Abstract
The intriguing deactivation of the cytochrome P450 (CYP) 2B4 enzyme induced by a mutation of a single residue, Phe429 to His, is explored by means of quantum mechanical/molecular mechanical (QM/MM) calculations of the O-OH bond activation of the (Fe3+OOH)− intermediate. It is found that the F429H mutant of CYP 2B4 undergoes homolytic, instead of heterolytic, O-OH bond cleavage. Thus, the mutant acquires the following characteristics of a heme oxygenase (HO) enzyme: (a) The donation by His429 of an additional NH---S H-bond to the cysteine ligand combined with the presence of the substrate retard the heterolytic cleavage and give rise to homolytic O-OH cleavage, and (b) the Thr302/water cluster orients the nascent OH• close to the meso position of the porphyrin, and ensures an efficient meso hydroxylation.
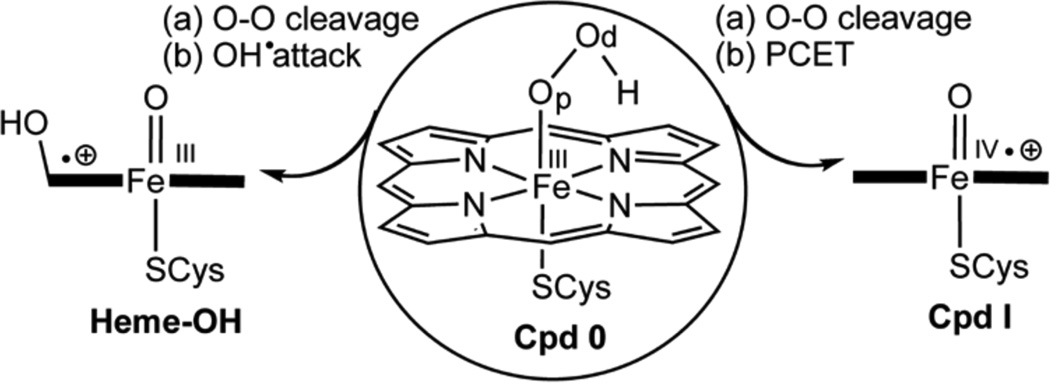
Cytochrome P450s (CYP) and heme oxygenase (HO) are O2 activating enzymes, which convert O2 to the ferrichydroperoxide species (Fe+3OOH−), so-called Compound 0 (Cpd 0), and then diverge in function. In the usual CYP function (see Scheme 1), Cpd 0 abstracts a proton, and splits off a water molecule to form the iron-oxo species, Compound I (Cpd I), which oxidizes a wide range of organic compounds and performs biosynthesis.1 By contrast, HO stops at Cpd 0, and uses it to activate the α-meso position of the porphyrin, which is subsequently degraded and frees iron, which is believed to be essential for iron homeostasis.2 In both cases, water molecules play a major role; in CYPs, they shuttle protons to Cpd 0,3a while in HO a water cluster safeguards Cpd 0 against conversion to Cpd I, and directs a homolytic O-OH cleavage and meso hydroxylation by the nascent OH radical.3b Considering the orthogonal functions of these enzymes, we report herein evidence that a single site mutation converts the CYP 2B4 enzyme to a hybrid CYP/HO enzyme.
Scheme 1.
The conversion of Cpd 0 to Cpd I vs. the HO option of generating a hydroxylated heme
Our interest in this problem was triggered by a recent experimental study,4 which reported that the cryoreduction of the oxyferrous complex in CYP 2B4 and its F429H mutant generated the same amounts of Cpd 0, and nevertheless, the F429H mutant (MT) exhibited only 4–5% product formation compared with the wild type (WT) enzyme.4a As such, a mutation of a single residue (Phe429 to His) that resides near the cysteinate ligand leads to deactivation of the enzyme, and apparently does so by disrupting the conversion of Cpd 0 to Cpd I. The position of the mutation near the cysteine ligand, and the fact that Phe429 is a conserved P450 residue that plays a role in the electron transfer pathway from the reductase to heme in CYP 2B4,4b makes the outcome of the mutation intriguing and requiring elucidation. Here, we studied the impact of this mutation using hybrid QM/MM calculations, which show that the mutation leads to HO activity of the F429H MT enzyme. As we shall demonstrate, this outcome is a result of a subtle interplay of electronic effects with changes caused by the substrate and the water structure in the active site.
