Abstract
Recently a fluorination enzyme was identified and isolated from Streptomyces cattleya, as the first committed step on the metabolic pathway to the fluorinated metabolites, fluoroacetate and 4-fluorothreonine. This enzyme, 5′-fluoro-5′-deoxy adenosine synthetase (FDAS), has been shown to catalyze C-F bond formation by nucleophilic attack of fluoride ion to S-adenosyl-L-methionine (SAM) with the concomitant displacement of L-methionine to generate 5′-fluoro-5′-deoxy adenosine (5′-FDA). Although the structures of FDAS bound to both SAM and products have been solved, the molecular mechanism remained to be elucidated. We now report site directed mutagenesis studies, structural analyses and isothermal calorimetry (ITC) experiments. The data establish the key residues required for catalysis and the order of substrate binding. Fluoride ion is not readily distinguished from water by protein X-ray crystallography, however using chloride ion (also a substrate) with mutants of low activity has enabled the halide ion to be located in non-productive co-complexes with SAH and SAM. The kinetic data suggest the positively charged sulfur of SAM is a key requirement in stabilizing the transition state. We propose a molecular mechanism for FDAS in which fluoride weakly associates with the enzyme exchanging two water molecules for protein ligation. The binding of SAM expels remaining water associated with fluoride ion and traps the ion in a pocket positioned to react with SAM, generating L-methionine and 5′-FDA. L-SAM then dissociates from the enzyme followed by 5′-FDA.
Introduction
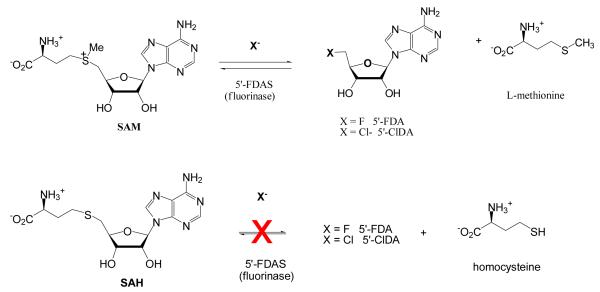
Biological halogenation has received considerable attention with recent and significant progress made in understanding the nature and mechanism of several enzymatic chlorination processes1-4. Although fluorine is the most abundant of the halogens in the earth’s crust, its biochemistry is very limited and only a few natural products containing fluorine have been reported. 5, 6 Zechel, Withers, and co-workers7, 8 first demonstrated enzymatic C-F bond formation in mutant β-glucosidase enzymes. Although an adventitious fluorination reaction of a mutant enzyme, this example demonstrated that fluoride ion can be desolvated on a protein7, 8. More recently the enzyme, 5′-fluoro-5′-deoxy synthetase (FDAS), was identified from the bacterium Streptomyces cattleya 9 an enzyme which catalyzes the reaction between S-adenosyl-L-methionine (SAM) and F− to form L-methionine and 5′-fluoro-5′-deoxyadenosine (5′-FDA) (Figure 1). The kcat of this reaction is slow and equilibrium for the reaction lies with products illustrating that the formation of the C-F bond is thermodynamically favored. The enzyme has been crystallized and its structure determined 10, 11 and it has also been shown to work with Cl− as well as F− ion 12. Interestingly there is no sequence match between FDAS and genomes from the plant kingdom. Several plants have been reported to accumulate high concentrations of fluoroacetate and in some cases fluorocitrate and ω-fluoro-oleic acid 5, 6 and this raises the possibility that biology may have evolved more than one catalytic strategy for F− incorporation.
Figure 1.
The reactions of F− and Cl− catalyzed by FDAS. SAH, the demethylated analogue of SAM, is an inhibitor of FDAS.
The structure of FDAS, complexed with SAM, shows it to be a novel fold 11. Complexes with 5′-FDA and L-methionine 11, 2′-deoxy-adenosine 13 and 5′-chloro-5′-deoxyadenosine 12 have subsequently been reported. These studies confirm the location of the components of SAM and have been used to predict a potential halogen binding site. Chemical labeling experiments show that fluoride incorporation proceeds with an inversion of the stereochemistry at the C5′ of SAM 14, experimentally supporting an SN2 process which requires at least partial desolvation of F−. The inferred location of the halide ion is consistent with an SN2 mechanism and would provide hydrogen bonds to the anion which could stabilize the desolvated F-. Experimentally, stripping F− of its last two remaining water molecules requires significantly more than half of the overall desolvation energy 15, suggesting a mechanism that leaves at least two hydrogen bonds to F− would be favored. Two recent theoretical studies have begun to reveal possible mechanisms 16, 17. QM/MM calculations 16 indicate that in solution the activation energy for the uncatalyzed process is approximately 100 kJ mol−1 and that the enzyme accelerates the reaction by between 106 and 108. These calculations proposed that the hydrogen of the hydroxyl of T80 adopts a different position from that in the ground state and coupled to slight adjustments in ligand and protein position now makes a crucial hydrogen bond to F− promoting its desolvation 16. A theoretical study in the gas phase indicated that only partial desolvation of the F− ion was required to create a potent nucleophile 17. The study highlighted the positive charge on SAM as an important factor in stabilizing the partially desolvated species 17. We present new experimental data that inform these key questions and identify the molecular mechanism of fluoride incorporation by FDAS.
