Abstract
Uridine diphosphogalactofuranose (UDP-Galf) is the precursor of the D-galactofuranose sugar found in bacterial and parasitic cell walls, including those of many pathogens. UDP-Galf is made from UDP-galactopyranose by the enzyme UDP-galactopyranose mutase. The enzyme requires the reduced FADH− co-factor for activity. The structure of the Mycobacterium tuberculosis mutase with FAD has been determined to 2.25Å. The structures of Klebsiella pneumoniae mutase with FAD and with FADH− bound have been determined to 2.2Å and 2.35Å resolutions respectively. This is the first report of the FADH− containing structure. Two flavin dependent mechanisms for the enzyme have been proposed, one which involves a covalent adduct being formed at the flavin and the other based on electron transfer. Using our structural data, we have examined the two mechanisms. The electron transfer mechanism is consistent with the structural data, not surprisingly since it makes fewer demands on the precise positioning of atoms. A model based on a covalent adduct FAD requires repositioning of the enzyme active site and would appear to require that the isoalloxazine ring of FADH− to buckle in a particular way. However, the FADH− structure reveals that the isoalloxazine ring buckles in the opposite sense, this apparently requires the covalent adduct to trigger profound conformational changes in the protein or to buckle the FADH− opposite to that seen in the apo structure.
Keywords: Crystal structure, mutase, TB, contractase
Introduction
Mycobacterium tuberculosis currently infects 1.7 billion people, a third of the world’s population, is responsible for more morbidity in humans than any other bacterial disease and accounts for over 1.8 million deaths/year in 2002 (World Health Organisation). In the U.S., the number of TB cases had dropped almost every year since records were kept until 1985 (Centre for Disease Control, USA). Also, in recent times, patient non-compliance has given rise to multi-drug resistant (MDR) strains of M. tuberculosis, which are particularly threatening. Klebsiellá pneumoniae is an opportunistic pathogen and tends to affect hospitalised, immunocompromised, or chronically ill patients. A recent study showed this bacterium was identified in about 20% (i.e. 3rd most prevalent overall) of surveyed blood cultures from intensive care units in the US1 and similar numbers have been reported in Europe2. This organism causes severe lung infections, urinary tract infections and bacteremia and Klebsiella isolates are generally resistant to many antibiotics.
Galactofuranose (Galf) is a vital component of the arabinogalactan that connects the peptidoglycan and the mycolic acids in the mycobacteria cell wall 3. The precursor of Galf is uridine diphosphate (UDP)-Galf, which is in turn synthesised from UDP-Galp by the reversible enzyme, UDP-galactopyranose mutase (mutase), which interconverts UDP-Galp and UDP-Galf (Figure 1a). Knockout mutants of M. smegmatis (a model for M. tuberculosis) show that the mutase gene is essential for cell viability4. D-galactofuranose Galf is found in the lipopolysaccharide (LPS) O antigens of a variety of Gram-negative bacteria, including K. pneumoniae and Escherichia coli5. The absence of Galf or mutase in higher eukaryotes makes the biosynthetic pathway of Galf an attractive target for inhibition by new antimicrobial compounds.
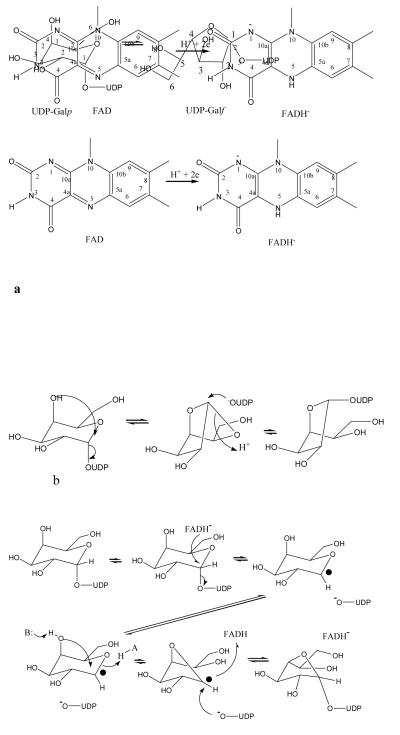
Figure 1.
The mutase enzyme function and structure.
a The chemical reaction catalysed by the mutase enzyme. The cofactor FAD and FADH− are shown. Ring positions numbers are referred to in the text.
b The bicyclic mechanism which does not require direct involvement of the FAD8.
c The redox mechanism in which one electron is transferred to the substrate11. The electron is transferred from FADH- to create a radical which then re-arranges to give product.
d The covalent intermediate mechanism, in the this N5 attacks C1 in a nucleophilic manner. The covalent intermediate then re-arranges to give product. The presence of the covalent intermediate was detected by mass specotrometry12.
The chemistry involved in this enzyme catalysed ring-contraction is unique and is of considerable interest. Early studies indicated mutase requires and consumes reduced nicotinamide adenosine dinucleotide (NAD(P)H) during turnover in spite of the reaction proceeding without a net gain or loss of electrons6, 7. Oxygen isotope experiments established that the anomeric C-O bond of UDP-Galp is cleaved during the reaction8. As a result a bicylic intermediate was proposed to occur during turnover8 (Figure 1b). Another study suggested that NAD(P)H was in fact not required by the enzyme and that enzyme with oxidised flavin was active9. However, a thorough study involving cycling between oxidised and reduced forms showed that NAD(P)H was not essential but that oxidised flavin was at least 106 fold less active than fully reduced flavin10. This critical role for reduced flavin suggested mechanisms invoking cryptic redox chemistry. A thermodynamic analysis of the K. pneumonia mutase11, showed that the fully reduced flavin is found in anionic form and that substrate stabilises a neutral flavosemiquinone radical. Such an anionic flavohydroquinone adenosine dinucleotide (FADH−) would allow rapid single electron transfer in a crypto-redox reaction but energetic considerations argued against a 2 electron transfer mechanism instead pointing to a one electron process (Figure 1c). In this process an electron is transferred to the substrate which then re-arranges. Recently another entirely novel mechanism has been proposed12. Using radiolabelling the work clearly showed the presence of a covalent adduct between flavin and the substrate. This new mechanism involves a nucleophilic attack by the nitrogen (N5) of the isoalloxazine ring on the substrate which then displaces the UDP (Figure 1d). The galactose ring attaches to the flavin, re-arranges and then dissociates from the flavin to give the final product 12 (Figure 1d). A hybrid of these differing mechanisms involving both a single electron transfer and covalent adduct has also been proposed13 based upon substitution of flavin with flavin analogues.