The QM/MM study was carried out in our usual software combination,5a–c and followed protocols,1b,5d as in previous studies.3b,5e–f Details are given in the Supporting Information (SI), and those more specific to the paper are given here briefly. Since Cpd 0 was experimentally shown to exist in both the WT and MT enzymes, we started from the WT PDB 1SUO,6a replaced the inhibitor by the OOH group, added missing hydrogen atoms, took care of the protonation states of acidic and basic residues,3b,5d,f and followed with geometry optimization and molecular dynamic (MD) simulations. The F429H mutant was initially generated by replacing Phe429 by His, followed by minimization of the protein. The so resulting WT and MT enzymes were both found to generate Cpd I with minor quantitative differences in the barriers of heterolytic cleavage (SI, Table S1). We therefore extended the MD simulation to 60 ns and extracted from the MD trajectories the most highly sampled conformational basins,6b termed hereafter as, LWi and LMi, where L stands for long MD, W for the WT enzyme and M for the MT, i is the number of the basin. The WT enzyme prefers conformation LW1 and LW3. Similarly, the MT enzyme prefers mostly LM3 and secondly LM1, while the unstable LM2 basin spontaneously decays to LM3 (Figure S1). These most sampled conformations were used subsequently. Docking in the substrate (benzphetamine) and reoptimizing the enzymes led to expulsion of water from the pocket (see SI, Figures S2–S3).4a,7 In LW1,3 and LM1-3 the water molecules mostly remained near the distal OH group of Cpd 0, near the Glu301/Thr302 residues, which can shuttle protons to cause heterolytic cleavage.5e,8 Thus, as expected, the entrance of the substrate disfavors the generation of H2O2 (“uncoupling”) via proximal-O protonation, and prefers the protonation of the distal OH group.9 The uncoupling barriers did not give clue about the incapacitation of the MT enzyme. We therefore relegate the uncoupling processes to the SI (see Figure S1, Tables S1–S2), and focus herein on the O-OH cleavage, using the conformations that retained water molecules in the pocket, near the residues Glu301/Thr302, which shuttle protons. The QM/MM geometry optimization involves the QM system of the WT and MT enzymes, and the residues and waters within 6Å of the heme (~645 atoms, see SI). As before,5e,8 the QM subsystem was computed with B3LYP10a using a split valence LACVP10b basis set (B1) for geometry optimization, and a triple-ζ valence polarized (TZVP)10c basis set (B2) for energy correction.
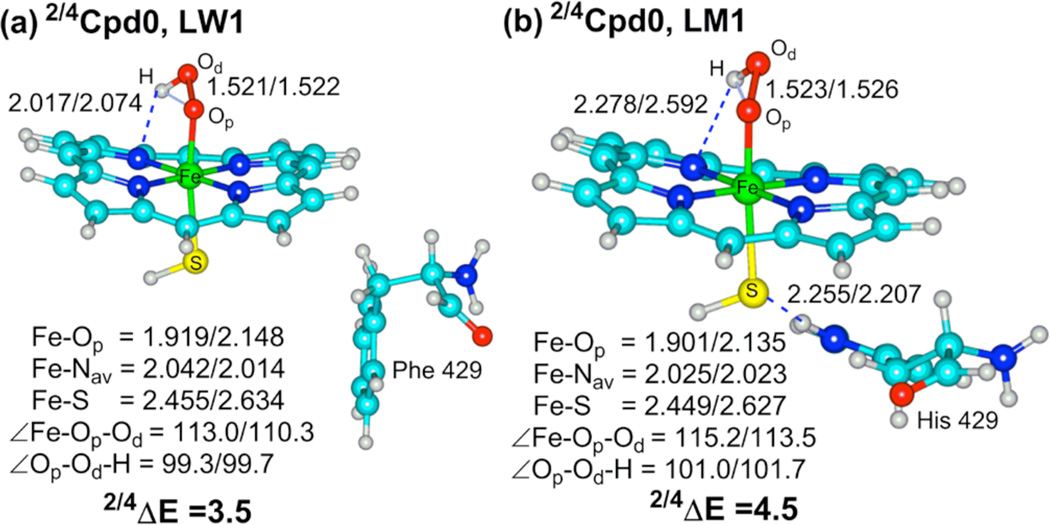
Figure 1 shows typical QM/MM optimized snapshots for Cpd 0 of the WT and MT enzymes. It is evident that the two species are similar in their geometries with the exception of a trans effect in the MT enzyme (shorter Fe-O distance), which is caused by the additional hydrogen-bonding (H-bonding)11a,b between the His429 with the sulfur ligand (Figure 1b) thereby weakening its “push effect”.11c In fact, the two Cpd 0 species have the same electronic structure and are similar in most respects, having doublet spin in the ground state and a quartet spin excited state.5e,f Thus, Cpd 0 gives no obvious clues about the mutation effect. Hence, we proceeded to consider the features of the O-OH cleavage mechanisms.
Figure 1.
B3LYP optimized geometries (Å and degrees), and doublet-quartet energy gap, 2,4ΔE (kcal/mol) of Cpd 0 in the conformations LW1 (a) and LM1 (b). Note the NH---S H-bond in (b).