Results
Binding order: In the reverse direction
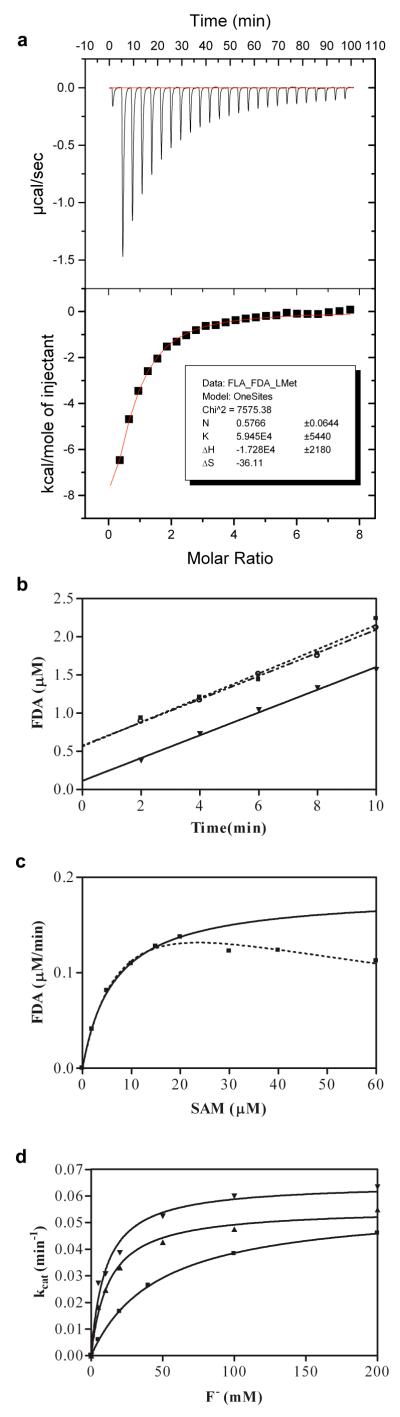
The binding affinities of SAM, 5′-FDA, L-methionine and the substrate analogues S-adenosyl homocysteine (SAH) and adenosine were measured by isothermal titration calorimetry (ITC) (Table 1 and Supporting Information Figure S1). The substrate SAM has the lowest affinity for the enzyme of the compounds studied, whereas SAH, an inhibitor of the enzyme, has the highest binding affinity and is 40 times more tightly bound than SAM. The weakness of SAM binding is reflected in the lower stoichiometry of binding. The binding of fluoride to the protein (either as apo or SAH complex) is too weak to measure by this technique. The product 5′-FDA (and its analogue adenosine) binds strongly to the enzyme suggesting product release may be slow. We do not detect L-methionine binding to apo-FDAS indicating weak binding (Ka < 1 ×103 M−1). However when FDAS is pre-incubated with 5′-FDA, L-methionine shows a very clear binding curve and Ka of 5.9×104 M−1(Figure 2a). Because measuring the thermodynamics of binding can be complicated by enzyme turn over, a control experiment with FDAS pre-incubated with adenosine, which does not turn over, was carried out. This gives an essentially identical result for L-methionine binding.
Table 1.
The binding affinities of ligands toward apo FDAS by isothermal titration calorimetry
| ligands | Stoichiometr y (N) |
Ka (app) (×105 M−1) |
ΔH (kcal/mol) |
S (cal/mol) |
|---|---|---|---|---|
| SAM | 0.56 ± 0.01 | 8.0 ± 0.5 | −25 ± 0.4 | −58 |
| SAH | 0.72 ± 0.01 | 302 ± 74 | −40 ± 0.5 | −98 |
| FDA | 1.16 ± 0.01 | 45.5 ± 6.5 | −14 ± 0.1 | −17 |
| Ade | 0.67 ± 0.01 | 12.2 ± 1.2 | −20 ± 0.4 | −40 |
| 3′-deoxy-Ade | 0.37 ± 0.02 | 4.5 ± 0.6 | −16 ± 0.9 | −27 |
| L-MetFDA* | 0.58 ± 0.07 | 0.59 ± 0.05 | −17 ± 2 | −36 |
| L-MetAde* | 0.48 ± 0.11 | 0.57 ± 0.07 | −13 ± 3 | −22 |
The binding affinity of L-MetFDA was obtained by titrating 1mM L-methionine into 0.08 mM FDA and 0.02 mM apo FDAS mixture, while the affinity of L-MetAde was obtained by titrating 1mM L-methionine into 0.024 mM Adenosine and 0.02 mM apo FDAS mixture
Figure 2.
Binding order of reactants (a) ITC determination of the binding affinity of L-methionine to FDAS. L-Methionine (1mM) was titrated into FDAS (0.02 mM) which had been pre-incubated with 5′-FDA (0.08 mM). (b) Active site titration of FDAS complexes: [FDAS◻F−] (◻), [FDAS◻SAM] (◻) and [FDAS] (◻), in which the titrations were initiated by adding the last reaction component in each case (e.g. SAM, F−, or SAM and F− ) in 20mM sodium phosphate buffer (pH 7.8). All titrations were performed at constant FDAS (2 μM) and F− (100mM) respectively with SAM (50 μM.) (c) The steady state kinetic analysis of apo FDAS and F− (200mM) towards SAM was calculated with (continuous line) and without (dotted line) substrate inhibition from the Michaelis-Menten equation. (d) The steady state kinetic analysis of apo FDAS towards [F−] at [SAM] of 20μM (◻), 60μM (◻) and 200μM (◻). The continuous lines describe the fitting of the initial velocity against [F−] to the Michaelis-Menten equation.