Here we report the structures of mutase from M. tuberculosis at 2.25Å resolution with flavin adenosine dinucleotide (FAD) bound, and K. pneumoniae at 2.2Å resolution with FAD and at 2.35Å resolution with FADH− bound. These structures represent the highest resolution for the mutase enzyme to date and the first with the active co-factor. We have used these structures to model both the covalent mechanism and the electron transfer mechanism.
Results and Discussion
Structure of mutase
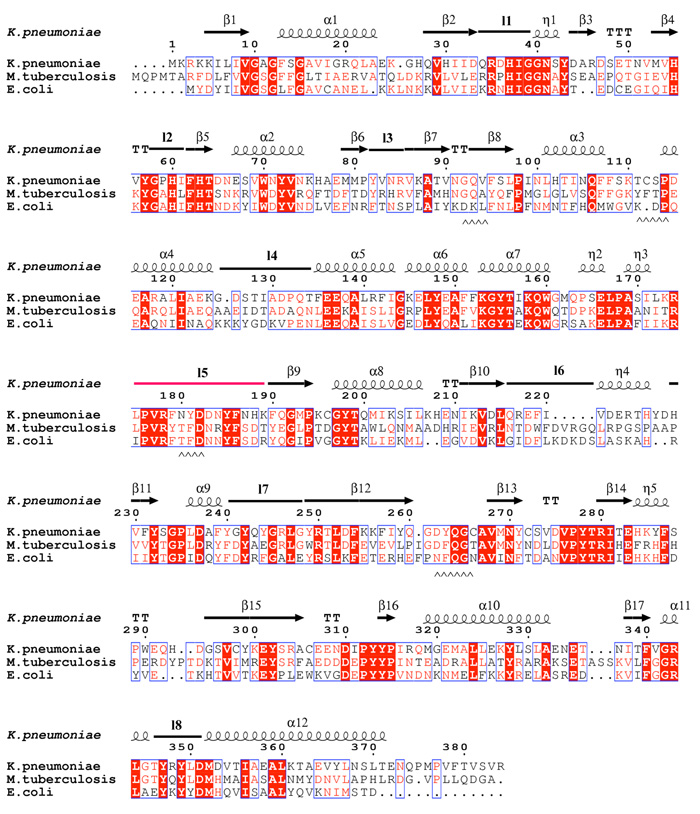
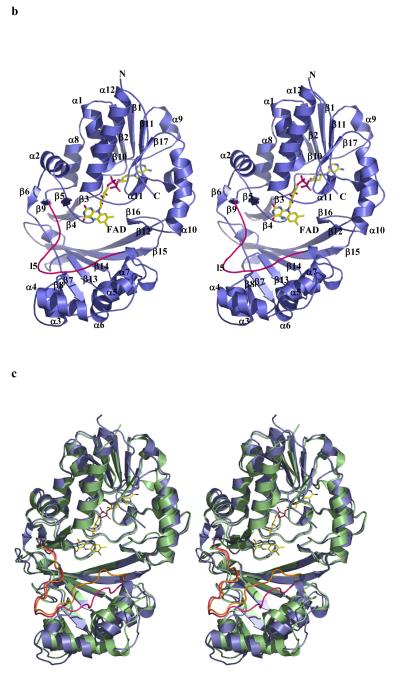
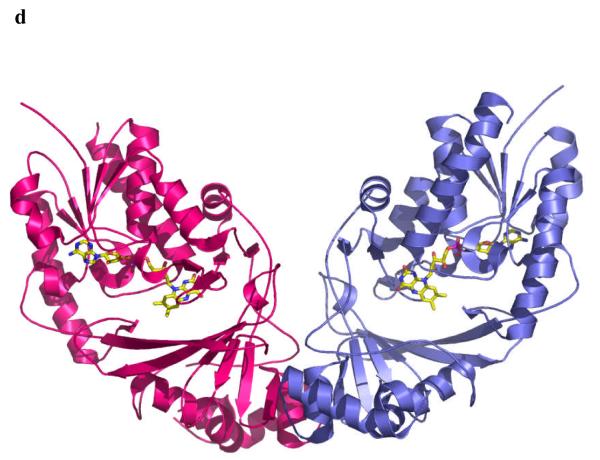
The enzyme from M. tuberculosis is 42% identical to that from E. coli and the enzyme from K. pneumoniae 37% identical (Figure 2a) to E. coli. As one would expect structures of the proteins from M. tuberculosis and K. pneumoniae are very similar to the E. coli homolog10 (Figure 2b). Mutase is a mixed α/β class of protein, and functions as a homodimer (Figure 2c), with each monomer binding one molecule of FAD. Although the dimer arrangement is conserved in all structures, the residues at the interface are not (Figure 2a, c).
