts to demonstrate release of LPS from membrane vesicles have been unsuccessful under conditions where LolCDE-mediate release of lipoproteins could be detected [27]. Instead, experimental evidence pointed to the involvement of export-active contact sites between the cytoplasmic and outer membranes. This is consistent with early electron microscopy (EM) investigations that identified specific sites for appearance of newly formed LPS on the cell surface [28]. These coincide with controversial ‘Bayer junctions’, regions where the cytoplasmic and outer membranes come into apposition [ [29] and [30]]. There is intriguing evidence for a helical arrangement of LPS-insertion sites on the cell surface, suggesting this is not a random process [31•]. Recent studies have identified an outer membrane protein (Imp) required for LPS export [ [32] and [33••]]. Imp interacts with a lipoprotein, RlpB [34•], suggesting obvious parallels to outer membrane protein export. Furthermore, genetic data suggests that YfgL plays a critical role in homeostasis of outer membrane assembly by balancing outer membrane protein and LPS assembly [8••]. Additional newly discovered components involved in export include a cytoplasmic ABC protein (LptB), a cytoplasmic membrane protein (YrbK), and a putative periplasmic component (LptA), reminiscent of lipoprotein assembly [35]. The identification of these various players now sets the stage for biochemical and structural biology initiatives to resolve the mechanisms.
More variety than we thought: Wza represents a new class of outer membrane protein
Wza is an outer membrane lipoprotein and is essential for expression of group 1 CPS on the surface of E. coli and it represents a new class of outer membrane protein [ [36] and [37]]. Group 1 CPSs are built on the polyisoprenoid carrier lipid (undecaprenol diphosphate; und-PP) by glycosyltransferases located in (or at) the inner leaflet of the inner membrane. These und-PP-linked intermediates are flipped across the inner membrane and polymerized at the periplasmic face in a reaction requiring an integral membrane protein, Wzy. Wzy represents the characteristic putative polymerase in a classical pathway first identified in the biosynthesis of LPS O antigens. The polymerization activity is regulated by an additional component, a member of the polysaccharide co-polymerase subfamily 2a (PCP-2a) [38] encoded by the wzc gene [ [39] and [40]]. The Wzc protein, however, plays an additional critical role in translocation of the polymer from the periplasm to the cell surface. It does this via critical interactions with the final component, the outer membrane protein, Wza.
Wza is a member of the OMA (outer membrane auxiliary [41]) protein family. Wza forms a remarkably stable multimer that maintains its higher-order state in SDS unless heated to ≥60 °C [36]. It is the only OMA protein for which a high-resolution structure is available. Sequential structural studies began with the soluble protein and 2D-arrays in proteolipids by electron microscopy (EM) [ [36], [37] and [42]] and culminated in a high-resolution (2.3 Å) crystal structure [43••]. These investigations have revealed a unique structure. Wza forms a large hollow barrel (interior volume ~15 000 Å3) with eightfold symmetry and is far too large to be contained within the outer membrane.
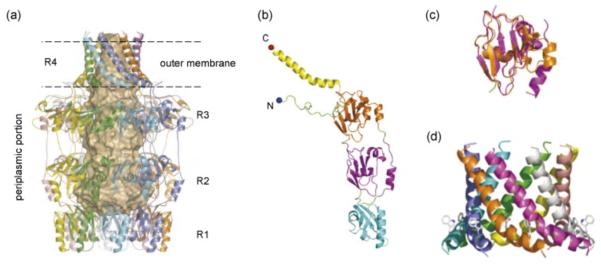
The shape of the octameric structure resembles a classical ‘amphora’ without the handles; it has a large internal cavity that is open at a narrow ‘neck’ and is closed at its base. The structure is built from four eightfold symmetrical rings (R1–R4) stacked one atop the other (Figure 1a). Each Wza monomer has four clearly identifiable domains (Figure 1b) contributing of the four rings. R1 presents a concave periplasmic base in the structure and its centre is filled by eight loops. This opens into an internal cavity with a diameter of 25–30 Å at R2 and R3. R1 and R2 contain the Pfam 02563 motif (PES; polysaccharide export sequence) that is found in all OMA proteins. Interestingly, R2 and R3 are probably gene duplications, both sharing the ubiquitin fold (Figure 1c). At the open end of R4, the internal diameter is reduced to 17 Å. Domain 4 is an amphipathic α-helix located at the C-terminus of the monomer and the helices from the eight monomers come together to create an α-helical barrel (Figure 1c). This provides the most striking feature of Wza; it represents the first example (and currently the only example) of an outer membrane protein whose transmembrane domain is α-helical. The Wza C-terminus is exposed at the cell surface confirming that the R4 barrel does indeed span the outer membrane [43••]. R1–R3 reside outside the membrane in the periplasm. The N-terminus of Wza is long loop, which is wrapped around the top of R3, placing the acyl chains in an ideal location for insertion into the inner leaflet of the outer membrane. Acylation plays a critical role in assembly; a non-acylated variant forms unstable octamers that are unable to direct CPS export [37]. However, it is not clear whether this is owing to an inability to form a stable interaction with the outer membrane, or problems with delivery to the outer membrane. The absence of β-sheet structure in Wza suggests YaeT is unlikely to be involved in its export. The Lol system seems a more likely route but this has yet to be examined.