Binding order: In the forward direction
FDAS was pre-mixed with water, SAM or F− to obtain three starting complexes: [FDAS], [FDAS SAM] and [FDAS F−]. The remaining reactant(s) was then added to titrate the number of active sites (burst size) (Figure 2b)18. The slope of the graph plotting 5′-FDA production against time reflects the reaction velocity at steady state, a rate independent of the order of the addition of the reactants. However the burst size, the intercept on the Y-axis, varies with the order of addition. When 50 μM SAM was mixed with FDAS and the reaction initiated with 100 mM F− addition, the burst size corresponds to 0.1 μM of active sites. Conversely, if 100 mM F− was pre-mixed with FDAS and the reaction initiated by the addition of 50 μM SAM, the burst represented 0.6 μM active sites. Adding both substrates simultaneously gave a (larger) burst size similar to the experiment where F− was pre-incubated with protein and SAM added. Similar results were obtained using 100 μM SAM (Supporting Information Figure S2).
Site directed mutagenesis and steady state kinetic analysis of FDAS
The mutants T80A, T80S, S158A, S158G, F156A, F156V, D16S, D16A and D16N were constructed, expressed in E. coli and purified to homogeneity. The near UV circular dichroism (CD) spectrum (260 – 320 nm) of each mutant was recorded and confirms that there are no gross conformational changes (Supporting Information Figure S3). Mutants were assessed in a two-step manner: firstly their turnovers were quantified at a fixed substrate concentration compared to the wild type (Table 2). Wild type FDAS and mutants T80S, T80A, S158A and S158G were selected for further detailed analysis. At SAM concentrations greater than 20 μM, FDAS displays very clear substrate inhibition (Figure 2c). The measured Km for F− is sensitive to SAM concentrations (Figure 2d), at 20 μM SAM the Km(F−) 10 ± 2 μM, at 60 μM SAM, the Km (F−) has increased to 16 ± 3 μM and at 300 μM SAM Km (F−) is 47 ± 2 mM. The measured kcat remains within error the same, 0.06 min−1. SAM is an apparent “competitive” inhibitor of F− ion binding. Kinetic parameters of F− for the mutants were evaluated holding the SAM concentration at 20 μM; 3 - 4 times it’s Km and thus are valid only for comparison of trends amongst the mutants.
Table 2.
Analysis of mutants
| % activity | SAM (200mM F-) | F- (20μM SAM) | |||
|---|---|---|---|---|---|
| kcat (min−1) | Km (μM) | kcat (min−1) | Km (mM) | ||
| Native | 100 | 0.07±0.001 | 6.5±0.3 | 0.06±0.003 | 10.2±1.9 |
| T80A* | 15 | 0.005±0.0002 | 3.0±0.4 | 0.004±0.0002 | 36.8±5.0 |
| T80S | 95 | 0.06±0.002 | 4.7±0.4 | 0.06±0.002 | 18.4±2.8 |
| S158A | 38 | 0.009±0.0006 | 9.2±1.2 | 0.006±0.0003 | 5.4±1.3 |
| S158G* | 8 | 0.0007±0.00003 | 0.8±0.5 | 0.0008±0.00003 | 1.36±0.22 |
| F156A | 3 | N.D. | N.D. | N.D. | N.D. |
| F156V | 25 | N.D. | N.D. | N.D. | N.D. |
| D16A^ | 0 | N.D. | N.D. | N.D. | N.D. |
| D16S^ | 0 | N.D. | N.D. | N.D. | N.D. |
| D16N | 3 | N.D. | N.D. | N.D. | N.D. |
These mutants contain residual SAM and the precise values are suspect. ^ No 5′-FDA found after 1.5 hours reaction by HPLC.
The kcat/Km of 104 min−1M−1, confirms that FDAS is a slow enzyme 9. Wild type, S158A, T80A and T80S all have similar Km’s for SAM. ITC measurements of SAM binding to the mutants are consistent (Supporting Information Table S1) in showing no real change in SAM binding. The exception is the weakly active S158G which purifies with bound SAM which cannot be entirely removed (removal relies on turnover) and thus may give misleading results. T80A and to a lesser extent T80S have increased Km’s for F− whereas S158G and S158A appear to bind F− more tightly than the native enzyme. T80S retains a similar turnover number to wild type, T80A and S158A are down by almost a factor of 10 and S158G is reduced by almost 100-fold.
Chemical reactivity of SAM vs. SAH
SAH is not a substrate of FDAS in the forward direction and it inhibits the reaction of SAM with F−. We have investigated the reverse reaction to exclude the possibility that this was in fact due the equilibrium position. Accordingly we have measured (by HPLC) SAH production from homocysteine and 5′-FDA (Figure 1) by the enzyme. There is no detectable production of SAH nor is any detected after incubation of 5-ClDA and homocysteine with the enzyme. Incubation of Cl− and SAH with the enzyme does not produce 5′-ClDA.
