Flavin-dependent halogenases have been shown to play a major role in biological halogenation reactions. For halogenating activity, flavin-dependent halogenases require reduced FAD, which is formed from FAD and NADH by a second enzyme, a flavin reductase. Although in a number of cases, a flavin reductase gene is present in the biosynthetic gene cluster of the halometabolites, it is unclear whether the corresponding flavin reductases interact directly with the halogenases. At least in a number of cases, flavin reductases from different bacterial strains can be used in combination with halogenases.1, 2–5 For the tryptophan 7-halogenase PrnA from Pseudomonas fluorescens BL915 which catalyzes the first step in pyrrolnitrin biosynthesis6 it could be shown that even chemically reduced FAD is used by the halogenase in the halogenation reaction.7 Based on the three-dimensional structure of PrnA, it was postulated that flavin hydroperoxide is formed by the reaction of halogenase-bound reduced flavin with oxygen. This flavin hydroperoxide then reacts with chloride ion leading to the formation of hypochlorous acid, which is then guided along a tunnel about 10 Å long towards the substrate tryptophan (Figure 1). A lysine residue (K79) was suggested to hydrogen bond with hypochlorous acid and thus position it to react with tryptophan.8 Yeh et al. demonstrated chloramine formation by the reaction of HOCl with the ε-amino group of lysine,9 suggesting that chloramine rather than HOCl is the active agent. The importance of K79 is undisputed; exchange of K79 against an alanine residue leads to total loss of halogenating activity as demonstrated for PrnA8 and for the tryptophan 7-halogenase RebH from rebeccamycin biosynthesis.9 However, other factors must also be at work, since chlorination of tryptophan cannot be accomplished by chloramine (or HOCl) in solution.10, 11 Chloramine is a weaker halogenating agent than HOCl,12 and according to quantum mechanical calculations, N-chloramine formation reduces the electrophilicity ofthe chlorine species; in other words, the charge Q(Cl) is reduced to −0.07 compared to Q(Cl)=+0.017 in free HOCl.
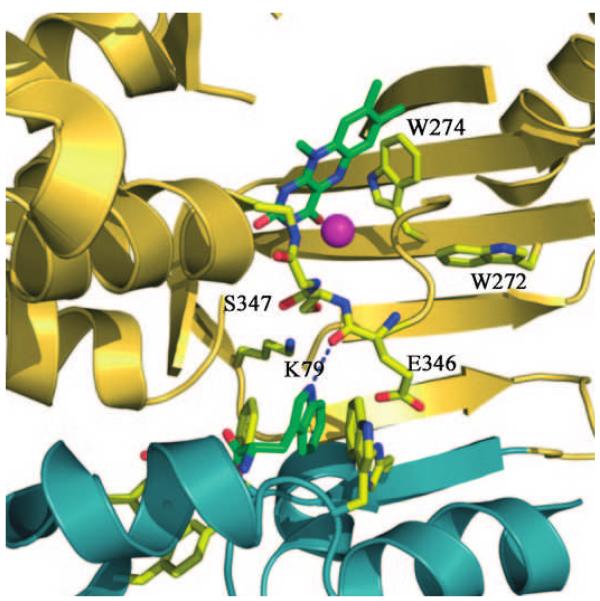
Figure 1.
Active site of PrnA: The amino acids residues that were exchanged are indicated by the corresponding numbers. The chloride ion bound in the active site is shown as a sphere near the isoalloxazine ring of FAD. The indole NH is hydrogen bonded to the oxygen of the peptide bond between E346 and S347.