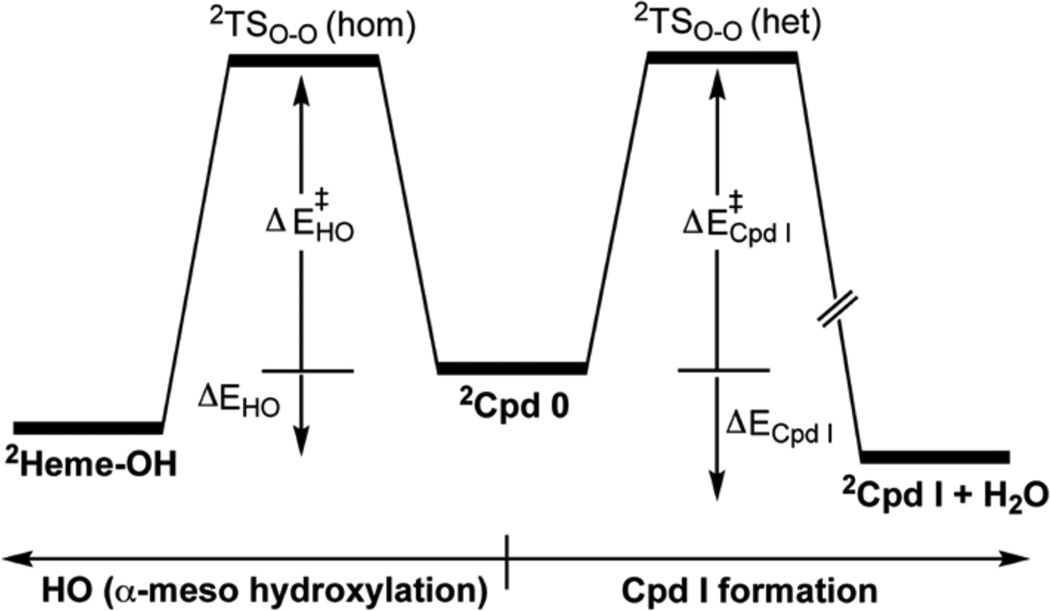
As before,5e,f,8a the O-OH cleavage process was scanned along the O-O distance and QM/MM optimization of all other parameters. The top of the scan was refined in a transition state (TS) search. Scheme 2 shows generic energy profiles of the two located processes for the LW and LM conformations; one pathway is Cpd I formation and the other one is HO activity leading to meso-hydroxylation of the heme.
Scheme 2.
Generic energy profiles starting from Cpd 0 (in its doublet ground state) and leading to Cpd I (to the right) and/or to meso-hydroxylation HO activity (to the left)
Table 1 shows the barriers and reaction energies for the two processes for the WT and MT enzyme conformations. The barriers are rather similar to ones computed before for Cpd I formation.5e,f,8 However, it is apparent that while the two conformations of the WT enzyme led to Cpd I formation, by contrast, in the MT enzyme two of the three conformations led to meso-hydroxylation, and the third one (LM2) to Cpd I. Comparison of the Cpd I formation barriers in the WT and MT enzymes shows that the process is not so much affected by the mutation. What is affected is the appearance of the HO mechanism in the MT enzyme in LM3 and LM1. Since LM2 spontaneously decays to LM3, it follows therefore that for the MT enzyme, the HO pathway competes favorably with Cpd I formation, while the WT enzyme favors Cpd I formation.
Table 1.
Barriers and Reaction Energies (kcal/mol) for the Conversion of 2Cpd 0 to 2Cpd I and/or to meso-Hydroxylated Heme.
| Conformation | ΔE‡Cpd I/ΔECpd I | ΔE‡HO/ΔEHO |
|---|---|---|
| LW1 | 9.7/−27.8 | N/Oa |
| LW3 | 16.0/−19.0 | N/Oa |
| LM1 | N/Oa | 14.9/−6.1 |
| LM3 | N/Oa | 17.4/−9.0 |
| LM2 | 13.3/−10.7 | N/Oa |
Not Observed
Importantly, the two processes compete along the same O-OH stretching coordinate. Thus, as before,5e,f,8a here too, we find that initially the O-OH bond breaks homolytically, and then the nascent hydroxyl radical accepts an electron from the heme and a proton, which is shuttled from the acidic residue Glu301 through the water chain and Thr302, thus leading to Cpd I in a proton-coupled electron transfer (PCET).5e,f When the PCET is interrupted by shortcutting the electron transfer, the nascent OH radical can attack the meso position of the heme before it diffuses away.3b Clearly, the F429H mutation affects the PCET process of the MT enzyme and prefers the homolytic cleavage pathway.2a,3a We shall now try to understand the root cause of this change.