The structure of apo FDAS
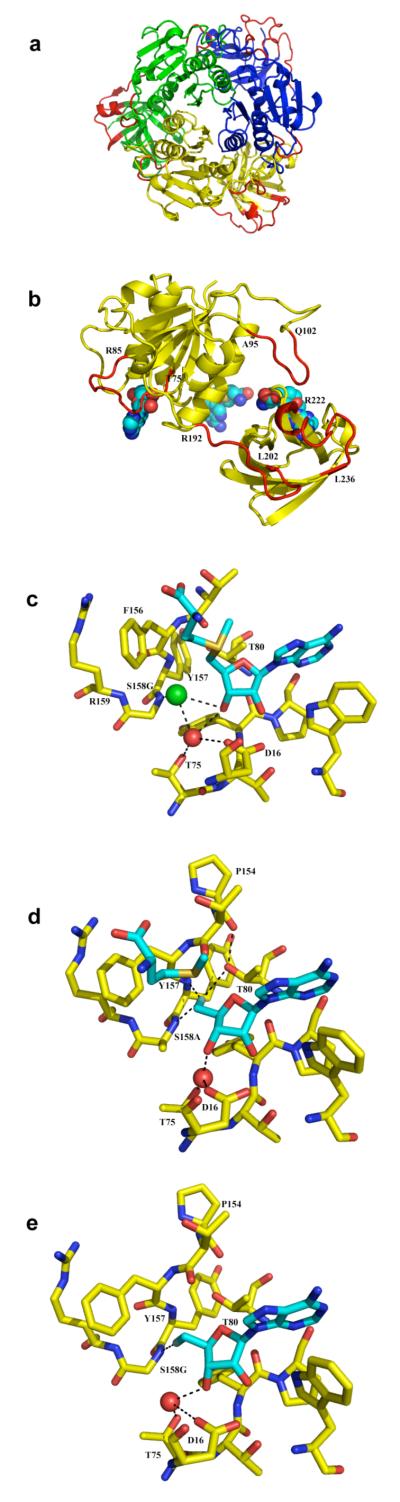
Apo FDAS was prepared as previously described 13 by incubating over-expressed protein with adenosine deaminase to remove and bound adenosine contamination. Crystallographic details are summarized in the Supporting Information Table S2. The folding of apo FDAS is essentially identical to the previous description of the wild type structure9. The structure is free of any bound adenosine component, but now four water molecules and a glycerol, which was used as a cryoprotectant, fill the active sites. The absence of a bound substrate does not significantly change the loop structure at the active site and as a result the active site remains shielded from the bulk solvent. Analysis of the thermal factors suggests that four loops (T75-R85, A95-Q102, R192-L202, R222-L236) in particular have high B-factors (indicative of distortion) relative to the average value (Figure 3a and 3b). The loop at R192-L202 is the linker between the two domains and the loop at R222-L236 is remote from the active site and both loops are flexible in all structures. Loop T75-R85 forms the binding site for the adenine ring and ribose of SAM and 5-FDA analogues. The loop at A95-Q102 has been highlighted as a key feature of the FDAS enzyme and is involved in formation of the L-methionine binding site, although it does not bind to L-methionine directly.
Figure 3.
Structures mapping the molecular mechanism of FDAS. (a) The trimeric structure of the apo-FDAS with chains differentially colored showing four flexible loops highlighted in red.(b) A detailed view of the flexible loops of the A subunit of apo FDAS, in which two SAM molecules are shown as they are found in the trimeric wild type FDAS binary complex (1rqp). This highlights the relationship between the loops and the related SAM. SAM is shown in spheres and colored by element with carbon-cyan, oxygen-red, nitrogen-blue and sulfur-yellow. (c) The binding site for the chloride ion determined from the S158G SAM Cl− ternary complex, the same location is observed in the SAH complex. This Cl− binding site is predicted to be immediately adjacent to the halide ion binding site at transition state. SAM and neighboring residues are shown in stick format and colored by element as Figure 3 (b) with the exception that the residue carbons are colored in yellow. The Cl− and a water molecule in the active site are indicated by spheres with Cl− green and water red. The possible hydrogen bonds of Cl− and the water molecule are indicated by dashed lines. (d) 5′-FDA bound to S158A mutant shows a conformation in which the C5′-fluorine appears to make a hydrogen bond to T80. A water molecule at the similar position to that in Figure 3 (c) is shown as a red sphere. (e) The binary complex of S158G with FDA viewed as Figure 3 (d) for comparison of the different orientations of the C-F bond between the (d) and (e) structures. Here the orientation of the fluoromethyl is essentially identical to the wild type FDAS ternary complex with FDA and L-Met, and the wild type FDAS binary complex with FDA and S158G binary complex structure with FDA. The conserved water molecule in this mutant, highlighted in figures 3 (c) and (d) could fulfill the positioning and hydrogen bonding role of the side chain hydroxyl group of S158.
The halogen anion binding site
The very weakly active mutant S158G was crystallized in the presence of SAM (2 mM) and separately with S-adenosylhomocysteine (SAH) (2 mM) in the presence of 200 mM Cl−. The data reduction and refinement statistics are summarized in Supporting Information Table S2. The presence of Cl− ion was identified by a strong peak in the Fo-Fc map (over 9 σ) in both the SAM and SAH structure (Supporting Information Figure S4d,f). A water molecule was first refined, but clear additional Fo-Fc density was observed (Supporting Information Figure S4e,g) and the water had an anomalously low thermal (B) factor. A Cl− ion refines with a B-factor close to the average for the structure (Supporting Information Figure S5d,e). Both structures are almost identical and for the purpose of discussion we focus on the A subunit of the SAM complex (Figure 3c). The Cl− ion is 3.0 Å from the backbone amide of S158G, the interaction predicted to be essential from the 5′-FDA complex. The Cl− is 3.1 Å from the O3 of SAM and 3.3 Å from C5′ of SAM and the angle between Cl−, C5 and SD of SAM is 129° (Figure 3c). In both structures, there is a water molecule 2.9 Å from away from the Cl− ion which occupies a similar position to the OG atom of S158 in the wild type structure (Figure 3c). The water molecule is held by a network of hydrogen bonds to O3 of SAM, the hydroxyl of T75 and the side chain of D16. The Cl− ion is 3.9 Å from the amides of R159 and 4.1 Å from Y157, although in both case the hydrogen is involved in inter protein hydrogen bonds and is unlikely to be available to contact the ion. The torsion angle of the C2-O2 and C3-O3 bonds in SAM is around 18° compared to 42° in the SAH structure. This difference results in the rupture of the D16 hydrogen bond to O3 in the SAH complex.