Figure 1.
Structure of the Wza octamer. (a) Structure of the octamer with each monomer drawn as a ribbon. The eightfold axis is parallel to the long axis of the molecule (top to bottom). The internal void of 15,000 Å3 is represented by the space-filling shape. The internal cavity has a polar surface and we believe this is the key to catalysing the passage of the polar carbohydrate. As the sugar molecule moves through the barrel water molecules can help mediate hydrogen bond interactions. Wza is over 140 Å in length from top to bottom and around 100 Å wide at the base. Antibodies to epitopes inserted at the C-terminus show it is located on the cell surface [43••]. (b) The monomer is composed of four domains. The cyan-coloured domain is the so-called PES domain and is relatively conserved. Domains two (purple) and three (orange) function as a spacers to form the internal barrel void. The long C-terminal helix (fourth domain) is coloured yellow. The eightfold symmetry means each domain forms an eightfold rotationally symmetric ring. These rings (R1–R4) stack on top of each other as can be seen in (a). (c) Domains 2 and 3 are probably gene duplications. Both share the ubiquitin fold but almost no sequence conservation. The lack of extensive sequence conservation is a feature of the Wza homologs. (d) The α-helical barrel has an exposed tryptophan residue, a hallmark of transmembrane regions. The exposed tryptophan acts much a like a barb helping to anchor the helices in the membrane. Each is helix is amphipathic and for this reason does not show up as a classic transmembrane helix and would be unlikely to insert on its own into a lipid bilayer.
It remains to be seen whether the transmembrane helical barrel of Wza is a unique feature confined to CPS export proteins or a more general but hitherto undiscovered transmembrane motif. Other candidates include the secretin proteins from protein secretion systems [44]. These are multimeric proteins and frequently are SDS-stable. PulD, a secretin from a type II secretion system, is a well-characterized representative. It lacks extensive β-sheet structure and is assembled in a YaeT-independent process [45]. Assembly of PulD into the outer membrane does require a lipoprotein ancillary protein (PulS) [46], so the Lol system may play an important role. Paradoxically, a close homologue (PilQ) apparently requires YaeT for insertion [47], but it is not dependent on the corresponding ancillary protein [48]. Clearly, more work is required before a unifying theme can be developed.
Inner membrane components: structure of the periplasmic domain of Wzz and its implications for Wzc
PCP proteins like Wzc possess a characteristic membrane topology with two transmembrane helices flanking a periplasmic domain that varies in size [ [38] and [49•]]. However, the group 1 CPS PCP-2a, proteins have an additional feature; they are tyrosine autokinases with a cytoplasmic C-terminal domain that is autophosphorylated at several tyrosine residues (reviewed in [49•]). Phosphorylation is mediated by a kinase domain with a Walker box motif. The closest sequence homolog (outside PCP proteins) for the kinase domain is found in MinD [50], an ATPase involved in bacterial cell division. Interestingly Wzc also has ATPase activity [51], suggesting that the unusual kinase mechanism of Wzc may be related to NTPases. C-terminal phosphorylation of Wzc is essential for high-molecular weight CPS assembly; in its absence, short oligosaccharides are produced in a phenotype identical to a wzc-null mutant [ [40] and [52]]. Paradoxically, CPS biosynthesis also requires dephosphorylation of Wzc by its cognate phosphatase [40], Wzb, and the current working model proposes that Wzc must cycle between its phosphorylated and non-phosphorylated forms to sustain CPS biosynthesis [ [52] and [53]]. Wzc forms a tetramer with extensive contact between the periplasmic domains of the monomers [54••] (Figure 2a). By contrast, the cytoplasmic kinase domains are well separated. This raises interesting mechanistic questions because it is well established that the C-termini of Wzc homologues are phosphorylated by intermolecular transfer [ [40] and [55]]. One interpretation is that the Wzc tetramer may undergo significant conformational changes (see below).
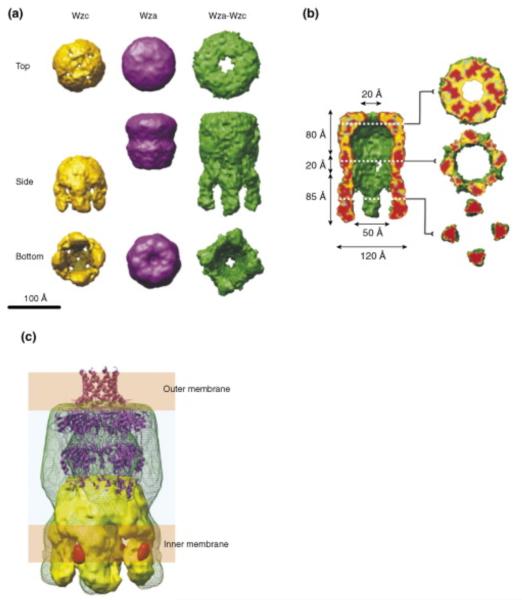
Figure 2.