In the active site, glutamate 346 (E346) is positioned across the tunnel from K79, and the positioning of the substrate tryptophan is supported by a hydrogen bond between the NH group of the indole ring and the peptide bond oxygen between E346 and serine 347 (S347) (Figure 1). Evidence that E346 could be involved in the catalytic cycle was the observation that E346Q is two orders of magnitude less active.8 Whereas K79 is absolutely conserved in all the flavin-dependent halogenases known so far, E346 and S347 are conserved only in flavin-dependent tryptophan halogenases and they are not present in halogenases acting on substrates with a phenol or pyrrole ring. Dong et al. suggested that E346 is required for the abstraction of a proton from and stabilization of the Wheland intermediate (Scheme 1).8 However, this generalized role is inconsistent with the lack of conservation of E346 in other non-trytophan halogenases. The important chemical difference is that substrates with a phenolic ring or a pyrrole ring are susceptible to chlorination by HOCl or chloramines, whereas tryptophan is not. Instead, the oxindole derivative of tryptophan is formed with HOCl.11
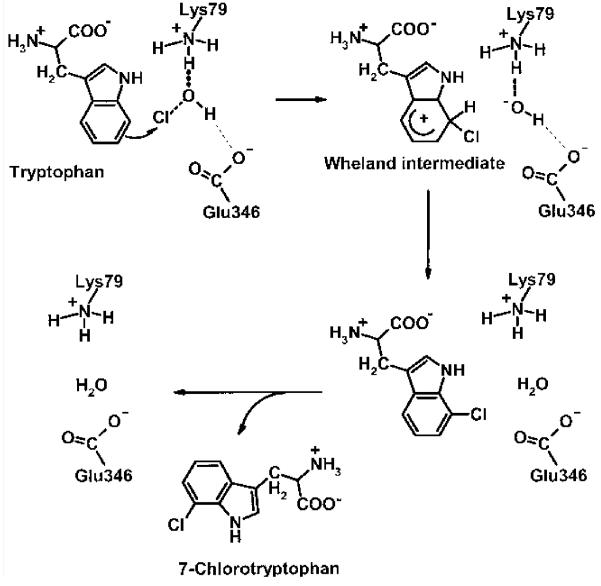
Scheme 1.
Mechanism proposed for the regioselective chlorination of tryptophan showing the involvement of K79 and E346 to enhance the electrophilicity of the chlorine species and for its correct positioning for incorporation into the 7-position. The strong interaction of the HOCl oxygen with the protonated e-amino group of K79 is indicated by a bold dashed line, and the weak interaction of the HOCl hydrogen with the carboxylate group of E346 is indicated by a thin dashed line.
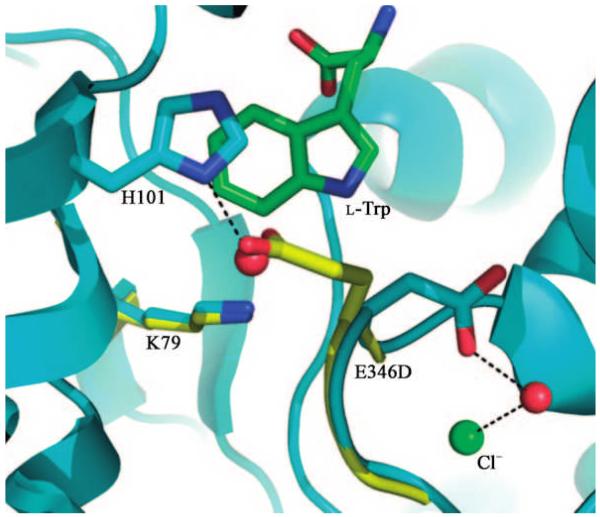
To analyze the importance of E346 for the halogenase reaction, it was exchanged against an aspartate residue by site-directed mutagenesis. X-ray structural analysis showed that the three-dimensional structure of the E346D mutant is essentially identical to that of the native enzyme, with the exception of the mutation site. The aspartate residue has a different orientation to the glutamate. An additional water molecule was found in the space created by loss of the glutamate. Crucially K79, the Cl– ion, tryptophan, and FAD are positioned identically as in the native enzyme (Figure 2). E346D, like K79A, is inactive, but we have shown that the mutation does not alter any other component of the reaction. A simple steric role in orienting substrates seems unlikely for three reasons. The additional water molecule in the mutant would achieve a similar effect, the tryptophan substrate location is unchanged, and the isosteric E346Q is almost inactive. These findings suggest that formation of chloramine (or HOCl) alone is not the sole key chemical step; rather we suggest that the spatial location of the negative charge is crucial to the mechanism. Since this residue is not conserved in all halogenases, it is crucial only for tryptophan halogenases. We suggest that the negative charge interacts with HOCl or chloramine, enhancing its electrophilicity. In doing so it would locate the chlorine species exactly at the 7-position of the indole ring system (Scheme 1).
Figure 2.
Active site of E346D mutant (C: green and cyan, O: red, N: blue). The native enzyme is shown in yellow. The mutation does not perturb K79, Cl−, tryptophan, or FAD. In the mutant structure the side chain of E346D has displaced a water molecule in comparison to the native structure.
Quantum chemical calculations suggest that a weak interaction of the HOCl hydrogen with the carboxylate group of glutamate would increase the charge of the chlorine species from +0.017 to +0.057. The corresponding interaction with the proton on the nitrogen of the chlorolysine is not possible since the separation in the native enzyme is over 5 Å. A strong interaction of the HOCl hydrogen with the carboxylate group would lead to an abstraction of the proton from HOCl, decreasing the charge of the chlorine species to –0.013. The E346D structure reveals that the shorter aspartate side chain is both too far away and in the wrong position to interact with the proton of HOCl.