Product complexes
Incubation of S158A with F− and SAM results in the ternary complex with 5′-FDA and L-Met. Crystallographic statistics are shown in Supporting Information Table S2. The protein structure of S158A is essentially identical to that of the wild type while the bound products 5′-FDA and L-methionine adopt only slightly different conformations compared to those in the wild type ternary complex structure11. A water molecule mimics the hydroxyl of S158. The most significant change is that the fluoromethyl group of FDA has rotated (Figure 3d). As a result, the fluorine atom retains its hydrogen bond to the amide NH of the mutated residue of S158A (2.8 Å) but now makes a polar link (geometry inconsistent with a hydrogen bond) with the amide of Y157 (2.6 Å). In this new position the fluorine (approximately 1.5 Å from its location in the native enzyme) could make a hydrogen bond to the hydroxyl of T80 (3.2 Å). However T80 remains within hydrogen bonding distance to the carbonyl at P154. Since the hydrogen of T80 hydroxyl is not experimentally located at this resolution we cannot establish the hydrogen bonding arrangement.
Two new binary complex structures of FDAS (native and S158G) with FDA bound have been determined at 1.9 Å resolution. These complement the previous 2.7 Å resolution FDA and L-methionine native11 and S158A ternary complexes. In both binary complexes the loop K96-Q102 is disordered. In the S158G mutant binary complex (Figure 3e), the O3 and O2 atoms of the ribose ring move by 0.5 and 0.3 Å with respect to their counterparts in wild type binary complex. This perturbs the hydrogen bonding interactions and again a water molecule occupies a similar position to the hydroxyl of the original S158 residue (Figure 3e). In the S158G mutant the ribose has a C2-O2 and C3-O3 torsion angle of 6° relative to 10° in the wild type binary structure.
Discussion
Order of binding of reactants
The FDAS assay is too inefficient to directly determine the order of binding and for this reason indirect methods were used. For the reverse reaction (removal of fluoride) crystal structures and ITC analysis clearly establish that FDAS can bind 5′-FDA and 5′-ClDA 12 in both the absence and presence of L-methionine. More importantly, the data demonstrate that L-methionine binds to FDAS with a measurable affinity, only after 5′-FDA is bound. Once bound 5′-FDA helps to form the binding site for L-methionine by ordering loop T75-85. This forces an obligate order of binding on the enzyme. L-methionine binding orders the A95-Q102 loop creating the fully functional enzyme.
The deduction of the order of the forward reaction is more complicated as ITC measurements cannot measure binding of the halide ion. This is consistent with steady state kinetic measurements which show a very weak binding of fluoride (Km for F− > 5mM). The slow rate of the forward reaction lends itself to measurement of the burst size resulting from the pre-steady state. The burst size equates to the number of functional active sites at zero time. The burst size is clearly smaller when the FDAS is pre-incubated with SAM. In the steady state, increasing SAM concentration increases the Km of fluoride ion. Taken together this data demonstrate that when SAM binds it inhibits the subsequent binding of fluoride ion. This is consistent with our structural analysis which suggests that once SAM is bound, the active site is inaccessible to solvent (and hence to anion binding), thus the Cl− ion has to bind first or at the same time. Our data do not rule out F− and SAM binding simultaneously, however the concentration of ion pairs is likely to be small and ion pair binding seems unlikely.
Location of the halogen binding site and key active site residues
The position of the halide ion had only been inferred from the structure of the 5′-FDA complex 9. Using the weakly active mutant S158G, chloride ion was located experimentally at a binding site in complexes with SAH and SAM. The trapped chloride is located at a 50° deviation from the ideal 180° angle between Cl−, C5′ and S required for an SN2 reaction trajectory. The structures show that the Cl− ion coordinates a water molecule. This water is present in only the S158 mutant structures and is close to the position of the original side chain hydroxyl of S158 in the native enzyme. We propose that this water functionally mimics the S158 hydroxyl in ligating fluoride ion and that S158 is in fact a key ligand to the halide. The residual activity of the S158A and S158G mutants results from the water molecule which compensates for the loss of the serine hydroxyl. The Cl− bound structures reveal a bifurcated hydrogen bond involving S158, ribose O3 and the halide ion. The 3-deoxy analogue of adenosine binds with a 3 fold reduction of affinity (Table 1) suggesting O3 is not critical in recognition but does not rule out a role in catalysis. ITC shows that D16 is important for substrate binding. Even for the relatively conservative D16N mutation, SAM does not bind with measurable affinity, we conclude that the correct positioning of the ribose ring by D16 is critical to enzyme activity.