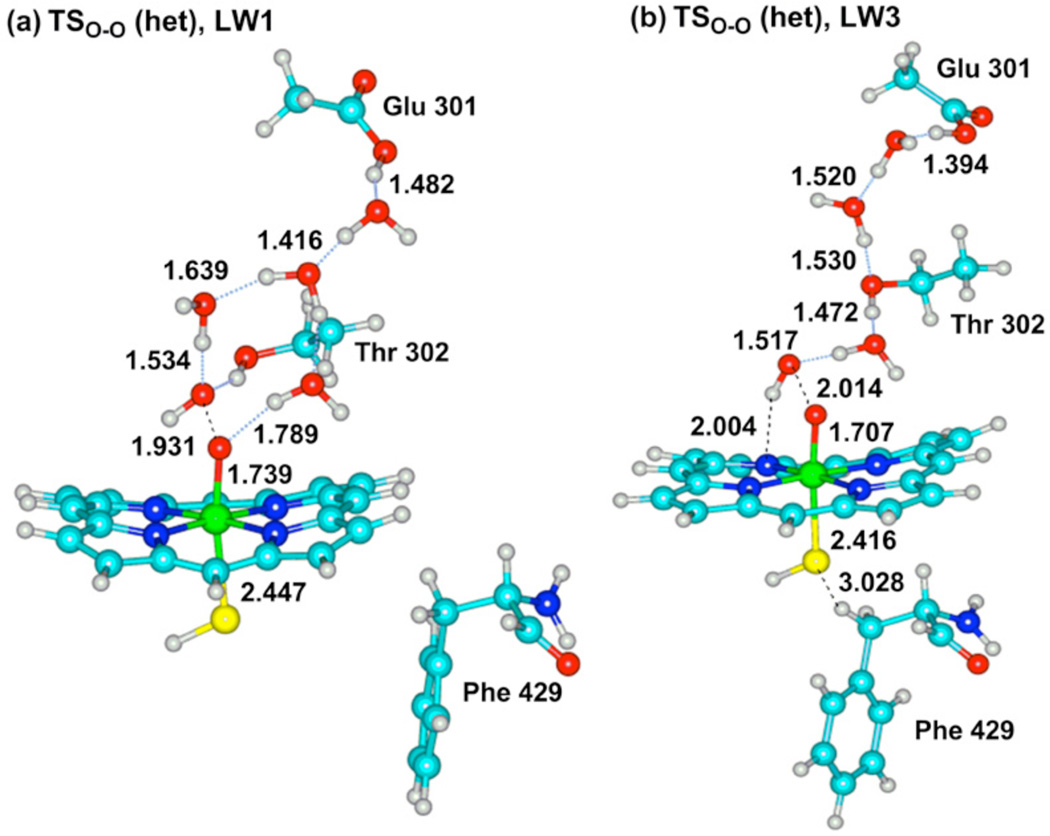
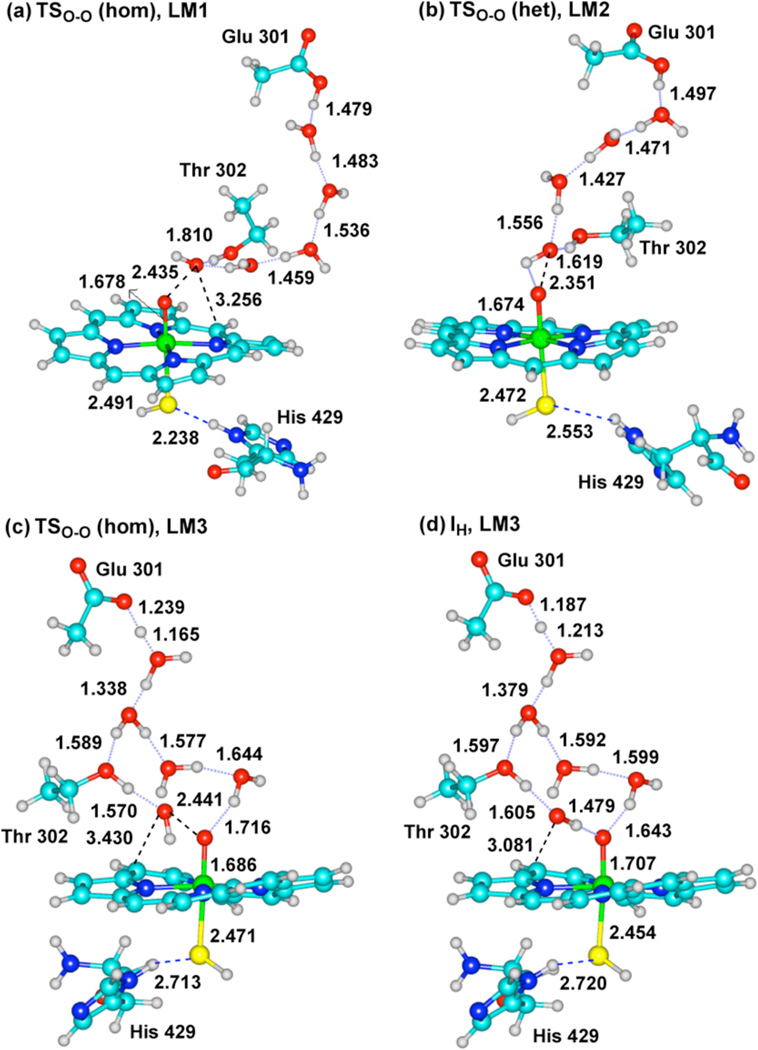
Figure 2 shows transition state structures, TSO-O(het), for the heterolytic process leading to Cpd I in the WT conformations LW1 and LW3. Figure 3 shows the two transition states for the homolytic process (TSO-O (hom)) leading to meso-hydroxylation, in the MT conformations LM1 and LM3, alongside TSO-O (het) for LM2. An additional structure, which is shown in Figure 3d and labeled as IH-LM3, is an intermediate point along the O-O scan and past the TSO-O (hom), just before collapse to the meso-hydroxylation product (a similar one, not shown here, is IH-LM1 in Figure S4).