An interaction between the chlorine of HOCl with the carboxylate group of E346 would be unfavorable (supported by quantum mechanical calculations). In an unconstrained environment, the chlorine atom would move away from the carboxylate group. However, in the confined volume of the active site, it cannot be ruled out that the negative charge of the carboxylate could serve to polarize the Cl atom of either HOCl or N-chlorolysine, but this seems rather unlikely.
S347, which is located between K79 and the flavin cofactor, might be the first amino acid residue to interact with HOCl and thus help to draw the HOCl into the tunnel and pass it along to K79. However, in contrast to K79, S347 is not absolutely essential to the activity, since an exchange against an alanine residue does not completely abolish halogenating activity. The enzyme was still active, although Vmax was reduced to about 25 % compared to the wild-type enzyme, whereas Km was not changed significantly (Table 1).
Table 1.
Km and Vmax values of native PrnA and of some mutants as obtained from Hanes plots.
| Enzyme | Km[μm] | Vmax [pmol min–1] |
|---|---|---|
| PrnA | 17.1±4.4 | 43±4 |
| PrnA_S347A | 10.0±3.4[a] | 17±1.2[a] |
| PrnA_W272F | 13.3±3.0 | 67±2 |
| PrnA_W274F | 22.3±2.9 | 45±3 |
| PrnA_W272F_W274F | 17.5±1.5 | 88±8 |
| PrnA_W272A | 29.6±2.1 | 74±5 |
| PrnA_W274A | no activity | no activity |
| PrnA_W272A_W274A | no enzyme | no enzyme |
Owing to the low activity of these mutants, the kinetic values were difficult to determine accurately.
The amino acid residues W272 and W274 are also absolutely conserved in all flavin-dependent halogenases detected to date.13 It was suggested by Dong et al. that these two large amino acid side chains prevent an organic substrate from binding in the vicinity of the isoalloxazine ring and thus prevent the enzyme from functioning as a monooxygenase.8 When these two amino acids were exchanged individually or together against phenylalanine residues, no change in halogenating activity could be detected. The Km values determined for the W272 to F272 and for the W274 to F274, and the W272F_W274F double mutant were determined to be 13.3, 22.3, and 17.5 μm, respectively. These values are almost identical to the value of 17.1 μm determined for the native enzyme (Table 1).
Exchange of W272 against the small amino acid alanine does also not result in any significant change of Km and Vmax (Table 1). When W274, which is closer to the isoalloxazine ring, was exchanged against an alanine residue, the enzyme was totally inactive. The conservation of the WXW motif in all halogenases is therefore puzzling as we can identify no in vivo role for W272, and only by radical mutation is any effect seen for W274. Interestingly, PltD, an enzyme originally suggested to function as a halogenase in pyoluteorin biosynthesis, contains a glycine residue at position 274.14 However, the involvement of PltD in the halogenation step of pyoluteorin biosynthesis has been excluded.2 Thus elucidation of the actual role of PltD in pyoluteorin biosynthesis could help to elucidate the function of the WxW motif in flavin-dependent halogenases.
For binding and correct positioning of the substrate tryptophan in the active site, the hydrogen bond between the indole NH and the oxygen atom of the peptide bond between E346 and S347 plays an important role but cannot be probed by mutagenesis. The amino acids that have been exchanged in our investigation described here are all conserved in tryptophan halogenases with different regioselectivities.1, 3, 6 It will be of great interest to see what the three-dimensional structures of a tryptophan 5- and a tryptophan 6-halogenase reveal. It can be assumed that the mechanism of these enzymes will be identical to that of PrnA, but the differences in structure could help in more clearly defining the mechanism. In particular, orientation of the substrate in the active site must be changed to correctly present the position to be halogenated to the chlorine species.
The results presented in this paper show that not only K79, but also E346, is absolutely essential for the chlorinating activity of tryptophan halogenases. This suggests that formation of chloramine by the reaction of K79 with hypochlorous acid alone is not sufficient to achieve chlorination of tryptophan. Since covalent attachment of chlorine in the active site was detected only in the absence of substrate,9 it could be that in the presence of substrate, free hypochlorous acid is re-formed as a result of the reversibility of the reaction of lysine with hypochlorous acid. Thus HOCl could interact with both amino acid residues, K79 and E346, such that the HOCl hydrogen interacts with the carboxylate group of E346. This would also explain why the E346Q mutant shows reduced activity,8 since the acid amide group could act as a donor as well as an acceptor for the formation of a hydrogen bond.