The F156A mutation severely attenuates activity whereas the larger F156V mutant retains much of the wild type activity. This residue provides the rear of the hydrophobic pocket necessary to assist desolvation of fluoride ion. The T80S mutant retains its activity whereas the activity of T80A is reduced more than 10 fold, consistent with an important role for T80. In the S158A mutant, the fluoromethyl group of 5′-FDA rotates towards T80 to come within hydrogen bonding distance supporting the possibility of such a hydrogen bond occurring in the transition state after a conformational change. In order to more fully probe the role of T80 we determined the structure of the T80A mutant with SAH. The loop containing T80A become disordered and significantly perturbs Y77 and Y157, both of which are at the halide binding site. The inactivity of T80A would seem more likely to result from significant conformational changes in the halide binding site than just the strategic loss of a hydrogen bond at the transition state, where residual water could play a compensating role.
Transition state stabilization
We observe that the ribose ring conformation varies significantly between complexes. Previous QM/MM calculations discount ring strain as a significant component of activating SAM for SN2 substitution 16. The structures of the SAH and SAM complexes reveal no structural reason for the much weaker binding of SAM, the common atoms of the two ligands superimpose. At the resolution of our studies we see no significant change in the protein structure in response to the methyl group. After the extra bulk of the methyl group, the positive charge on the sulfur atom is the obvious difference between the molecules. The desolvation energy of charged SAM could be higher than neutral SAH accounting for their different affinities, but binding affinity does not explain the lack of reactivity. Homocysteine does not react with either 5′-ClDA or 5′-FDA establishing that the transition state involving SAH is not sufficiently stabilized by the enzyme. The presumed transition state involves a partially negatively charged desolvated F− ion (Figure 4) where desolvation of F− is the principle kinetic barrier to reaction. The methyl group of SAM and / or the positive charge on sulfur must be essential in lowering this energy barrier. It is difficult to see why the steric bulk of the methyl group per se would influence the stability of the transition state therefore this leaves the positive charge on the sulfur atom as crucial to transition state stabilization, a finding echoed by gas phase theoretical studies 17.
Figure 4.
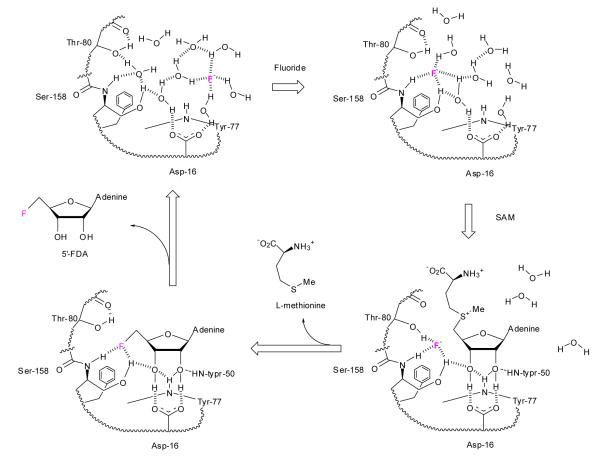
Cartoon illustration of the sequential mechanism of FDAS. (a) Shows an open binding site with a low affinity for hydrated fluoride ion; (b) illustrates the position of fluoride ion diffusing into the developing S158 binding site with hydrogen bonding to water being replaced by contacts to the residues of the protein. (c) Specific SAM binding, with contacts (not shown) between the protein and the adenine base and L-methionine residue forces out water and traps fluoride ion in a reactive position on the protein bound to S158 and possibly T80 and stabilized by charge neutralization (between F− and S+). (d). A nucleophilic reaction occurs and L-methionine is released followed by 5′-FDA.
Conclusions: A molecular mechanism for FDAS
Combining these findings allows us to propose a detailed molecular mechanism for FDAS. The solvated halide ion binds to an unoccupied pocket with a low affinity and exchanges water molecules for hydrogen bonds to polar groups of the protein (the amide NH and the side chain OH of S158). The sequential exchange of water for protein ligands is a low energy process and is likely to be compensated by entropy gains as water molecules are displaced. Upon binding SAM the remaining water molecules become dissociated from F− ion as it becomes wedged into the hydrophobic binding pocket. This desolvation is driven by SAM binding and in particular the high affinity of the adenosine ring for the enzyme. With fluoride ion trapped and partially desolvated it now acts as a nucleophile, although T80 may assist it in this stabilization it does not seem crucial. The transition state is significantly stabilized by the positive charge on SAM in an example of substrate assisted catalysis. The SN2 attack of F− then occurs at the C5′ of SAM with displacement of L-methionine. L-Methionine is then released by the enzyme after a conformational change in the A95-Q102 loop and then in the final step, 5′-FDA is released with a conformational change of loop T75-R85 (Figure 4).