Three-dimensional (3D) reconstruction of the Wza–Wzc complex from cryo-EM [58]. (a) Comparison of a surface rendered structure of the complex with the isolated Wza and Wzc components. (b) Fifty percent of the foremost volume is removed to reveal the central cavity and the (upper) exit pore. The slices provide detail through the structure. Note that lipid from the cytoplasm would fill the centre of the Wzc tetramer precluding a contiguous connection from the cytoplasm to the exterior of the cell. (c) Proposed organization of the complex (green wire frame) in the cell envelope. The Wzc EM structure (yellow) and the ribbon structure of Wza are included. The orange densities are regions occupied by cytoplasmic N-terminal hexahistidine tags (labeled with nanogold). Indicating the approximate location of the Wzc kinase domains The EM structure of the complex suggests Wza undergoes major conformational changes and broadening in the region encompassing the PES domain, where it interacts with Wzc. Note that the α-helical outer membrane barrel in Wza is destabilized under the pH conditions of the EM experiments (R.C. Ford, J.H. Naismith and C. Whitfield, unpublished results).
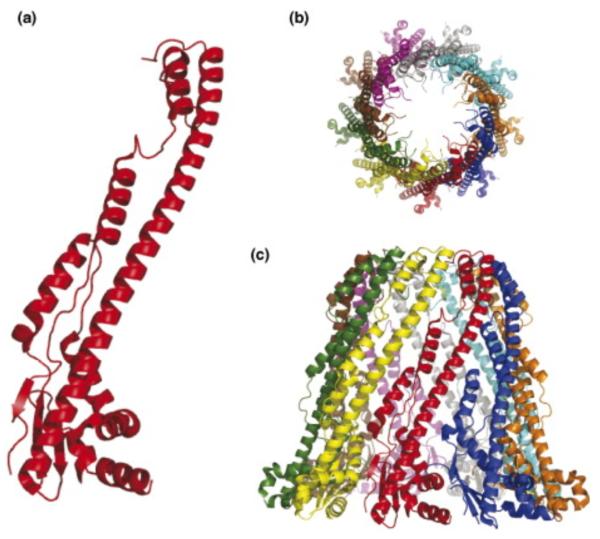
Recently, a crystal structure was reported for the periplasmic domains of several PCP-1 (Wzz) proteins [56]. PCP-1 members participate in lipopolysaccharide O-antigen biosynthesis by a Wzy-dependent pathway similar to group 1 CPSs. Although PCP-1 proteins share overall membrane topology features with Wzc, they have a smaller periplasmic domain and lack the cytoplasmic tyrosine kinase domain. Like Wzc, the periplasmic domains of the PCP-1 proteins form multimers, although the physiological oligomerization state and packing state is less certain [ [56••] and [57]]. The PCP-1 protomer consists of a membrane-proximal mixed α/β base domain from which extends a long (~100 Å) α helical hairpin (Figure 3a). Crystal structures and cryo-EM images of oligomers of one of the full-length PCP-1 proteins reveals a cone-like shape (Figure 3b and c). Although this structure is substantially more extended than what is seen in the Wzc oligomer, the predicted secondary structure for the periplasmic region of Wzc also indicates a mixture α/β structure and an extended α-helical domain, suggesting there may be some similarities in the structures of PCP proteins at an atomic level.
Figure 3.
Structure of the periplasmic domain of the PCP-1 protein (FepE) from E. coli O157:H7 [56••]. (a) The protomer has an N-terminal α/β base domain from which a long α-helix extends. (b) The protomers assemble into a nonamer structure that extends ~100 Å into the periplasm. (c) The nonamer is open at the top generating a solvent-filled cavity with the lipid of the cytoplasmic membrane at its base.
A periplasmic machine: Wza and Wzc form a molecular scaffold spanning the cell envelope
Interactions between Wza and Wzc were initially identified by chemical cross-linking methods [37] but the complex formed by the oligomeric partners is sufficiently specific and stable that it can be reconstituted using purified components [58]. The 3D-structure of the resulting heterocomplex was determined using negative staining cryo-EM, combined with single particle analysis (~12 Å resolution). The long axis (height) of overall complex is 170 Å and the width is 100–120 Å, sufficient to span the cell envelope and providing a contiguous scaffold across the periplasm. All available structural evidence points to the Wza–Wzc heterocomplex bringing the inner and outer membranes into close proximity at the site of carbohydrate export. The solute accessible periplasmic thickness of ~60 Å (or less) calculated from the cryo-EM structure, is considerably narrower than the 200 Å measured in electron micrographs of frozen thin-sections [59]. These structures are consistent with in vivo studies that identified specific sites (Bayer junctions) of membrane apposition where group 1 CPS export occurs under permissive conditions in conditional mutants [ [30] and [60]]. The local compression of the periplasm would serve as a site for recruitment of other machinery required for carbohydrate export.