Figure 2.
a Sequence alignment of K. pneumoniae with the M. tuberculosis and E. coli mutase enzymes. The alignment is guided by structure and the secondary structure elements for the K. pneumoniae enzyme are shown. The absolutely conserved residues are boxed in red, conservatively substituted residues are shown in red text. Stretches of conservation are shown boxed. The flexible loop 5 discussed in the text is highlighted in the figure in pink. The level of identity is approximately 40%. Residues at the dimer interface are underlined with ^^ symbols.
b The monomer structure of the K. pneumoniae enzymes, secondary structure elements are labelled (consistent with Figure 2a). Loop 5 is shown highlighted in pink. FAD sits facing into a large open cleft.
c Superposition of K. pneumoniae (coloured as Figure 2b) enzyme with E. coli enzyme shown in green. The flexible loop in E. coli is coloured orange.
d The dimer structure of the M. tuberculosis enzyme. The dimeric arrangement is conserved in all structures of the enzyme from different organisms.
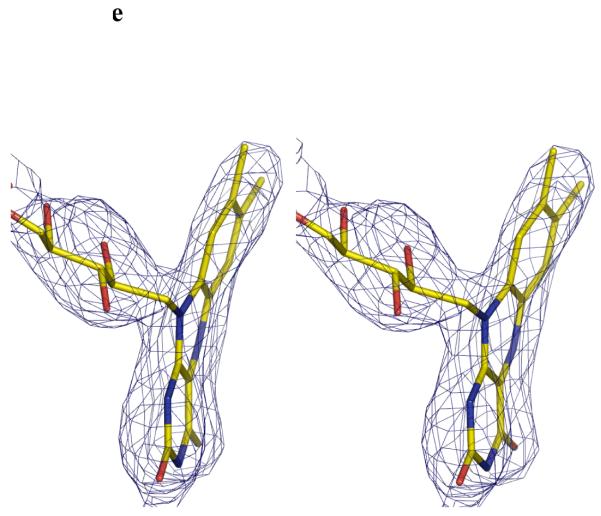
e Stereo image FADH− isoalloxazine ring. The unbiased Fo-Fc map contoured at 3σ for FADH−. FADH− is shown in stick representation. The curvature is towards the si-face. The N5 atom is moved towards the protein and out of the cleft.
The four monomers found in the asymmetric unit of M. tuberculosis superimpose with each other with an rmsd of 0.2Å for all ordered Cα atoms. The loops in this structure do not show the variation between the monomers observed in the E. coli structure10. Monomer A of mutase from M. tuberculosis can be superimposed with the monomer A of E. coli and K. pneumoniae with a Cα root mean square deviation (rmsd) of 1.16Å and 1.15Å over 330 Cα atoms, respectively. Monomer A of oxidised mutase from K. pneumoniae can be superimposed with the monomer of the reduced structure with a Cα root mean square deviation (rmsd) of 0.24Å over 383 Cα atoms, indicating that the chemical reduction of flavin does not induce significant changes in the conformation of the protein and no movement in side chains is detected upon visual inspection.
A structure based sequence alignment of the three structures shows that the residues of the β-sheets and α-helices are highly conserved, whereas the identity of the residues in loops is more variable (Figure 2a). As a result there are small variations in peripheral loop structure. However, loop 5 shows considerable structural change. In E. coli mutase it folds in the cleft between domains 1 and 3, whereas in M. tuberculosis and K. pneumoniae it is found facing the solvent, almost perpendicular to the conformation in E. coli (Figure 2b). This loop is significant as it lines the open cleft near the FAD (Figure 2c).
Flavin binding site
The enzyme binds FAD through a combination of hydrogen bonds (some water mediated) and by van der Waals interactions. Many of the side chains involved in the FAD binding are absolutely conserved. The discussion focuses on the K. pneumoniae structure as it has been determined in both oxidised and reduced form. The adenine moiety of the FAD is bound in a pocket formed by the side chains of Val9 and Gln34 and the main chains of Phe219 and Gly234. The only direct hydrogen bond is to the backbone amide of Phe219. Stabilisation of the negative charge of the pyrophosphoryl groups is achieved by interactions with Arg343, Arg35 and backbone amides (Ser14 & Asn41). The ribosyl moiety is recognised through hydrogen-bonds between its hydroxyl groups and the side chain of Glu33.
In each oxidised structure, the FAD isoalloxazine ring is planar as it is a conjugated aromatic system. This to be expected as the oxidised ring is a single conjugated system enforcing sp2 hybridisation on all ring atoms. The isoalloxazine ring sits between Tyr349 and His60 with the re-face of the isoalloxazine ring open to the cleft (Figures 2c and 3a). N10 of FAD is 3.1Å from the hydroxyl group of an absolutely conserved tyrosine, Tyr349 but as the N10 is sp2 hybridised, a hydrogen bond is not possible. Rather it is possible that the proton attached to Tyr349 could participate in a hydrogen π interaction with the isoalloxazine ring14. The separation between the centre of the isoalloxazine ring and the OH of Tyr 349 is 3.1Å. The N1 atom is within 3.5Å from the N-terminus of helix α11 and the N5 atom is within hydrogen bonding distance (3.0Å) of the carbonyl group of Pro59. For a bond to exist that N5 would be require to be protonated15, an unusual occurrence16. The N5 “hydrogen bond” to the backbone carbonyl is observed in all three oxidised structures; however the identity of the residue making contact is not conserved. At the resolution of our studies we cannot identify whether a hydrogen bond exists. The backbone amide nitrogen of a conserved methionine, Met352, forms a hydrogen-bond with the O2 atom of FAD. The methyl groups of the isoalloxazine ring in FAD are stabilised by hydrophobic contacts from a conserved tyrosine, Tyr314.
