Abstract
Embryonic stem (ES) and induced pluripotent stem (iPS) cells self-renew and are pluripotent. Differentiation of these cells can yield over 200 somatic cell types, making pluripotent cells an obvious source for regenerative medicine. Before the potential of these cells can be maximally harnessed for clinical applications, it will be necessary to understand the processes that maintain pluripotentiality and signal differentiation. Currently, three unique molecular properties distinguish pluripotent stem cells from somatic cells. These include a unique transcriptional hierarchy that sustains the pluripotent state during the process of self-renewal; a poised epigenetic state that maintains chromatin in a form ready for rapid cell fate decisions; and a cell cycle characterized by an extremely short gap 1 (G1) phase and the near absence of normal somatic cell checkpoint controls. Recently, B-MYB (MYBL2) was implicated in the gene regulation of two pluripotency factors and normal cell cycle progression. In this article, the three pluripotency properties and the potential role of B-Myb to regulate these processes will be discussed.
Keywords: Pluripotency, Stem cells, Transcription Factors, Epigenetics, Cell Cycle, B-Myb
Introduction
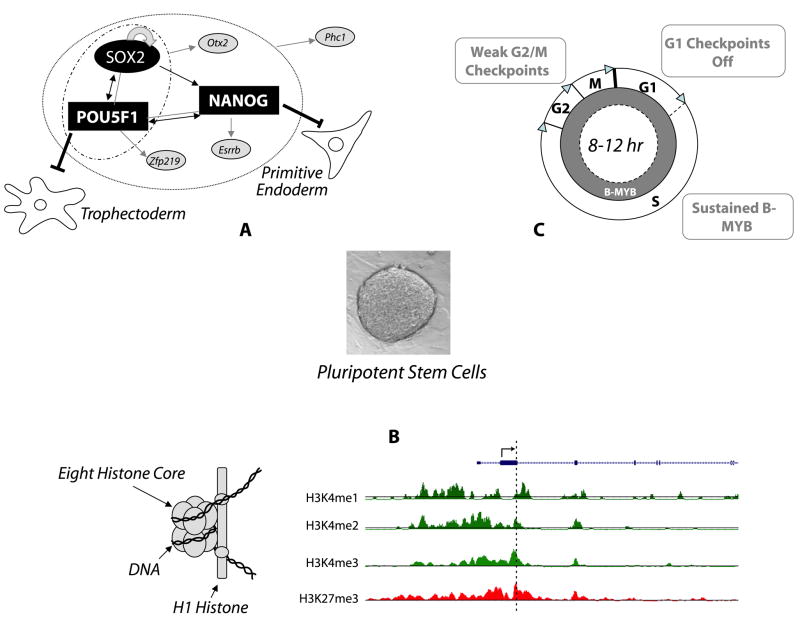
Embryonic stem (ES) cells are derived from the inner cell mass (ICM) of pre-implantation embryos. When differentiated in vitro, ES cells give rise to all cell types derived from the three primary germ layers and, following tetraploid aggregation, mouse ES cells can generate a viable embryo (Wobus and Boheler, 2005). Since ES cells do not readily generate all extra-embryonic tissues associated with normal embryo development, these cells are most aptly described as pluripotent and not as totipotent. The ability to maintain stem cell pluripotency in culture and differentiate these cells on demand has led to their widespread use as an in vitro model of development. More importantly, these cells and other pluripotent lines have been proposed as a possible source for cell replacement therapy to treat diseases or syndromes like Parkinsonism, spinal cord injury, diabetes, and heart failure [for review see (Wobus and Boheler, 2005)]. To achieve the goal of clinical applications, ES cells ultimately need to be differentiated efficiently to specific cell types, and potential problems associated with tumor formation or immunological rejection must be overcome. Tumor formation may be obviated through differentiation to “mature” phenotypes or through appropriate selection paradigms to remove any undifferentiated pluripotent stem cells. Immunologic limitations may be overcome through the generation of induced pluripotent stem (iPS) cells that can be created in vitro through transcription factor reprogramming of adult cells isolated directly from patients (Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Yu et al., 2007). Other than confounding integration dependent events, these pluripotent cells are very similar to or indistinguishable from blastocyst-derived ES cells (Maherali et al., 2007). Continued research and a thorough understanding of the molecular mechanisms that underlie the pluripotency of stem cells are, however, prerequisites before the goal of therapeutics can be realized. Three unique molecular properties distinguish pluripotent stem cells from somatic cells, including a unique transcriptional hierarchy, a poised epigenetic state, and a short cell cycle transit time (Figure 1).