Features corresponding to the individual Wza and Wzc oligomers are clearly evident in the Wza–Wzc heterocomplex but there are some notable differences in conformation of the individual oligomers when compared individually or in the complex (Figure 3a). The most profound changes are evident in the Wzc–Wza interface; the periplasmic ring structure of the Wzc oligomer appears to separate and the base of the Wza octamer seems to widen, creating putative portals in the side of the complex. The portal has a maximum height of ~20 Å and connects the periplasm to the central cavity (Figure 2b). Computational analyses of the Wza octamer indicate that R2 is the least stable of the rings (ΔG of dissociation of −14 kJ mol–1). By contrast, R1 and R3 have calculated ΔG values of +9 and +180 kJ mol–1, respectively (R.C. Ford, J.H. Naismith and C. Whitfield, unpublished data). The simplest export model invokes the nascent polymer entering portals created from an opening up of R2. The atomic model of the periplasmic domain of the PCP-1 protein allows some interesting speculation as to the potential process. An attractive model is that the long helices of Wzc monomers occupy the large grooves on the outside of the R1 (the conserved PES domain). This would account for the apparent broadening in the base of Wza seen in cryo-EM images of the complex (described above). The formation of the complex is independent of the phosphorylation state of Wzc. This feature of the Wzc protein may have more to do with modulating phosphorylation than export, but it is also conceivable that changes in phosphorylation state influence conformation to drive the export process. The force for polymer extrusion could be generated by extension at the reducing end (proximal to the inner membrane), or by conformational changes in the complex. From the central cavity the polymer then exits from the periplasm and passes to the extracellular space through the α-helical barrel provided by R4. The measured diameters are sufficient to accommodate a polymer in an extended conformation [43••]. In summary, the ‘open’ conformation of the translocon therefore appears to need the association of the Wza and Wzc oligomers and this interdependence is consistent with comparable acapsular phenotypes for wza and wzc mutants [ [36] and [40]].
Perspectives
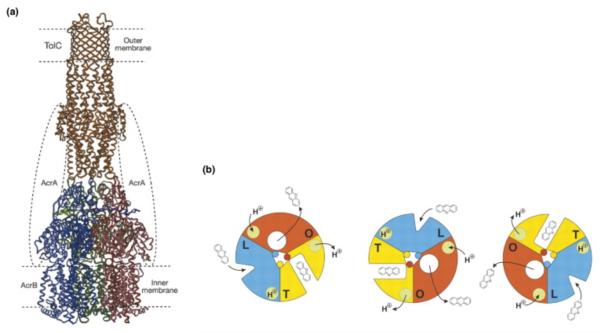
There are a growing number of trans-periplasmic machines under detailed investigation. Perhaps the one most relevant to Wza–Wzc is the TolC AcrA/AcrB drug efflux pump. TolC is a β-barrel protein that uses a periplasmic coiled α-helical region to both ‘gate’ the channel [ [21] and [61•]] and dock to its inner membrane pump AcrB [62]. The relatively weak interactions in this complex are stabilized by the periplasmic adaptor (or membrane-fusion) protein AcrA in a process that also recruits TolC to an active pump [63•] (Figure 4a). The dynamics of this machine are being unravelled in a series of studies. AcrA (and its homolog MexB [64]) possesses a flexible hairpin domain that may participate in TolC opening [ [63•] and [65]], in a process involving a twisting of the coiled α-helical region [21]. This proposal has been verified by producing inactive ‘locked’ mutants [66]. AcrB undergoes a series of large conformation changes, akin to a peristaltic pump [ [67] and [68••]]. In the working model, this pulls in material from the outer leaflet of the cytoplasmic membrane/periplasm and pushes it into the central cavity of TolC (Figure 4b). Support for this model has been provided by engineering of disulfides into AcrB that were designed to lock the conformation of AcrB and, as predicted, these do indeed inhibit efflux [68••].
Figure 4.
Structure and function of the TolC-AcrB-AcrA drug efflux complex. (a) A model for the periplasmic machine formed by TolC, AcrB and AcrA [62]. TolC was manually docked to AcrB. It is thought that AcrA stabilizes the complex between TolC and AcrB. Although TolC lacks the signature C-terminal β-strand, it is a β barrel protein. It is assumed that one of the β-strands interacts with YaeT. (Reprinted by permission from Macmillan Publishers Ltd.: Nature, copyright 2002.) (b) The peristaltic pump mechanism proposed to operate in the AcrB-AcrA-TolC machine [67]. Each subunit of AcrB undergoes a series of changes. In the L form, AcrB binds the drug. This leads to the T state where the drug is tightly bound. In the final step, AcrB forms an O state in which the drug is released into the central cavity of TolC and is exported out of the cell. (Reprinted by permission from Science, copyright 2006.)