Figure 2.
Key bond distances (Å) of the QM region in TSO-O (het) of the WT conformations LW1 (a) and LW3 (b).
Figure 3.
Key bond distances (Å) in TSO-O (hom) and TSO-O (het) of the MT conformations, LM1 (a) LM2 (b) LM3 (c), and the homolytic intermediate point IH of LM3 (d).
The heterolytic mechanism of the MT enzyme in Figure 3b is similar, with the exception that in the WT enzyme (Figure 2) the O-O distances, 1.931 and 2.041Å, in the TSO-O (het) structures, are significantly shorter than the same distance, 2.351Å, in the TSO-O (het) of the MT enzyme. This long O-O bond already shows the diminished push effect due to the H-bonds to thiolate. Indeed, the long O-O distance typifies also the two TSO-O (hom) structures in the MT enzyme (Figs. 3a and 3c). Moreover, in the IH-LM3 structure in Figure 3d, the nascent OH has spin density of −0.99, and is clearly a radical. Thus, it appears that the proton- and electron-transfer events in the MT enzyme lag behind the O-OH cleavage, so that the nascent OH radical can either attack the meso position of the heme (Figures 3a,c) or be captured by the electron + proton to form Cpd I. On the other hand, as the O-OH cleavage begins in the WT enzyme, the electron + proton capture the nascent OH• and release it as water, giving it no chance to attack the heme. It is this change that we ought to analyze.
Origins of the Mechanistic Switch in the MT enzyme
The appearance of a competitive HO pathway in the mutant originates from the interplay of factors that adversely affect the PCET mechanism: An electronic factor that delays the electron transfer to the nascent hydroxyl, and a structural factor that holds the OH• moiety close to the meso position of the heme. The structural factor is apparent from Figure 3, where it is seen that the Thr302 and the surrounding three water molecules hold the nascent OH• quite close to the heme by an array of H-bonding (e.g., in Fig. 3a the OH---Cmeso distance is 3.256Å), precisely as observed in the original HO enzyme, where an (H2O)6 cluster orients the OH radical, and forces it to stay close to the α-meso position of the porphyrin.2b,c,3b
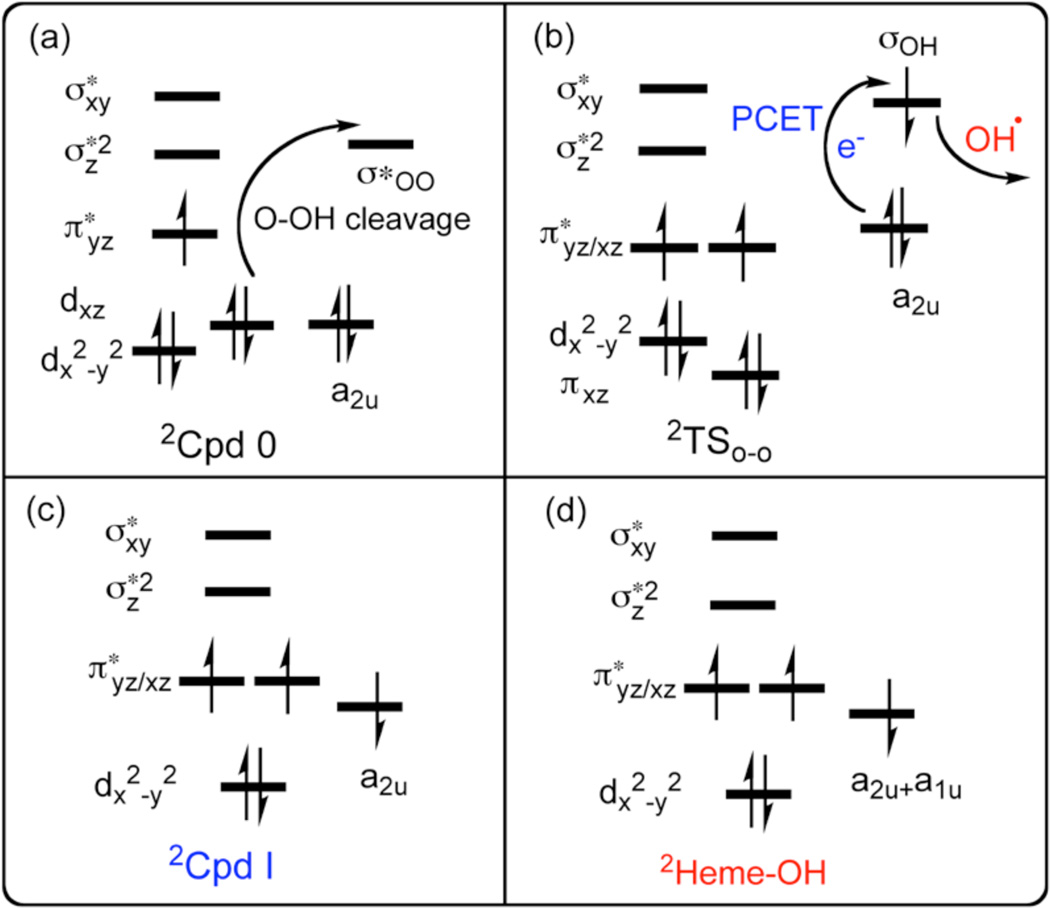