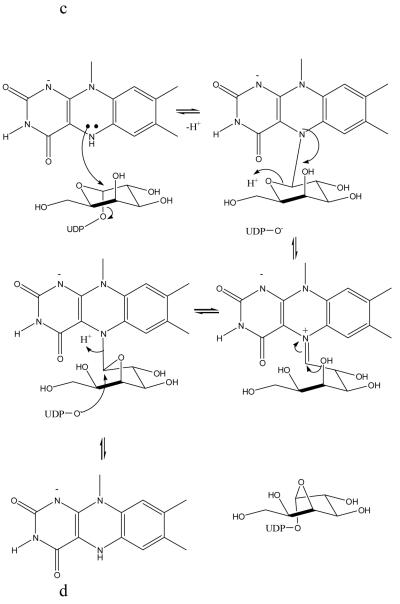
Figure 3.
Stereo images of models of substrate with mutase
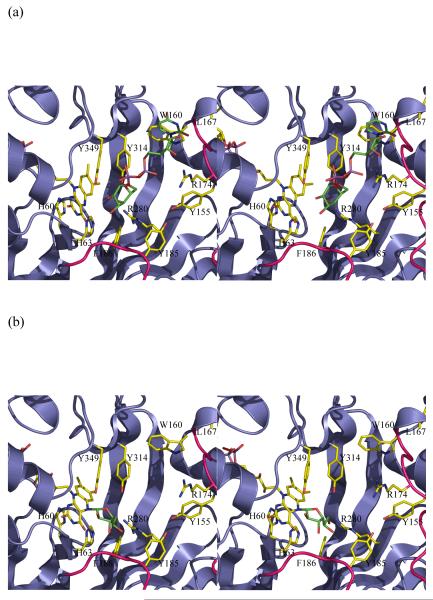
(a) The initial complex between UDP-galactose of the active reduced form of the K. pneumoniae enzyme. This complex is predicted to occur in a mechanism involving electron transfer or a covalent intermediate. No significant re-arrangements are required to accommodate the substrate. The structurally diverse loop 5 is shown in pink.
(b) A model of the covalent adduct with the re-face buckle of isoalloxazine ring. The re-face buckled isoalloxazine ring is taken from a thioredoxin structure 17. This model allows interactions with key conserved residues. The model would require conformation changes in side chain positions only to avoid steric clashes.
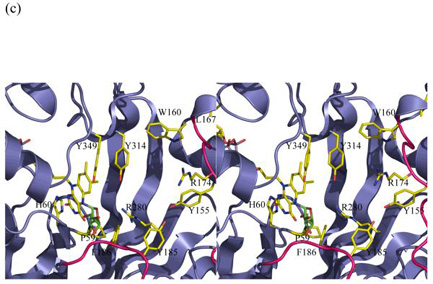
(c) The covalent adduct based on the experimental K. pneumoniae FADH− structure. The sugar is interpenetrating with the protein structure. Either FADH− adopts a different buckle in the presence of substrate or the protein undergoes a profound conformation change. His 63 has been omitted for clarity and Pro59 added to this figure.
In the reduced K. pneumoniae mutase structure, the FADH− isoalloxazine ring has a pronounced buckle (Figure 2e) which is due N5 adopting sp3 hybridisation. In the FADH− structure, the hydrophobic contacts between Tyr314 and the methyl groups of the isoalloxazine ring are lost. The N10 of the FADH− is 3.2Å from the OH group of Tyr349. In our dictionary we restrain the N10 atom to be sp2 hybridised (null hypothesis), any sp3 character could point the lone pair towards to Tyr349 making a hydrogen bond. Higher resolution (<1.5Å) would be required to reliably identify any sp3 character of this atom. The buckling of the ring increases the separation of the Tyr349 OH from the isoalloxazine ring to 3.4Å. N1, where the full negative charge of FADH− would reside, remains close to the positively charged dipole of α11. The distance between N5 and the backbone carbonyl is now shorter (2.7Å). In FADH− (unlike FAD) N5 normally carries a proton. To make the hydrogen bond, the proton would require to point towards the carbonyl dictating that the lone pair on the sp3 hybridised N5 points into the cleft.
Due to its chiral nature there are two distinct possibilities for ring buckling in FADH−, either re- or si- face bending. In contrast to other FADH2-containing structures, thioredoxin reductase from E. coli (1C10)17, polyamine oxidase (1B37)18, and cholesterol oxidase (1COY)19, which have the isoalloxazine ring buckled in the re-face, the isoalloxazine ring of FADH− in mutase from K. pneumoniae, is buckled in the si-face (Figure 2e). The si conformation has also been observed in the isoalloxazine ring of the flavin mononucleotide (FMN) in nitroreductase from Enterobacter cloaca (1QKD) 20.
Putative ligand binding site
Attempts to either co-crystallise or soak UDP-Galp in the active site of both the K. pneumoniae and M. tuberculosis have been unsuccessful. As the bicyclic mechanistic hypothesis does not involve FAD (Figure 1b), the binding site could be located in any part of the protein8 and our results do not inform a discussion of this mechanism. If flavin is involved in the mechanism either by electron transfer (Figure 1c) or covalent bond formation (Figure 1d), the substrate requires to be close to the flavin isoalloxazine ring system. A large cavity containing water molecules is located adjacent to the isoalloxazine ring in all the structures of enzyme. Site-directed mutagenesis studies indicated residues in the cleft adjacent to the flavin perturbed enzyme activity10. Our thermodynamic characterisation of the enzyme indicated that substrate binding influenced the spectrum of the flavin 11, the first direct evidence that flavin was indeed adjacent to substrate. Access to, or from the cavity in the E. coli structure would require movement of loop 5. In M. tuberculosis and K. pneumoniae this loop does not occlude the active site. The variation in structure of this loop is consistent with a role in substrate binding and catalysis. The cleft into which the flavin faces is lined by the absolutely conserved residues His63, Tyr155, Gln159, Trp160, Tyr185, Phe186, Arg250 (although the side chain conformation points away from the cleft in K. pneumoniae but in M. tuberculosis Arg261 points into the cleft), Asn270, Arg280, Tyr314 and Asp 351. The Tyr and Trp residues have been shown to be required for efficient catalysis10. The charge density in this region is consistent with the binding of a negatively charged substrate.