What is clear is that the principles underlying the Wza–Wzc system have clear analogies to other transmembrane machines. The molecular mechanisms that control their function may prove tractable to a combination of single particle EM, crystallography and novel spectroscopy methods. It is a measure of the progress of structural biology that these complex multi-protein machines are now coming into view.
Acknowledgements
CW holds a Canada Research Chair. Work in the authors’ laboratories is supported by the Canadian Institutes of Health Research (CW), the Wellcome Trust (CW and JHN) and the Biotechnology and Biological Sciences Research Council (JHN).
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Micr Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schulz GE. The structure of bacterial outer membrane proteins. Biochim Biophys Acta. 2002;1565:308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- 3•.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Ann Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [A review describing the state of the art in capsule biosynthesis, focusing on paradigms from E. coli prototypes.] [DOI] [PubMed] [Google Scholar]
- 4.Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 5•.Bos MP, Robert V, Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Ann Rev Biochem. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [Detailed review of the state of the art in outer membrane assembly.] [DOI] [PubMed] [Google Scholar]
- 6.Gold VA, Duong F, Collinson I. Structure and function of the bacterial Sec translocon. Mol Membr Biol. 2007;24:387–394. doi: 10.1080/09687680701416570. [DOI] [PubMed] [Google Scholar]
- 7.Mogensen JE, Otzen DE. Interactions between folding factors and bacterial outer membrane proteins. Mol Microbiol. 2005;57:326–346. doi: 10.1111/j.1365-2958.2005.04674.x. [DOI] [PubMed] [Google Scholar]
- 8••.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [The existence of a multiprotein complex containing YaeT and the lipoproteins YfiO, NlpB, and YfgL is established by elegant genetic and biochemical approaches. Genetic data assigns an intriguing role for YfgL in homeostasis of outer membrane assembly, coordinating outer membrane protein and LPS assembly.] [DOI] [PubMed] [Google Scholar]
- 9.Xu X, Wang S, Hu YX, McKay DB. The periplasmic bacterial molecular chaperone SurA adapts its structure to bind peptides in different conformations to assert a sequence preference for aromatic residues. J Mol Biol. 2007;373:367–381. doi: 10.1016/j.jmb.2007.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–2484. doi: 10.1101/gad.1581007. [Describes a depletion strategy to assess the roles and relationships of a chaperone proteins in the assembly of outer membrane proteins. Identifies SurA as the major chaperone.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korndorfer IP, Dommel MK, Skerra A. Structure of the periplasmic chaperone Skp suggests functional similarity with cytosolic chaperones despite differing architecture. Nat Struct Mol Biol. 2004;11:1015–1020. doi: 10.1038/nsmb828. [DOI] [PubMed] [Google Scholar]
- 12.Walton TA, Sousa MC. Crystal structure of Skp, a prefoldin-like chaperone that protects soluble and membrane proteins from aggregation. Mol Cell. 2004;15:367–374. doi: 10.1016/j.molcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 13.Bulieris PV, Behrens S, Holst O, Kleinschmidt JH. Folding and insertion of the outer membrane protein OmpA is assisted by the chaperone Skp and by lipopolysaccharide. J Biol Chem. 2003;278:9092–9099. doi: 10.1074/jbc.M211177200. [DOI] [PubMed] [Google Scholar]
- 14.Qu J, Mayer C, Behrens S, Holst O, Kleinschmidt JH. The trimeric periplasmic chaperone Skp of Escherichia coli forms 1:1 complexes with outer membrane proteins via hydrophobic and electrostatic interactions. J Mol Biol. 2007;374:91–105. doi: 10.1016/j.jmb.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 15.Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stegmeier JF, Andersen C. Characterization of pores formed by YaeT (Omp85) from Escherichia coli. J Biochem. 2006;140:275–283. doi: 10.1093/jb/mvj147. [DOI] [PubMed] [Google Scholar]
- 17••.Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [The structure of the periplasmic domain of YaeT and its POTRA-domain fold is described. This paper proposes a model for the function of YaeT and the multiprotein complex it resides in. A dimer, thought to be a crystallographic artefact provides insight into a possible β-strand augmentation principle for interaction with export substrate proteins.] [DOI] [PubMed] [Google Scholar]