However, the structural factor by itself would be insufficient if there were no electronic effects that disrupted the PCET process, by preventing the electron transfer that converts the OH to OH− which easily abstracts a proton. This effect is elucidated with aid of the electron-shift diagram in Scheme 3. Thus, as the O-OH bond stretches (Scheme 3a), an electron initially in the iron dxz orbital shifts to the nascent OH moiety, while the dxz orbital becomes π*xz (FeO), thus forming a Compound II (Cpd II) ferryl species with a triplet configuration in the Fe=O bond (Scheme 3b). This is in fact what we find for all the TSOO species be they heterolytic or homolytic types; the electronic structure involves a triplet Cpd II antiferromagnetically coupled to an electron in a nascent OH species. What matters is, what transpires immediately afterwards. Thus, at this junction (Scheme 3b), if the a2u orbital of the porphyrin is sufficiently high-lying, an electron will shift to the orbital (σOH) of the nascent OH• which will simultaneously accept a proton from the nearby Glu/Thr-proton channel and form Cpd I and a water molecule as shown in Scheme 3c. By contrast, if the a2u orbital is low lying, and the protein electric field is improperly oriented, the electron will not shift to σOH, and the OH radical (in red) will attack the porphyrin and generate a hydroxylated porphyrin, as in Scheme 3d.3b What determines whether the OH• radical attacks the porphyrin or diffuses away, is the arrangement of the water cluster and Thr302 residue, which in this MT enzyme of 2B4 in the presence of the benzphetamine substrate, appears to assume a directing effect as does the protein2 in the HO enzyme.
Scheme 3.
Electronic shift diagrams during O-OH cleavage of 2Cpd 0 in (a), during PCET and OH expulsion past 2TSO-O in (b), and the electronic configuration of 2Cpd I (c) and 2Heme-OH products (d).
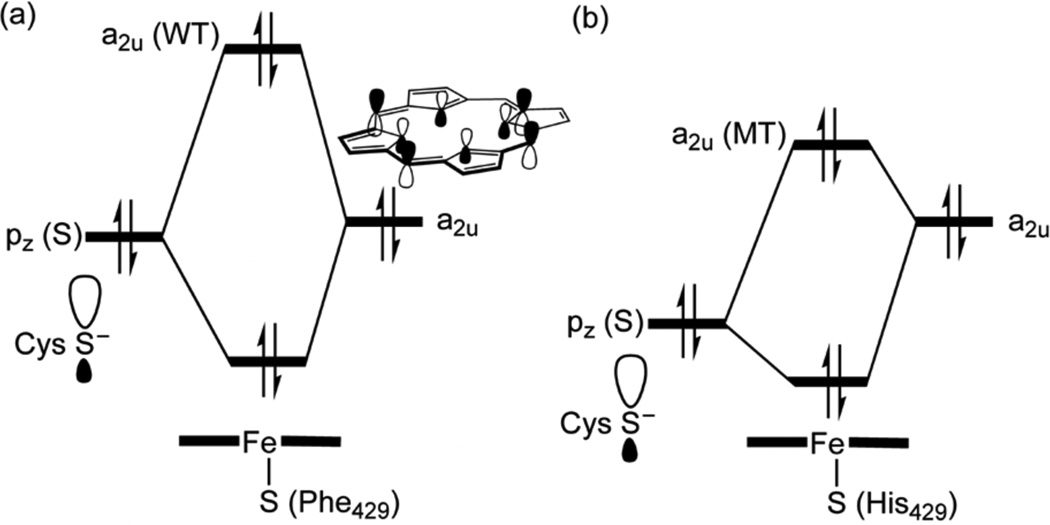
What are the factors that stabilize the a2u orbital in the MT enzyme and retard the electron transfer? As we have mentioned already, in Figure 1b, the His429 residue maintains an NH----S H-bond to the sulfur ligand. As shown in Scheme 4, this interaction lowers the 3pσ orbital of sulfur,1b,12 and minimizes its mixing with the a2u orbital, which thereby remains low in energy and hence it requires significant O-OH cleavage to transfer the requisite electron to the nascent OH•. Additionally, the entrance of the substrate, which expels and reorients water molecules, changes the direction of the intrinsic electric field that the MT protein exerts on the FeOOH moiety, such that the negative pole of the field is oriented now in LM3 and LM1 in the direction of the nascent OH radical (Figure S5–S6), thereby retarding the electron flow.13
Scheme 4.