UDP-Galp modelling
Whether covalent attack or electron transfer, the C1′ of the substrate should be close to N5 of the isoalloxazine ring and the whole substrate should make reasonable contacts with the protein. Using the structures determined in this work, we have modelled in the substrate (UDP-Galp) into the enzyme (Figure 3a) by placing it in the large cleft adjacent to the FAD. We have used this model to examine the mechanistic hypotheses (electron transfer or nucleophilic attack or hybrid) that require flavin. We stress the limitations of modelling, in particular we are modelling using an apo structure and UDP-galactose is a relatively large flexible molecule. The model is being employed to gain low resolution insight rather than molecular details. However, the model we have produced does seem plausible. UDP-galactose is modelled in an elongated conformation in the cleft with the uridine ring stacks between Trp160 on one side with Ile171 and Leu167 on the other, Leu167 and Trp160 are absolutely conserved (Figure 2a) in all mutase sequences reported. The side chains of Arg250 and Arg280 are close by and with some re-arrangement could make contacts with the pyrophosphoryl groups, a common feature of protein sugar nucleotide interactions21. The model places the C1′ of the carbohydrate about 4.8Å from the N5 of the isoalloxazine ring. Although this is longer than the distance one expects for electron transfer given the approximations inherent in modelling substrate into an apo structure, we think the model indicates that electron transfer to the C1′ position of galactose is plausible. Some minor re-arrangements of structure of the sort normally seen on binding ligand would be required to bring the C1′ closer to N5. Loop5 which is known to be flexible sits alongside the substrate. Galactose is placed close to Tyr314, Tyr349 and Arg280. The simple electron transfer mechanism (Figure 1c) has no steric requirement for any particular conformation (si- or re- face) of FADH− and the structure presents no problems for this mechanism.
As with electron transfer, 4.8Å is also too far for a covalent attack. Some re-arrangements would be required to bring N5 and C1′ to the appropriate separation for covalent attack. The task of constructing models involving a covalent intermediate is quite simple, as there is a defined chemical structure at the experimentally located flavin. Both the hybrid mechanism and the simple covalent mechanism require both the furanose and pyranose rings are attached to flavin at various stages (Figure 1d). We assumed that the isoalloxazine ring orientation relative to sugar ring was determined by the requirement of an SN2 attack at C1′. Our first model was based on the FAD containing structures, however the N5 is planar and the proposed covalent intermediates interpenetrate with the protein backbone. The configuration of N5 depends on the extent to which the lone pair is involved in the molecular orbital of the isoalloxazine ring, whether it remains sp2 or becomes sp3 hybridised. In the short chain dehydrogenase (SDR) system it has been shown that NADH is significantly buckled the ring nitrogen becomes sp3 hybridised22. There are several examples of FAD-binding proteins that have their reduced isoalloxazine ring deviating from planarity, for example thioredoxin17. In this structure the isoalloxazine ring has a re-face buckle and the N5 deviates significantly from planarity. In the context of the mutase structure, a re-face buckle presents a concave surface to the cleft and pushes the N5 atom into the cleft. This model of the covalent intermediate of re-face buckled FADH− and galactose indicates that such a compound could be accommodated, but there would require some re-arrangement of protein side chains to avoid steric clashes.
Cleavage of the O5′ - C1′ bond in galactose is required during turnover and is favoured by a trans arrangement of the N5 lone pair and O5′. In this orientation there are no steric clashes and several potential hydrogen bonds to the carbohydrate (Figure 3b). Importantly, the hydroxyls of the conserved and required Tyr314 and Tyr349 are close to O5′ and may supply the proton required for C5′ - O5′ cleavage. No major re-organisation is required to accommodate the intermediates. Significantly our model of UDP-galactose at the protein active site orients UDP-galactose correctly for a nucleophilic (SN2) attack at C1′ by N5 of isoalloxazine which would displace UDP. The angle between N5 C1′ and O(UDP) is 150°, close to the required 180°.
In order to provide further experimental data for the model, the reduced form of mutase was obtained by chemical reduction with sodium dithionite. This method of reducing the flavin has also been reported by Lennon et al. 17. In their study, they observed that the chemically reduced structure (re-face bend of the isoalloxazine ring), is in agreement with theoretical23 and NMR studies24, thus chemical reduction of apo proteins is not known to induce conformational artefacts per se. The result obtained was therefore surprising, the reduced mutase structure shows that the isoalloxazine ring has a pronounced si-face buckle and the N5 atom moves towards the protein and out of the cleft (Figure 2e). The two aromatic rings are offset by approximately 8° relative to each other. This value may be an underestimate as we restrained N10 to be trigonal during refinement. The bending of the ring is notably less than the 27° predicted to occur for free flavin16. A 27° buckle would cause the isoalloxazine ring to clash with side chains. There is no obvious steric barrier which is preventing a re-face buckling of the isoalloxazine ring. We therefore conclude that the apo structure actively favours si-face over re-face buckling. Although the lone pair points towards the cleft, as required for nucleophilic attack, our structural observation presents a significant difficulty for the covalent model. With a si-face buckle of FADH−, the clashes of the covalent intermediate with protein are exacerbated relative to the flat FAD. The atoms of galactose interpenetrate with the main- and side-chain atoms of the turn between β4 and β5 strands (Figure 3c). If the covalent intermediate is indeed formed with a si-face buckled flavin, then the protein would have to undergo a profound conformation change and major re-arrangement of backbone structure. An alternative to a conformational change in the protein would be movement of the isoalloxazine ring into the cleft. While possible, this would also require substantial changes in the protein, as the isoalloxazine ring is pinned to the protein by an absolutely conserved Tyr residue. Again such a movement would require large conformational changes in protein backbone structure. It therefore seems unlikely that the covalent intermediate forms with a si-face buckled FADH−. The intermediate must form between re-face buckled FADH− and galactose.