- 18.Bos MP, Robert V, Tommassen J. Functioning of outer membrane protein assembly factor Omp85 requires a single POTRA domain. EMBO Rep. 2007;8:1149–1154. doi: 10.1038/sj.embor.7401092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Robert V, Volokhina EB, Senf F, Bos MP, Van Gelder P, Tommassen J. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 2006;4:e377. doi: 10.1371/journal.pbio.0040377. [In vitro reconstitution of the interaction between YaeT and a substrate β-barrel protein. Begins to establish the principles involved in YaeT recognition of its substrates.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werner J, Misra R. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol Microbiol. 2005;57:1450–1459. doi: 10.1111/j.1365-2958.2005.04775.x. [DOI] [PubMed] [Google Scholar]
- 21.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 22.Tommassen J. Biochemistry. Getting into and through the outer membrane. Science. 2007;317:903–904. doi: 10.1126/science.1146518. [DOI] [PubMed] [Google Scholar]
- 23.Tokuda H, Matsuyama S. Sorting of lipoproteins to the outer membrane in E. coli. Biochim Biophys Acta. 2004;1694:1–9. IN. [PubMed] [Google Scholar]
- 24.Miyamoto S, Tokuda H. Diverse effects of phospholipids on lipoprotein sorting and ATP hydrolysis by the ABC transporter LolCDE complex. Biochim Biophys Acta. 2007;1768:1848–1854. doi: 10.1016/j.bbamem.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Narita S, Tokuda H. An ABC transporter mediating the membrane detachment of bacterial lipoproteins depending on their sorting signals. FEBS Lett. 2006;580:1164–1170. doi: 10.1016/j.febslet.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 26.Taniguchi N, Matsuyama S, Tokuda H. Mechanisms underlying energy-independent transfer of lipoproteins from LolA to LolB, which have similar unclosed β-barrel structures. J Biol Chem. 2005;280:34481–34488. doi: 10.1074/jbc.M507388200. [DOI] [PubMed] [Google Scholar]
- 27.Tefsen B, Geurtsen J, Beckers F, Tommassen J, de Cock H. Lipopolysaccharide transport to the bacterial outer membrane in spheroplasts. J Biol Chem. 2005;280:4504–4509. doi: 10.1074/jbc.M409259200. [DOI] [PubMed] [Google Scholar]
- 28.Mühlradt PF, Menzel J, Golecki JR, Speth V. Outer membrane of Salmonella. Sites of export of newly synthesized lipopolysaccharide on the bacterial surface. Eur J Biochem. 1973;35:471–481. doi: 10.1111/j.1432-1033.1973.tb02861.x. [DOI] [PubMed] [Google Scholar]
- 29.Kellenberger E. The Bayer bridges confronted with results from improved electron microscopy methods. Mol Microbiol. 1990;4:697–705. doi: 10.1111/j.1365-2958.1990.tb00640.x. [DOI] [PubMed] [Google Scholar]
- 30.Bayer ME. Zones of membrane adhesion in the cryofixed envelope of Escherichia coli. J Struct Biol. 1991;107:268–280. doi: 10.1016/1047-8477(91)90052-x. [DOI] [PubMed] [Google Scholar]
- 31•.Ghosh AS, Young KD. Helical disposition of proteins and lipopolysaccharide in the outer membrane of Escherichia coli. J Bacteriol. 2005;187:1913–1922. doi: 10.1128/JB.187.6.1913-1922.2005. [Identifies a helical pattern of LPS insertion sites on the surface of E. coli and discusses the issues associated with lateral diffusion of large macromolecuales in the outer membrane.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braun M, Silhavy TJ. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol. 2002;45:1289–1302. doi: 10.1046/j.1365-2958.2002.03091.x. [DOI] [PubMed] [Google Scholar]
- 33••.Bos MP, Tefsen B, Geurtsen J, Tommassen J. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc Natl Acad Sci USA. 2004;101:9417–9422. doi: 10.1073/pnas.0402340101. [By exploiting the unusual ability of N. meningitidis to survive without LPS biosynthesis, the authors provide the first direct evidence that Imp is involved in the assembly of LPS in the outer membrane. Imp is the first component of the LPS assembly pathway to be defined.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34•.Wu T, McCandlish AC, Gronenberg LS, Chng SS, Silhavy TJ, Kahne D. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2006;103:11754–11759. doi: 10.1073/pnas.0604744103. [Describes the involvement of an outer membrane lipoprotein, RlpB, in the assembly of LPS and its interactions with Imp. The parallels to the YaeT/YfiO/NlpB/SmpA/YfgL complex for outer membrane protein assembly are striking.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sperandeo P, Cescutti R, Villa R, Di Benedetto C, Candia D, Deho G, Polissi A. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J Bacteriol. 2007;189:244–253. doi: 10.1128/JB.01126-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drummelsmith J, Whitfield C. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli (O9a:K30) requires a multimeric complex in the outer membrane. EMBO J. 2000;19:57–66. doi: 10.1093/emboj/19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nesper J, Hill CM, Paiment A, Harauz G, Beis K, Naismith JH, Whitfield C. Translocation of group 1 capsular polysaccharide in Escherichia coli serotype K30. Structural and functional analysis of the outer membrane lipoprotein Wza. J Biol Chem. 2003;278:49763–49772. doi: 10.1074/jbc.M308775200. [DOI] [PubMed] [Google Scholar]