Experimental
The PrnAE346D mutant was constructed by using Strategene’s QuickChange Site-Directed Mutagenesis Kit following the manufacturer’s instruction using pET28b-prnA as the template.8 The mutagenic sites are underlined in the primers. The two primers used were 5′-TGCTTTCTGGAGCCCCTGGACTCGACG-3′ (sense) and 5′-TCGAGTCCAGGGGCTCCAGAAAGCACG-3′ (antisense).
The PrnA_S347A, PrnA_W272F, PrnA_W272A, PrnA_W274F, PrnA_W272A, and the corresponding double mutants were constructed by overlap extension polymerase chain reaction using pUC-prnA as the template.15 The mutagenic sites are underlined in the primers and restriction sites are in bold. The primers used for PrnA_S347A were: primer a: 5′-CTGCAGGATCCCTAAGGAGATTCCACCATGAAC-3′ (sense), primer b: 5′-GTAGATGAAGTAGATCCCCGTGGCTTCCAGG-3′ (antisense), primer c: 5′-CCTGGAAGCCACGGGGATCTACTTCATCTAC-3′ (sense), primer d: 5′-GGATCCTCCAAGCTTCGTTCCACTACAGGC-3′ (antisense).
The PrnA_W272F, PrnA_W272A, PrnA_W274F, PrnA_W272A, and the corresponding double mutants were created by amplifying an XhoI fragment of prnA containing the mutation and ligation to the rest of the prnA gene. The primers used were:
For PrnA_W272F: primer a: 5′-CTGCAGGATCCCTCAGGAGATTCCACCATGAACAAGCCGATCAAGAATATCGTCATC-3′ (sense), primer b: 5′-ATGGCTCGAGAAGACGTAGCCGCTGCCGAACCGGCCCAGCATCGGAATCTTCCAGGTGAATCC-3′ (antisense).
For PrnA_W274F: primer a: 5′-CTGCAGGATCCCTCAGGAGATTCCACCATGAACAAGCCGATCAAGAATATCGTCATC-3′ (sense), primer b: 5′-ATGGCTCGAGAAGACGTAGCCGCTGCCGAACCGGCCCAGCATCGGAATCTTGAAGGTCCATCC-3′ (antisense).
For PrnA_W272F_W274F: primer a: 5′-CTGCAGGATCCCTCAGGAGATTCCACCATGAACAAGCCGATCAAGAATATCGTCATC-3′ (sense), primer b: 5′-ATGGCTCGAGAAGACGTAGCCGCTGCCGAACCGGCCCAGCATCGGAATCTTGAAGGTGAATCC-3′ (antisense).
For PrnA_W272A: primer a: 5′-CTGCAGGATCCCTCAGGAGATTCCACCATGAACAAGCCGATCAAGAATATCGTCATC-3′ (sense), primer b: 5′-ATGGCTCGAGAAGACGTAGCCGCTGCCGAACCGGCCCAGCATCGGAATCTTCCAGGTCGCTCCCGAGTT-3′ (antisense).
For PrnA_W274A: primer a: 5′-CTGCAGGATCCCTCAGGAGATTCCACCATGAACAAGCCGATCAAGAATATCGTCATC-3′ (sense), primer b: 5′-ATGGCTCGAGAAGACGTAGCCGCTGCCGAACCGGCCCAGCATCGGAATCTTCGCGGTCCATCCCGAGTT-3′ (antisense).
For the double-mutant PrnA_W272A_W274A: primer a: 5′-CTGCAGGATCCCTCAGGAGATTCCACCATGAACAAGCCGATCAAGAATATCGTCATC-3′ (sense), primer b: 5′-ATGGCTCGAGAAGACGTAGCCGCTGCCGAACCGGCCCAGCATCGGAATCTTCGCGGTCGCTCCCGAGTT-3′ (antisense).