The only explanation consistent with the published data regarding the covalent mechanism is that substrate binding perturbs the structure of the active site of mutase in such a way that favours re-face or prevents si-face buckling of the FADH− ring. Our spectroscopic characterisation of the protein indicated that substrate (and UDP) binding did significantly perturb the redox potential of the flavin (FAD to FADH−) relative to apo protein. It appears that the difference in the redox potential reflects that in the complex structure the redox potential measures FAD to FADH− (si) where as in the apo it measures FAD to FADH−(re). Nucleotide sugars have been seen in other systems to cause a conformation changes which perturbs the redox potential of the cofactors relative to the apo form.
Our structural results extend our understanding of this fascinating enzyme and suggest areas for further research. In particular, the structural effects which force re or si flavin bending require further study.
Materials and Methods
Purification and crystallisation
Mutase from M. tuberculosis was overexpressed in E. coli BL21(DE3) cells transformed with the pET-29b(+) plasmid. The protein was overexpressed and purified in a similar manner to that described for the E. coli enzyme 25, 26.
Purified protein was concentrated to 15 mg ml−1 and 7 mM DTT final concentration was added prior to crystallisation. Large yellow orthorhombic crystals (oxidised FAD) were grown at 277K from a solution containing 4 μl of protein sample and 4 μl of precipitant (0.1M bicine pH 9.0 and 1.6 M ammonium sulphate) using the sitting drop vapour diffusion method. Crystals grew within three weeks to final dimensions of 2.0 × 0.5 × 0.5 mm.
One data set was collected from a crystal rapidly cooled to 100K. The crystal was cryoprotected by soaking in a mother liquor solution containing 4M sodium formate for 5 seconds. The resulting crystals belonged to space group P21212 with cell dimensions a=137.6 Å, b=153.7 Å, c=137.7 Å and α=β=γ=90°.
The K. pneumoniae mutase was overexpressed and purified as previously described 11. Large yellow tetragonal crystals were grown at 293K from a solution containing 4 μl of protein sample (20mM Tris-HCl pH 7, 9mg ml−1) and 4 μl of precipitant (0.1M bis-tris pH 6.5, 50mM CaCl2, and 30% PEGMME 550) using the sitting drop vapour-diffusion method. Crystals appeared within 3 days. One data set was collected from a crystal rapidly cooled to 100K, the precipitant acts as its own cryo protectant. The crystals belonged to the space group P3221 with cell dimensions a=b=85.7 Å, c=99.8 Å and α=β=90°, γ=120.0°.
Chemical reduction of oxidised crystals
The crystals of oxidized K. pneumoniae mutase (yellow) were soaked in a solution containing 100mM sodium dithionite. The crystals were observed under a microscope and when they turned colourless they were flash-frozen in a cold nitrogen stream for data collection. The resolution of our studies cannot identify a mixed population of reduced and oxidised flavin. However, for enzyme in solution the conditions we employed for preparation of the crystals completely reduce the enzyme. We therefore assume any amount of oxidised flavin in the structure is insignificant.
Data collection
Data to 2.25Å for M. tuberculosis was collected at ESRF ID14-2 at a wavelength of 0.933Å using an ADSC Quantum 4 CCD detector with 0.25° oscillations. Data to 2.0Å (oxidized crystals) and 2.35Å (reduced crystals) for K. pneumoniae were collected with a Rigaku MicroMax-007 rotating-anode generator operated at 40 kV and 20 mA with 0.5° oscillations. All data were processed with the program MOSFLM 27 and merged and scaled using the program SCALA 28 from the CCP4 program suite. Statistics are summarized in Table 1.
Table 1.
X-ray data collection statistics (values in parentheses refer to the highest resolution shell)
| Protein | M. tuberculosis |
K. pneumoniae FAD |
K. pneumoniae FADH− |
|---|---|---|---|
| Data collection | ESRF ID14-2 | Inhouse | FADH- |
| Wavelength (Å) | 0.933 | 1.54178 | 1.54178 |
| Spacegroup | P21212 | P3221 | P3221 |
| Resolution (Å) | 24.5-2.25 (2.37-2.25) |
39.2-2.00d (2.12-2.00)d |
28.1-2.35 (2.48-2.35) |
| Cell (Å) | a=137.6, b=153.73, c=137.68 α=β=γ=90.0° |
a=85.7, b=85.7, c=99.8 α=β=90°, γ=120.0° |
a=86.0, b=86.0, c=100.8 α=β=90°, γ=120.0° |
| Unique reflections | 1188969 | 24352 | 17405 |
| I/σ | 23 (1.1) | 25 (4.3) | 12 (1.9) |
| Multiplicity | 6.2 (5.3) | 17 (9.1) | 2.5 (2.5) |
| Data completeness (%) | 89 (85.2) | 88 (49)d | 100(99.1) |
| Rmerge (%)a | 10.2 (61.4) | 4.6 (17.7) | 6.4 (47.9) |
| Refinement | |||
| Rfactorb (%) | 24.2 (34.9) | 19.3 (29.0) | 18.7 (27.6) |
| Rfreec (%) | 28.3 (36.1) | 24.7 (34.1) | 24.3 (34.1) |
| Rmsd bond distances (Å) | 0.014 | 0.010 | 0.011 |
| Rmsd angle ° | 1.453 | 1.203 | 1.230 |
| In Ramachandran % Core e % disallowed e |
91 0 |
91 0 |
89 0 |
| PDB code31 | 1VOJ | 2B17 | 1USJ |
Rmerge = Σhkl Σl|Ii − <I>|/ Σhkl Σl <I>, where Ii is an intensity for the ith measurement of a reflection with indices hkl and <I> is the weighted mean of the reflection intensity.