- 38.Morona R, Van Den Bosch L, Daniels C. Evaluation of Wzz/MPA1/MPA2 proteins based on the presence of coiled-coil regions. Microbiology. 2000;146:1–4. doi: 10.1099/00221287-146-1-1. [DOI] [PubMed] [Google Scholar]
- 39.Drummelsmith J, Whitfield C. Gene products required for surface expression of the capsular form of the group 1 K antigen in Escherichia coli (O9a:K30) Mol Microbiol. 1999;31:1321–1332. doi: 10.1046/j.1365-2958.1999.01277.x. [DOI] [PubMed] [Google Scholar]
- 40.Wugeditsch T, Paiment A, Hocking J, Drummelsmith J, Forrester C, Whitfield C. Phosphorylation of Wzc, a tyrosine autokinase, is essential for assembly of group 1 capsular polysaccharides in Escherichia coli. J Biol Chem. 2001;276:2361–2371. doi: 10.1074/jbc.M009092200. [DOI] [PubMed] [Google Scholar]
- 41.Paulsen IT, Beness AM, Saier MHJ. Computer-based analyses of the protein constituents of transport systems catalysing export of complex carbohydrates in bacteria. Microbiology. 1997;143:2685–2699. doi: 10.1099/00221287-143-8-2685. [DOI] [PubMed] [Google Scholar]
- 42.Beis K, Collins RF, Ford RC, Kamis AB, Whitfield C, Naismith JH. Three-dimensional structure of Wza, the protein required for translocation of group 1 capsular polysaccharide across the outer membrane of Escherichia coli. J Biol Chem. 2004;279:28227–28732. doi: 10.1074/jbc.M402913200. [DOI] [PubMed] [Google Scholar]
- 43••.Dong C, Beis K, Nesper J, Brunkan-Lamontagne AL, Clarke BR, Whitfield C, Naismith JH. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature. 2006;444:226–229. doi: 10.1038/nature05267. [This paper reports the first crystal structure of an OMA protein and provides insight into possible mechanisms for the export of capsular polysaccharide. Wza is the only outer membrane protein reported to date that contains an α-helical outer membrane channel.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collins RF, Derrick JP. Wza: a new structural paradigm for outer membrane secretory proteins? Trends Microbiol. 2007;15:96–100. doi: 10.1016/j.tim.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 45.Collin S, Guilvout I, Chami M, Pugsley AP. YaeT-independent multimerization and outer membrane association of secretin PulD. Mol Microbiol. 2007;64:1350–1357. doi: 10.1111/j.1365-2958.2007.05743.x. [DOI] [PubMed] [Google Scholar]
- 46.Guilvout I, Chami M, Engel A, Pugsley AP, Bayan N. Bacterial outer membrane secretin PulD assembles and inserts into the inner membrane in the absence of its pilotin. EMBO J. 2006;25:5241–5249. doi: 10.1038/sj.emboj.7601402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 48.Carbonnelle E, Helaine S, Prouvensier L, Nassif X, Pelicic V. Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occurring after pilus assembly, essential for fibre stability and function. Mol Microbiol. 2005;55:54–64. doi: 10.1111/j.1365-2958.2004.04364.x. [DOI] [PubMed] [Google Scholar]
- 49•.Grangeasse C, Cozzone AJ, Deutscher J, Mijakovic I. Tyrosine phosphorylation: an emerging regulatory device of bacterial physiology. Trends Biochem Sci. 2007;32:86–94. doi: 10.1016/j.tibs.2006.12.004. [One subfamily of PCP proteins involved in capsule assembly in Gram-negative and Gram-positive bacteria contains novel tyrosine autokinase activities. This review provides a detailed overview of the structures and diverse functions of these proteins.] [DOI] [PubMed] [Google Scholar]
- 50.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 51.Soulat D, Jault JM, Geourjon C, Gouet P, Cozzone AJ, Grangeasse C. Tyrosine-kinase Wzc from Escherichia coli possesses an ATPase activity regulated by autophosphorylation. FEMS Microbiol Lett. 2007;274:252–259. doi: 10.1111/j.1574-6968.2007.00841.x. [DOI] [PubMed] [Google Scholar]
- 52.Paiment A, Hocking J, Whitfield C. Impact of phosphorylation of specific residues in the tyrosine autokinase, Wzc, on its activity in assembly of group 1 capsules in Escherichia coli. J Bacteriol. 2002;184:6437–6447. doi: 10.1128/JB.184.23.6437-6447.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Obadia B, Lacour S, Doublet P, Baubichon-Cortay H, Cozzone AJ, Grangeasse C. Influence of tyrosine-kinase Wzc activity on colanic acid production in Escherichia coli K12 cells. J Mol Biol. 2007;367:42–53. doi: 10.1016/j.jmb.2006.12.048. [DOI] [PubMed] [Google Scholar]