METHODS
Site directed mutagenesis, protein expression and protein purification
The fla ORF was subcloned into pHisTev expression vector (courtesy from Dr H. Liu, Centre for Bimolecular Sciences, University of St. Andrews) the original pET28-flA expression vector by using the following two primers, 5′-GTAGCCCCATGGCTGCCAAC-3′ and 5′-GCTAGTTATTGCTCAGCGGTG-3′. The resultant plasmid pFLA_HT was used for the expression of FDAS and the site-directed mutagenesis as the template. The QuickChange II XL site-directed mutagenesis kit (Stratagene) was used to introduce D16A/S/N, T80A/S, F156A/V, S158A/G mutations into the new constructed FDAS expression vector pFLA_HT, using the following primers as listed in Supporting Information Table S3. The complete coding regions of the mutated plasmids were sequenced to ascertain the fidelity of the mutant construct. The pFLA_HT vectors encoding the wild type or each of the mutants were used to transform Escherichia coli C43 (DE3) cells. The overexpressed FDAS or the mutants were purified as previously described9. Although native and active mutants contain Ade, the weakly active mutants contain SAM. We suggest this is because the active mutants and native turnover any bound SAM during expression or purification and the resultant 5-ClDA is displaced by Ade. The bound adenosine is converted to inosine by adenosine deaminase (Type X, Sigma-Aldrich Co), which does not bind and can be dialyzed away to obtain the apo form of enzyme. For the mutants with very low FDAS activity we incubated for prolonged periods with F− to decompose as much SAM as possible and then removed 5-FDA using the deaminase. The purified proteins was verified by SDS-PAGE and the quantified by UV absorbance at 280 nm with the extinction coefficient ε = 35900 M−1*cm−1, which is calculated by the ProtParam tool on the ExPASy Proteomics Server (www.expasy.org).
Isothermal calorimetric analysis on the ligand binding of FDAS and its mutants
ITC experiments were carried out using a VP-ITC device (microCal, Northampton, MA). All solutions were degassed. Protein samples were dialyzed against 20mM sodium phosphate buffer (pH 7.8) and all the ligands were dissolved in the above buffer as well and quantified by the adenosine extinction coefficient of 15400 M−1*cm−1at 260 nm if it contains the adenosine moiety.. The ligand solutions were injected at 25 °C into the sample cell containing ~1.4 ml of FDAS or the mutants with the concentration around 20 μM. Each titration consisted of a first 1 μl injection followed by up to 25 subsequent 10 μl injections of the ligands with 10-fold molar concentration of the protein concentration. Calorimetric data were analyzed using MicroCal ORIGIN software using a single binding site model.
Discontinuous assay of FDAS
Enzymatic activity was assayed at 25 °C by monitoring 5′-FDA production using analytic HPLC (Varian 9012 UV-Vis detector at 260 nm and 9050 solvent delivery module). 0.5 mg/ml apo FDAS or the mutants were incubated with the indicated SAM and KF in 20mM sodium phosphate buffer (pH 7.8), in a final volume of 0.7 ml. 100 μl sample was taken out at every 30 minutes and mixed with 0.5 ml ice cold ethanol to terminate the reaction by precipitating the protein and the fluoride ion. Before further treatment, the samples were kept on dry ice. Precipitated material was removed by centrifugation (13000 rpm 10 min 4 °C) before the reaction mixtures were dried in a Savant vacuum concentrator (Stratech Scientific, U.K.). The residues were dispersed in 100 μl KH2PO4 (50 mM) and acetonitrile (95:5 v/v) and the samples were clarified by centrifugation (13000 rpm 10min 4 °C) before 20 μl portions were subjected to analytic HPLC, which was done as previously described. For the aim of active site titration and the steady state kinetic studies , 2-20 μM apo FDAS or the mutants were assayed and 5 reaction samples were taken at every 2 min (or as indicated in Supporting Information Figure S6-S17) to monitor the amount of 5′-FDA product during the time course. The velocity of each reaction was reflected by the slope of linear fitting of the FDA production against the time, while the intercept reflects the burst size. The steady state kinetic parameters were obtained by least square fitting the initial velocity against the substrate concentrations according to the Mechaelis-Menten equation.
Crystallography of the FDAS and the mutants
The crystals of FDAS or mutants complexes with SAM or FDA or FDA and L-methionine bound were grown as previously described under 0.1 M citrate-phosphate buffer (pH 4.6), 18-28 % (w/v) PEG 1000 and 0.2 M Li2SO4. The crystals of ternary complex of S158G with SAM/SAH and Cl− bound were obtained with the presence of 2mM SAM or SAH under 0.1 M citrate-phosphate buffer (pH 4.6), 18-28 % PEG 1000 and 0.2 M LiCl. The crystals were cryoprotected by increasing the concentration of PEG 1000 towards 40%. The apo form of FDAS was crystallized under a different condition including 0.2 M sodium thiocyanate, 20 % (w/v) PEG 3350 and cryoprotected by complement with 20 % (v/v) glycerol. Crystals were mounted in Hampton Research cryoloops and flash-frozen at 100 K. Data were collected from a single crystal in a single pass using the 0.25 ° oscillations by using ESRF ID14 or Daresbury SRS station 9.6 or in house RigakuMicromax-007 rotating anode with Osmic mirrors on a Rigaku RaxisIV++ image plate. Data were indexed and integrated using MOSFLM 19 and scaled using SCALA 20, 21. The data of the complexes of FDAS or the mutants were reduced in the same space group C2221 as the previously described structure of FDAS complex with SAM (1rqp). The structures were solved by rigid body refinement 22 and the reflections for Rfree calculation were copied from the structure factor of 1rqp. The apo form of FDAS was indexed and integrated in P212121 and the structure was solved by molecular replacement with MOLREP 23 using FDAS complex structure (1rqp) without SAM as the search model. The further rebuild and refinement were carried out until R free failed to decrease using Coot 24 and Refmac5 22. The data processing and refinement statistics are summarized in Supporting Information Table S2. Unbiased Fo-Fc electron density maps for all ligands are shown in supporting materials (Supporting Information Figure S4).