Figure 1.
Unique characteristics of pluripotent cells. A) Regulatory network controlled at least partially by the master transcriptional regulators POU5F1, SOX2 and NANOG. The network illustrated here describes known interactions among the core regulators (black arrows) and their protein interaction partners (grey lines). Grey arrows depict targets regulated either by individual master regulators or by the combinations of regulators located in the ellipses. Specific target examples are taken from (Zhou et al., 2007). Moreover, NANOG inhibits formation of primitive endoderm and POU5F1 inhibits formation of trophectoderm. B) Central to epigenetic control is chromatin, and its fundamental unit the nucleosome, which is subject to modifications through a variety of enzymes and pathways. In the example to the right, the methylation states of histone 3 at residues lysine 4 and 27 are given. Methylation of lysine 4 is generally associated with gene activation (trithorax group proteins) and lysine 27 with gene repression (polycomb group proteins); however, the simultaneous trimethylation of both residues (H3K4 and H3K27, denoted by dashed line) is associated with a “poised” state, where transcription is generally low, but upon differentiation, gene transcription either can be rapidly activated or repressed. Additional epigenetic traits that are unique to pluripotent cells involve other histone modifications and DNA methylation modifications, which are not discussed here. C) Pluripotent cells generally do not have a G0 phase, have very short G1, and G2/M phases, and lack robust checkpoint controls. Consequently, several cell cycle-related proteins are maintained at high levels throughout the cell cycle. These include Cyclins E and A, hyper-phosphorylated pRB proteins, and sustained levels of B-MYB.
Pluripotency Factors and Regulatory Hierarchies
Transcriptional control contributes to the maintenance of pluripotency, and three transcription factors, including the homeodomain proteins POU5F1 (OCT3/4) and NANOG, and the SRY-related HMG box containing protein SOX2, have been directly implicated in the maintenance of pluripotent ES cells (Figure 1A). POU5F1 is a member of the POU transcription factor family and is expressed in the mouse embryo from the 4-to 8-cell stage until the epiblast begins to differentiate. Its presence is essential for the initial development of pluripotentiality in the inner cell mass (ICM), and consequently, high levels of this transcription factor are largely restricted to pluripotent lineages (Wobus and Boheler, 2005). Mouse embryos lacking POU5F1 protein die following implantation due to a lack of an ICM (Nichols et al., 1998). Importantly, POU5F1 must be present at appropriate levels to maintain pluripotency, because even a two-fold increase in expression causes differentiation into primitive endoderm and mesoderm, while loss of POU5F1 induces the formation of trophectoderm concomitant with a loss of pluripotency (Niwa et al., 2000). The latter is dependent upon interactions between POU5F1 and the definitive trophectodermal marker and transcription factor CDX2 (Niwa et al., 2005). Thus POU5F1 represses differentiation to trophoblast, and it works with other factors to maintain the undifferentiated state (Figure 1A). During early differentiation, however, POU5F1 exhibits pleiotrophic and dose-dependent functions that direct differentiation of ES cells to some cell lineages (Zeineddine et al., 2006).