Qualitative molecular orbital diagrams of mixing of the sulfur hybrid into the a2u orbital in WT (a) and MT (b)
Thus, the combination of the H-bond of His429 in F429H CYP 2B4, and the reorientation of the protein electric field in LM1,3 disrupt the PCET process, and enables the homolytic O-OH cleavage to take over and produce the hybrid HO/CYP activity, predicted here. This proposed impact of the H-bonding of His429 matches the recently reported blue shift in the absorption spectra for ferrous and ferric NO, and ferrous CO complexes of the F429H mutant of CYP 2B4.14 It may well be that the presence of Phe429 in the proximal side of CYP 2B4 (Fig. 1a) near the cysteine ligand was evolved to protect the sulfur from assuming too many hydrogen bonds,11a,b and thereby it regulates its “push effect”.11c The fact that a proximal Phe is a conserved residue in CYPs may supports the suggestion that Phe performs a regulatory function in modulating the redox potential of the heme14,15 so it can participate in the PCET process that leads to Cpd I formation, rather than allowing a homolytic O-OH cleavage. This fine-tuning of CYPs’ function, by a single residue, is an ever-intriguing feature of these enzymes. This delicate balance is highlighted by findings that the peroxide complexes of CYPs exhibit a mixture of homolytic and heterolytic bond cleavages,3a,16 and even a propensity towards heme degradation.16b
In conclusion, our QM/MM results show that the MT enzyme F429H of CYP 2B4 develops characteristics of a HO enzyme: (a) its NH---S H-bond to the cysteine ligand, and the changes in the protein electric field induced by the entrance of the substrate, retard the heterolytic cleavage and prefer homolytic O-OH cleavage, and (b) its Thr302/water cluster and the bulky substrate structurally orient the nascent OH• close to the porphyrin, and enforce meso hydroxylation. However, we have not explored other competitive processes, like diffusion of the OH radical followed by H-abstraction from the surrounding protein residues, nor did we consider the H-abstraction by the sulfur from His429 and generation of the inactive P420 form.14 Still however, the meso hydroxylation is barrier free, and hence the predictable HO activity should be observable.
Supplementary Material
Acknowledgments
Funding Sources
SS and LW are supported by an NIH grant, No RO1-GM-094209, and by Veterans Administration Review Grant CC103 to LW. C. Z thanks Hewlett Packward for computational facilities
Footnotes
ASSOCIATED CONTENT
Supporting information: Tables S1–S6, Figures S1–S6, Cartesian coordinates, computational details, and full reference of 5a and 13a. This material is available at http://pubs.acs.org.
Contributor Information
Lucy Waskell, Email: waskell@med.umich.edu.
Sason Shaik, Email: sason@yfaat.ch.huji.ac.il.
REFERENCES
- 1.(a) Ortiz de Montellano PR, editor. Cytochrome P450: structure, mechanism and biochemistry. 3rd ed. New York: Kluwer Academic/Plenum Publishers; 2004. [Google Scholar]; (b) Shaik S, Cohen S, Wang Y, Chen H, Kumar D, Thiel W. Chem. Rev. 2010;110:949–1017. doi: 10.1021/cr900121s. [DOI] [PubMed] [Google Scholar]
- 2.(a) Colas C, Ortiz de Montellano PR. Chem. Rev. 2003;103:2305–2332. doi: 10.1021/cr0204303. [DOI] [PubMed] [Google Scholar]; (b) Unno M, Matsui T, Ikeda-Saito M. Nat. Prod. Rep. 2007;24:553–570. doi: 10.1039/b604180a. [DOI] [PubMed] [Google Scholar]; (c) Matsui T, Unno M, Ikeda-Saito M. Acc. Chem. Res. 2010;43:240–247. doi: 10.1021/ar9001685. [DOI] [PubMed] [Google Scholar]
- 3.(a) Denisov IG, Makris TM, Sligar SG, Schlichting I. Chem. Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]; (b) Chen H, Moreau Y, Derat E, Shaik S. J. Am. Chem. Soc. 2008;130:1953–1965. doi: 10.1021/ja076679p. [DOI] [PubMed] [Google Scholar]