Supplementary Material
Acknowledgements
We thank the funding from BBSRC BBS/B/04579. We thank Drs Changjiang Dong and Hai Deng for assistance and helpful discussions.
Footnotes
Supporting Information Available: Table S1 showing the binding affinities of ligands towards FDAS and mutants by isothermal titration calorimetry, Table S2 of crystallography statistics for the structures, Table S3 containing the Primers used to generate the mutations in FDAS, Figure S1 showing ITC determination of the binding affinity of ligands towards FDAS or the mutants, Figure S2 of active site titration of FDAS complexes at 100 μM SAM, Figure S3 showing the near UV CD spectra of mutants, Figure S4 of the unbiased Fo-Fc map for all ligands in the related structures, Figure S5 of the 2Fo-Fc map for all ligands in the related structures, Figure S6-S17 showing the steady state kinetic analysis of wild type FDAS and the mutants as SAM or F− is the variable substrate.
References
- 1.Vaillancourt FH, Yeh E, Vosburg DA, Garneau-Tsodikova S, Walsh CT. Chem. Rev. 2006;106:3364–3378. doi: 10.1021/cr050313i. [DOI] [PubMed] [Google Scholar]
- 2.Dong C, Flecks S, Unversucht S, Haupt C, van Pee KH, Naismith JH. Science. 2005;309:2216–2219. doi: 10.1126/science.1116510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaillancourt FH, Yeh E, Vosburg DA, O’Connor SE, Walsh CT. Nature. 2005;436:1191–1194. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- 4.Yeh E, Blasiak LC, Koglin A, Drennan CL, Walsh CT. Biochemistry. 2007;46:1284–1292. doi: 10.1021/bi0621213. [DOI] [PubMed] [Google Scholar]
- 5.O’Hagan D, Deng H, Schaffrath C. Nat. Prod. Rep. 2004;21:773–784. doi: 10.1039/b415087m. [DOI] [PubMed] [Google Scholar]
- 6.O’Hagan D, Harper DB. J. Fluorine Chem. 1999;100:127–133. [Google Scholar]
- 7.Zechel DL, Reid SP, Nashiru O, Mayer C, Stoll D, Jakeman DL, Warren PAJ, Withers SG. J. Am. Chem. Soc. 2001;123:4350–4351. doi: 10.1021/ja005855q. [DOI] [PubMed] [Google Scholar]
- 8.Nashiru O, Zechel DL, Stoll D, Mohammadzadeh T, Warren RAJ, Withers SG. Angew. Chemie, Int. Ed. Engl. 2001;40:417–420. doi: 10.1002/1521-3773(20010119)40:2<417::AID-ANIE417>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 9.O’Hagan D, Schaffrath C, Cobb SL, Hamilton JTG, Murphy CD. Nature. 2002;416:279–279. doi: 10.1038/416279a. [DOI] [PubMed] [Google Scholar]
- 10.Dong CJ, Deng H, Dorward M, Schaffrath C, O’Hagan D, Naismith JH. Acta Cryst. Section D-Biol. Cryst. 2003;59:2292–2293. doi: 10.1107/s0907444903019826. [DOI] [PubMed] [Google Scholar]
- 11.Dong CJ, Huang FL, Deng H, Schaffrath C, Spencer JB, O’Hagan D, Naismith JH. Nature. 2004;427:561–565. doi: 10.1038/nature02280. [DOI] [PubMed] [Google Scholar]
- 12.Deng H, Cobb SL, McEwan AR, McGlinchey RP, Naismith JH, O’Hagan D, Robinson DA, Spencer JB. Angew Chem. Int. Ed. Engl. 2006;45:759–762. doi: 10.1002/anie.200503582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cobb SL, Deng H, McEwan AR, Naismith JH, O’Hagan D, Robinson DA. Org. Biomol. Chem. 2006;4:1458–1460. doi: 10.1039/b600574h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cadicamo CD, Courtieu J, Deng H, Meddour A, O’Hagan D. ChemBioChem. 2004;5:685–90. doi: 10.1002/cbic.200300839. [DOI] [PubMed] [Google Scholar]
- 15.Arshadi M, Yamdagni R, Kebarle P. J. Phys. Chem. 1970;74:1475–1482. [Google Scholar]
- 16.Senn HM, O’Hagan D, Thiel W. J. Am. Chem. Soc. 2005;127:13643–13655. doi: 10.1021/ja053875s. [DOI] [PubMed] [Google Scholar]
- 17.Vincent MA, Hillier IH. Chem. Comm. 2005:5902–5903. doi: 10.1039/b510477g. [DOI] [PubMed] [Google Scholar]
- 18.Fersht A. Enzyme structure and mechanism. second edition W. H. Freeman and Co; 1984. [Google Scholar]
- 19.Leslie AGW. Joint CCP4 and ESF-EAMCB Newsletter on Protein Crystallography. 1992;26:1–10. [Google Scholar]
- 20.Collaborative Computational Project Number 4, The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 21.Evans PR. SCALA. Joint CCP4 and ESF-EAMCB Newsletter on Protein Crystallography. 1997;33:22–24. [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. Acta Cryst. D. Biol Cryst. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.Vagin A, Teplyakov A. MOLREP: J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 24.Emsley P, Cowtan K. Coot. Acta Cryst. D Biol Cryst. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.