Rfactor = Σhkl|Fo(hkl)| − |Fc(hkl)||/ Σhkl| Fo(hkl)|, where Fo and Fc are the observed and calculated structure factors, respectively.
Rfree is the crystallographic Rfactor calculated with 5% of the data that were excluded from the structure refinement.
Data extend to 2.0Å and were used in refinement. Due to a technical problem with the detector which prevented its distance being changed, these data were collected in the corners and are incomplete. The structure is quoted to 2.2Å as this is the last shell (2.3Å-2.2Å) where completeness is 90% and the overall completeness to 2.2Å is 98%.
Core and disallowed areas are defined by PROCHECK32.
Structure solution and refinement
All structures were solved by molecular replacement (with AMoRe 28). The phasing model used was the 2.4Å refined structure of mutase from E coli (PDB code: 1I8T). For M. tuberculosis mutase, there are four monomers in the asymmetric unit with a Matthews’s coefficient of 4.0 Å3 Da−1 (70% water). K. pneumoniae mutase solution has one monomer in the asymmetric unit with the crystallographic two fold creating a dimer, the Matthews coefficient of 2.3 Å3 Da−1 (47% water).
A random subset of data (5%) was omitted from all refinement calculations to provide an assessment of the refinement process. All structures were refined using REFMAC 5 29. During early stages of refinement and model building FAD/FADH− was left out from the model and placed when the difference electron density was clear. Water molecules were added to complete the refinements. Residues Met1, Gln2, Pro3, Asp137, Ala138, Gln139, Asn140, Leu141, Gln396, Asp397, Gly398, and Ala399 were not experimentally located in the M. tuberculosis enzyme. Residues Met1, Lys2 were not experimentally located in the K. pneumonia enzyme. Table 1 summarises the refinement statistics. We report the FAD structure as 2.2Å resolution based on the low completeness of the final shell (2.2-2.0Å) but all data were included in the refinement. Structures and experimental data have been deposited. Crystal decay prevented a complete data set being collected on the M. tuberculosis structure, although with four molecules in the asymmetric unit, we feel the structure is robust.
Model building
The construction of the models was carried out using the program O30. For the construction of the Galf covalent adduct with FADH− buckled at the re-face, the FADH− from thioredoxin reductase (1C10) 17 was modelled in the active site of K. pneumoniae mutase. For construction of the same adduct with a si-face buckled FADH− the experimental structure from K. pneumoniae was used (current study).
Acknowledgements
We would like to thank M. Dorward for technical assistance. CW is Canada Research Chair whose work is supported by the Canadian Bacterial Diseases Network (NCE program). JHN is a BBSRC career development fellow. This work is supported by the BBSRC through a grant to JHN. We thank the reviewers for their valuable criticism of the manuscript which has much improved it.
References
- 1.Neuhauser MM, Weinstein RA, Rydman R, Danziger LH, Karam G, Quinn JP. Antibiotic resistance among gram-negative bacilli in US intensive care units - Implications for fluoroquinolone use. Jama-Journal of the American Medical Association. 2003;289:885–888. doi: 10.1001/jama.289.7.885. [DOI] [PubMed] [Google Scholar]
- 2.Hanberger H, Garcia-Rodriguez JA, Gobernado M, Goossens H, Nilsson LE, Struelens MJ. Antibiotic susceptibility among aerobic gram-negative bacilli in intensive care units in 5 European countries. Jama-Journal of the American Medical Association. 1999;281:67–71. doi: 10.1001/jama.281.1.67. [DOI] [PubMed] [Google Scholar]
- 3.Besra GS, Khoo KH, McNeil MR, Dell A, Morris HR, Brennan PJ. A new interpretation of the structure of the mycolyl-arabinogalactan complex of Mycobacterium tuberculosis as revealed through characterization of oligoglycosylalditol fragments by fast-atom bombardment mass spectrometry and 1H nuclear magnetic resonance spectroscopy. Biochemistry. 1995;34:4257–66. doi: 10.1021/bi00013a015. [DOI] [PubMed] [Google Scholar]
- 4.Pan F, Jackson M, Ma Y, McNeil M. Cell wall core galactofuran synthesis is essential for growth of mycobacteria. J Bacteriol. 2001;183:3991–8. doi: 10.1128/JB.183.13.3991-3998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knirel’ Iu A, Kochetkov NK. Structure of lipopolysaccharides from gram-negative bacteria. III. Structure of O-specific polysaccharides. Biokhimiia. 1994;59:1784–851. [PubMed] [Google Scholar]
- 6.Nassau PM, Martin SL, Brown RE, Weston A, Monsey D, McNeil MR, Duncan K. Galactofuranose biosynthesis in Escherichia coli K-12: identification and cloning of UDP-galactopyranose mutase. J Bacteriol. 1996;178:1047–52. doi: 10.1128/jb.178.4.1047-1052.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koplin R, Brisson JR, Whitfield C. UDP-galactofuranose precursor required for formation of the lipopolysaccharide O antigen of Klebsiella pneumoniae serotype O1 is synthesized by the product of the rfbDKPO1 gene. J Biol Chem. 1997;272:4121–8. doi: 10.1074/jbc.272.7.4121. [DOI] [PubMed] [Google Scholar]