- 4.(a) Davydov R, Razeghifard R, Im SC, Waskell L, Hoffman BM. Biochemistry. 2008;47:9661–9666. doi: 10.1021/bi800926x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hamdane D, Xia C, Im S-C, Zhang H, Kim J-JP, Waskell L. J. Biol. Chem. 2009;284:11374–11384. doi: 10.1074/jbc.M807868200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Sherwood P, et al. J. Mol. Struct. THEOCHEM. 2003;632:1–28. [Google Scholar]; (b) Brooks BR, Burccoleri RE, Olafson BD, States DJ, Karplus M. J. Comput. Chem. 1983;4:187–217. [Google Scholar]; (c) Ahlrichs R, Bär M, Häser M, Horn H, Kölmel C. Chem. Phys. Lett. 1989;162:165–169. [Google Scholar]; (d) Senn HM, Thiel W. Angew. Chem. Int. Ed. 2009;48:1198–1229. doi: 10.1002/anie.200802019. [DOI] [PubMed] [Google Scholar]; (e) Zheng J, Wang D, Thiel W, Shaik S. J. Am. Chem. Soc. 2006;128:13204–13214. doi: 10.1021/ja063439l. [DOI] [PubMed] [Google Scholar]; (f) Chen H, Hirao H, Derat E, Schlichting I, Shaik S. J. Phys. Chem. B. 2008;112:9490–9500. doi: 10.1021/jp803010f. [DOI] [PubMed] [Google Scholar]
- 6.(a) Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. J. Biol. Chem. 2004;279:27294–27301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]; (b) Zazza C, Amadei A, Palma A, Sanna N, Tatoli S, Aschi M. J. Phys. Chem. B. 2008;112:3184–3192. doi: 10.1021/jp0774692. [DOI] [PubMed] [Google Scholar]
- 7.(a) Mak PJ, Im SC, Zhang H, Waskell L, Kincaid JR. Biochemistry. 2008;47:3950–3963. doi: 10.1021/bi800034b. [DOI] [PubMed] [Google Scholar]; (b) Mak PJ, Zhang H, Hollenberg PF, Kincaid JR. J. Am. Chem. Soc. 2010;132:1494–1495. doi: 10.1021/ja910276s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Altarsha M, Benighaus T, Kumar D, Thiel W. J. Am. Chem. Soc. 2009;131:4755–4763. doi: 10.1021/ja808744k. [DOI] [PubMed] [Google Scholar]; (b) Sen K, Hackett JC. J. Phys. Chem. B. 2009;113:8170–8182. doi: 10.1021/jp902932p. [DOI] [PubMed] [Google Scholar]
- 9.(a) Makris TM, von Koeing K, Schlichting I, Sligar SG. Biochemistry. 2007;46:14129–14140. doi: 10.1021/bi7013695. [DOI] [PubMed] [Google Scholar]; (b) Vatsis KP, Peng HW, Coon MJ. J. Inorg. Biochem. 2002;91:542–553. doi: 10.1016/s0162-0134(02)00438-5. [DOI] [PubMed] [Google Scholar]; (c) Raag R, Poulous T. Biochemistry. 1989;28:7586–7592. doi: 10.1021/bi00445a013. [DOI] [PubMed] [Google Scholar]
- 10.(a) Becke AD. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]; (b) Hay PJ, Wadt WR. J. Chem. Phys. 1985;82:299–310. [Google Scholar]; (c) Schäfer A, Huber C, Ahlrichs R. J. Chem. Phys. 1994;100:5829–5835. [Google Scholar]
- 11.(a) Poulos TL. J. Biol. Inorg. Chem. 1996;1:356–359. [Google Scholar]; (b) Yoshioka S, Takahashi S, Ishimori K, Morishima I. J. Inorg. Biochem. 2000;81:141–151. doi: 10.1016/s0162-0134(00)00097-0. [DOI] [PubMed] [Google Scholar]; (c) Dawson JH. Science. 1988;240:433–439. doi: 10.1126/science.3358128. [DOI] [PubMed] [Google Scholar]
- 12.(a) Ogliaro F, de Visser SP, Shaik S. J. Inorg. Biochem. 2002;91:554–567. doi: 10.1016/s0162-0134(02)00437-3. [DOI] [PubMed] [Google Scholar]; (b) Shaik S, Hirao H, Kumar D. Nat. Prod. Rep. 2007;24:533–552. doi: 10.1039/b604192m. [DOI] [PubMed] [Google Scholar]
- 13. Cho KB, et al. J. Phys. Chem. A. 2008;112:13128–13138. doi: 10.1021/jp806770y. (b) See eq. in Table S1 of Lai WZ, Chen H, Cho KB, Shaik S. J. Phys. Chem. Lett. 2010;1:2082–2087.
- 14.Perera R, Sono M, Kinloch R, Zhang H, Tarasev M, Im S-C, Waskell L, Dawson JH. Biochimic Biophysic Acta. 2011;1814:69–75. doi: 10.1016/j.bbapap.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ost TWB, Miles CS, Munro AW, Murdoch J, Reid GA, Chapman SK. Biochemistry. 2001;40:13421–13429. doi: 10.1021/bi010716m. [DOI] [PubMed] [Google Scholar]
- 16.For example, (a) See Ref. 3a and discussions therein. White RE, Sligar SG, Coon MJ. J. Biol. Chem. 1980;255:11108–11111. Shimizu T, Murakami Y, Hatano M. J. Biol. Chem. 1994;269:13296–13304.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.