All mutants were confirmed by DNA sequencing. The native and mutant proteins were produced in Pseudomonas fluorescens BL915 Δ ORF 1–4 with and without a His tag. His-tagged enzymes were purified using a Ni-chelating sepharose FF column and non-His-tagged enzymes were partially purified using a Q-sepharose FF column (Amersham Biosciences) as described by Keller et al.5 The activity of PrnA was determined according to the method described previously.5, 7 The reaction mixture was composed of 55 μL PrnA-containing protein solution, 10.8 mU FAD reductase, 10 μm FAD, 2.4 mm NADH, 12.5 mm MgCl2, 100 U catalase, and 0.6 mm tryptophan in a total volume of 200 μL in 10 mm potassium phosphate buffer, pH 7.2. After incubation at 30 °C for 30 min, the reaction was stopped and the assay mixture was analyzed by HPLC. For determination of Km values, tryptophan concentrations were varied between 5 and 50 μm, since higher substrate concentrations inhibit the reaction. However, especially in crude extracts, higher substrate concentrations were used, because these extracts contained other enzymes using tryptophan as a substrate. The mutant enzyme was crystallized, and structure data to 2.3 Å were recorded in analogy to the structural investigation of the wild-type enzyme.8 Structure and data have been deposited (PDB code 2jkc).
For an evaluation of the electrophilic character of HOCl, the Mulliken charges at the chlorine were calculated within the semiempirical AM1 model. The HOCl molecule was located between the protonated amino group of the lysine and carboxylate group of glutamate. The charge at the chlorine was then calculated as a function of the length of the hydrogen bond between the proton at the amino group of the lysine (R(NH···O)) and between the proton of HOCl and the oxygen of the carboxylate group of glutamate (R(OH···O)). The distance between the nitrogen of the protonated amino group and the carbon of the acid group (R(NC)) was kept fixed at 7.4 Å in agreement with the crystal structure.8
References
- 1.Zehner S, Bister B, Süssmuth RD, Méndez C, Salas JA, van Pée K-H. Chem. Biol. 2005;12:445–452. doi: 10.1016/j.chembiol.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Dorrestein PC, Yeh E, Garneau-Tsodikova S, Kelleher NL, Walsh CT. Proc. Natl. Acad. Sci. USA. 2005;102:13843–13848. doi: 10.1073/pnas.0506964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seibold C, Schnerr H, Rumpf J, Kunzendorf A, Hatscher C, Wage T, Ernyei AJ, Dong C, Naismith JH, van Pée K-H. Biocatal. Biotransform. 2006;24:401–408. [Google Scholar]
- 4.Lin S, Van Lanen SG, Shen B. J. Am. Chem. Soc. 2007;129:12432–12438. doi: 10.1021/ja072311g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keller S, Wage T, Hohaus K, Hölzer M, Eichhorn E, van Pée K-H. Angew. Chem. 2000;112:2380–2382. doi: 10.1002/1521-3773(20000703)39:13<2300::aid-anie2300>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 6.Hammer PE, Hill DS, Lam ST, van Pée K-H, Ligon JM. Appl. Environ. Microbiol. 1997;63:2147–2154. doi: 10.1128/aem.63.6.2147-2154.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Unversucht S, Hollmann F, Schmid A, van Pée K-H. Adv. Synth. Catal. 2005;347:1163–1167. [Google Scholar]
- 8.Dong C, Flecks S, Unversucht S, Haupt C, van Pée K-H, Naismith JH. Science. 2005;309:2216–2219. doi: 10.1126/science.1116510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeh E, Blasiak LC, Koglin A, Drennan CL, Walsh CT. Biochemistry. 2007;46:1284–1292. doi: 10.1021/bi0621213. [DOI] [PubMed] [Google Scholar]
- 10.Yeh E, Garneau S, Walsh CT. Proc. Natl. Acad. Sci. USA. 2005;102:3960–3965. doi: 10.1073/pnas.0500755102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrison M, Schonbaum GR. Annu. Rev. Biochem. 1976;45:861–867. doi: 10.1146/annurev.bi.45.070176.004241. [DOI] [PubMed] [Google Scholar]
- 12.Pattison DI, Davies MJ. Biochemistry. 2005;44:7378–7387. doi: 10.1021/bi0474665. [DOI] [PubMed] [Google Scholar]
- 13.Hornung A, Bertazzo M, Dziarnowski A, Schneider K, Welzel K, Wohlert S-E, Holzenkämpfer M, Nicholson GJ, Bechthold A, Süssmuth RD, Vente A, Pelzer S. ChemBioChem. 2007;8:757–766. doi: 10.1002/cbic.200600375. [DOI] [PubMed] [Google Scholar]
- 14.Nowak-Thomson B, Chaney N, Wing JS, Gould SJ, Loper JE. J. Bacteriol. 1999;181:2166–2174. doi: 10.1128/jb.181.7.2166-2174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higuchi R, Krummel B, Saiki RK. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]