- 54••.Collins RF, Beis K, Clarke BR, Ford RC, Hulley M, Naismith JH, Whitfield C. Periplasmic protein-protein contacts in the inner membrane protein Wzc form a tetrameric complex required for the assembly of Escherichia coli group 1 capsules. J Biol Chem. 2006;281:2144–2150. doi: 10.1074/jbc.M508078200. [The interaction of an octamer of the outer membrane protein, Wza, and a tetramer of the inner membrane protein, Wzc, cretes a molecular machine for export of group 1 capsular polysaccharides. Cry-EM structures reveal conformational changes in Wza mediated by the interactions and provide additional insight into the capsule export pathway.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grangeasse C, Doublet P, Cozzone AJ. Tyrosine phosphorylation of protein kinase Wzc from Escherichia coli K12 occurs through a two-step process. J Biol Chem. 2002;277:7127–7135. doi: 10.1074/jbc.M110880200. [DOI] [PubMed] [Google Scholar]
- 56••.Tocilj A, Munger C, Proteau A, Morona R, Purins L, Ajamian E, Wagner J, Papadopoulos M, Van Den Bosch L, Rubinstein JL, et al. Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat Struct Mol Biol. 2008;15:130–138. doi: 10.1038/nsmb.1374. [Proteins in the PCP family participate in polymerization of many different polysaccharide structure in Gram-negative and Gram-positive bacteria with diverse biologies. This study provides the first detailed structural view of a periplasmic domain from PCP members. The structure reveals a novel fold with an extended α-helical domain and opens the way to biochemical approaches to probe PCP interactions and functions.] [DOI] [PubMed] [Google Scholar]
- 57.Whitfield C, Larue K. Stop and go: regulation of chain length in the biosynthesis of bacterial polysaccharides. Nat Struct Mol Biol. 2008;15:121–123. doi: 10.1038/nsmb0208-121. [DOI] [PubMed] [Google Scholar]
- 58.Collins RF, Beis K, Dong C, Botting CH, McDonnell C, Ford RC, Clarke BR, Whitfield C, Naismith JH. The 3D structure of a periplasm-spanning platform required for assembly of group 1 capsular polysaccharides in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:2390–2395. doi: 10.1073/pnas.0607763104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matias VR, Al-Amoudi A, Dubochet J, Beveridge TJ. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J Bacteriol. 2003;185:6112–6118. doi: 10.1128/JB.185.20.6112-6118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bayer ME, Thurow H. Polysaccharide capsule of Escherichia coli: microscope study of its size, structure, and sites of synthesis. J Bacteriol. 1977;130:911–936. doi: 10.1128/jb.130.2.911-936.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61•.Andersen C, Koronakis E, Bokma E, Eswaran J, Humphreys D, Hughes C, Koronakis V. Transition to the open state of the TolC periplasmic tunnel entrance. Proc Natl Acad Sci USA. 2002;99:11103–11108. doi: 10.1073/pnas.162039399. [The TolC protein from drug efflux systems has an extended periplasmic domain formed from 12 helices. The authors use mutations designed to disrupt interactions within the helical domain to provide supporting evidence for a model where TolC opens by an ‘iris-like’ realignment in the helices.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 63•.Lobedanz S, Bokma E, Symmons MF, Koronakis E, Hughes C, Koronakis V. A periplasmic coiled-coil interface underlying TolC recruitment and the assembly of bacterial drug efflux pumps. Proc Natl Acad Sci USA. 2007;104:4612–4617. doi: 10.1073/pnas.0610160104. [This paper describes experiments designed to test predictions molecular modelling of interactions between TolC and the periplasmic adaptor protein, AcrA. The working model addresses the accommodation of these interactions during transition of TolC to the open state, a critical element in drug efflux pump action.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Higgins MK, Bokma E, Koronakis E, Hughes C, Koronakis V. Structure of the periplasmic component of a bacterial drug efflux pump. Proc Natl Acad Sci USA. 2004;101:9994–9999. doi: 10.1073/pnas.0400375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure. 2006;14:577–587. doi: 10.1016/j.str.2005.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eswaran J, Hughes C, Koronakis V. Locking TolC entrance helices to prevent protein translocation by the bacterial Type I export apparatus. J Mol Biol. 2003;327:309–315. doi: 10.1016/s0022-2836(03)00116-5. [DOI] [PubMed] [Google Scholar]
- 67.Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 68••.Seeger MA, von Ballmoos C, Eicher T, Brandstatter L, Verrey F, Diederichs K, Pos KM. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat Struct Mol Biol. 2008;15:199–205. doi: 10.1038/nsmb.1379. [From X-ray crystal data, AcrB is proposed to act like a ‘periplasmic pump’ with several conformational states contributing to the pump action. Here, the authors test this prediction by introducing cysteines at precise positions. Disufide bond formation inhibited function and this effect could be reversed by reducing agents, confirming a functional rotation mechanism.] [DOI] [PubMed] [Google Scholar]