- 8.Barlow JN, Girvin ME, Blanchard JS. Positional isotope exchange catalyzed by UDP-galactopyranose mutase. Journal of the American Chemical Society. 1999;121:6968–6969. [Google Scholar]
- 9.Zhang Q, Liu H. Mechanistic investigation of UDP-galactopyranose mutase from Escherichia coli using 2- and 3-fluorinated UDP-galactofuranose as probes. J Am Chem Soc. 2001;123:6756–66. doi: 10.1021/ja010473l. [DOI] [PubMed] [Google Scholar]
- 10.Sanders DA, Staines AG, McMahon SA, McNeil MR, Whitfield C, Naismith JH. UDP-galactopyranose mutase has a novel structure and mechanism. Nat Struct Biol. 2001;8:858–63. doi: 10.1038/nsb1001-858. [DOI] [PubMed] [Google Scholar]
- 11.Fullerton SW, Daff S, Sanders DA, Ingledew WJ, Whitfield C, Chapman SK, Naismith JH. Potentiometric analysis of UDP-galactopyranose mutase: stabilization of the flavosemiquinone by substrate. Biochemistry. 2003;42:2104–9. doi: 10.1021/bi027077f. [DOI] [PubMed] [Google Scholar]
- 12.Soltero-Higgin M, Carlson EE, Gruber TD, Kiessling LL. A unique catalytic mechanism for UDP-galactopyranose mutase. Nat Struct Mol Biol. 2004;11:539–43. doi: 10.1038/nsmb772. [DOI] [PubMed] [Google Scholar]
- 13.Huang ZH, Zhang QB, Liu HW. Reconstitution of UDP-galactopyranose mutase with 1-deaza-FAD and 5-deaza-FAD: analysis and mechanistic implications. Bioorganic Chemistry. 2003;31:494–502. doi: 10.1016/j.bioorg.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Burley SK, Petsko GA. Amino-Aromatic Interactions in Proteins. Febs Letters. 1986;203:139–143. doi: 10.1016/0014-5793(86)80730-x. [DOI] [PubMed] [Google Scholar]
- 15.Fraaije MW, Mattevi A. Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem Sci. 2000;25:126–32. doi: 10.1016/s0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
- 16.Zheng YJ, Ornstein RL. A theoretical study of the structures of flavin in different oxidation and protonation states. Journal of the American Chemical Society. 1996;118:9402–9408. [Google Scholar]
- 17.Lennon BW, Williams CH, Jr., Ludwig ML. Crystal structure of reduced thioredoxin reductase from Escherichia coli: structural flexibility in the isoalloxazine ring of the flavin adenine dinucleotide cofactor. Protein Sci. 1999;8:2366–79. doi: 10.1110/ps.8.11.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Binda C, Coda A, Angelini R, Federico R, Ascenzi P, Mattevi A. A 30-angstrom-long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase. Structure Fold Des. 1999;7:265–76. doi: 10.1016/s0969-2126(99)80037-9. [DOI] [PubMed] [Google Scholar]
- 19.Yue QK, Kass IJ, Sampson NS, Vrielink A. Crystal structure determination of cholesterol oxidase from Streptomyces and structural characterization of key active site mutants. Biochemistry. 1999;38:4277–86. doi: 10.1021/bi982497j. [DOI] [PubMed] [Google Scholar]
- 20.Haynes CA, Koder RL, Miller AF, Rodgers DW. Structures of nitroreductase in three states: effects of inhibitor binding and reduction. J Biol Chem. 2002;277:11513–20. doi: 10.1074/jbc.M111334200. [DOI] [PubMed] [Google Scholar]
- 21.Field RA, Naismith JH. Structural and mechanistic basis of bacterial sugar nucleotide-modifying enzymes. Biochemistry. 2003;42:7637–7647. doi: 10.1021/bi0345079. [DOI] [PubMed] [Google Scholar]
- 22.Beis K, Allard ST, Hegeman AD, Murshudov G, Philp D, Naismith JH. The structure of NADH in the enzyme dTDP-d-glucose dehydratase (RmlB) J Am Chem Soc. 2003;125:11872–8. doi: 10.1021/ja035796r. [DOI] [PubMed] [Google Scholar]
- 23.Zheng Y-J, Ornstein R. A Theoretical Study of the Structures of Flavin in Different Oxidation and Protonation States. J Am Chem Soc. 1996;118:9402–9408. [Google Scholar]
- 24.Eisenreich W, Kemter K, Bacher A, Mulrooney SB, Williams CH, Jr., Muller F. 13C-, 15N- and 31P-NMR studies of oxidized and reduced low molecular mass thioredoxin reductase and some mutant proteins. Eur J Biochem. 2004;271:1437–52. doi: 10.1111/j.1432-1033.2004.04043.x. [DOI] [PubMed] [Google Scholar]
- 25.Weston A, Stern RJ, Lee RE, Nassau PM, Monsey D, Martin SL, Scherman MS, Besra GS, Duncan K, McNeil MR. Biosynthetic origin of mycobacterial cell wall galactofuranosyl residues. Tuber Lung Dis. 1997;78:123–31. doi: 10.1016/s0962-8479(98)80005-1. [DOI] [PubMed] [Google Scholar]
- 26.McMahon SA, Leonard GA, Buchanan LV, Giraud MF, Naismith JH. Initiating a crystallographic study of UDP-galactopyranose mutase from Escherichia coli. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 2):399–402. doi: 10.1107/s0907444998010877. [DOI] [PubMed] [Google Scholar]
- 27.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 and ESF-EAMCB Newsletter on Protein Crystallography. 1992;26 [Google Scholar]
- 28.Evans PR. “SCALA”. Joint CCP4 and ESF-EAMCB Newsletter on Protein Crystallography. 1997;33:22–24. [Google Scholar]
- 29.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 1):247–55. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 30.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 31.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, Fagan P, Marvin J, Padilla D, Ravichandran V, Schneider B, Thanki N, Weissig H, Westbrook JD, Zardecki C. The Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:548–558. [Google Scholar]