Expression of the homeodomain protein NANOG in pre-implantation embryos is restricted to the inner cells of the compacted morula and blastocyst, and it is largely absent from differentiated cells (Chambers et al., 2003; Mitsui et al., 2003). Elevated levels of NANOG confer constitutive self-renewal of mouse ES cells in the absence of Leukemia Inhibitor Factor (LIF)/gp130 stimulation, and NANOG over-expressing cells can be propagated in serum-free media in the absence of Bone Morphogenetic Protein (BMP) (Ying et al., 2003). Over-expression of NANOG promotes high levels of Inhibitor of DNA Binding (Id) expression, which may serve to inhibit neuroectoderm formation. Nanog knockout mice develop to the blastocyst stage, but when cultured, the ICM differentiates to parietal endoderm-like cells. Thus, NANOG not only maintains pluripotency, it appears to inhibit the transition of undifferentiated ES cells to primitive endoderm (Figure 1A). In human ES cells, NANOG over-expression furthermore enables the cells to grow without feeder cells. Activation of p53, however, induces the differentiation of DNA-damaged ES cells by directly suppressing the expression of Nanog (Lin et al., 2005).
The sox2 gene encodes a transcription factor with a single HMG DNA binding domain that generally functions to bend DNA. It is expressed in the pluripotent lineages of early mouse embryos, but unlike POU5F1, it is also expressed in the multipotent cells of the extra-embryonic ectoderm. Sox2-null mutant mice fail to develop beyond implantation (Avilion et al., 2003), and down-regulation of Sox2 is associated with differentiation. Moreover, knockdown of Sox2 induced polyploidy (8C) and trophectoderm differentiation in mouse ES cells (Li et al., 2007). These latter results suggest that SOX2 may cooperate with POU5F1 to prevent trophectoderm differentiation in ES cells.
From chromosomal immunoprecipitation (ChIP)-on-chip or ChIP-paired-end ditag (PET) analyses (Eisenstein, 2006), these three transcription factors were found to co-occupy a substantial portion of target genes (Boyer et al., 2005; Loh et al., 2006). In both mouse and human, POU5F1 and NANOG co-occupy transcriptionally active and silent genes, including some that are transcriptional regulators implicated in lineage specification and cell fate determination during development (Boyer et al., 2005; Kim et al., 2008; Loh et al., 2006; Wang et al., 2006). Through RNA interference-mediated depletion of Pou5f1 and Nanog transcripts coupled with microarray expression profiling, it was determined that these factors can indeed activate or suppress gene transcription (Loh et al., 2006). Several other features suggest that the NANOG and POU5F1 protein interactions are actively implicated in the maintenance of ES cell pluripotency (Wang et al., 2006). First, the network is enriched in proteins required for survival or differentiation of the ICM. Second, most genes encoding the proteins in this network are co-regulated or down-regulated upon differentation. Some examples of overlap include genes for Hoxa1, Foxd3, Msx2 and Hexb. Third many of the proteins in the interactome were independently identified by ChIP-on-chip or ChIP-PET analyses, suggesting that these factors are common to pluripotent cell lines. Fourth, the network was linked to several cofactor pathways involved in transcriptional repression; and fifth, multiple interactions between key factors within the network may positively or negatively affect the stoichiometry within the complexes that maintain pluripotency. This latter finding may provide a framework for understanding the reprogramming of differentiated cells to a pluripotent state. Thus, these two factors may act in concert to regulate a common set of downstream pathways and developmental hierarchies. Octamer and Sox elements are also required for transcriptional cis-regulation of both nanog (Kuroda et al., 2005; Rodda et al., 2005) and pou5f1 gene expression (Okumura-Nakanishi et al., 2005). Moreover, POU5F1 modulates an upstream enhancer in the sox2 gene (Catena et al., 2004).
Additional transcription factors implicated in the maintenance of ES cell pluripotentiality include those encoded by c-Myc (Cartwright et al., 2005), Essrb (van den Berg et al., 2008; Zhang et al., 2008), Fgf4 (Ambrosetti et al., 1997), Foxd3 (Genesis)(Sutton et al., 1996), Gbx2 (Chapman et al., 1997), Pem (Fan et al., 1999), Rif1 (Loh et al., 2006), Sall4 (Zhang et al., 2006), Tbx3 (Ogawa et al., 2007), Utf1 (Okuda et al., 1998), Zfp42 (Rex1)(Hosler et al., 1993), Zfx (Galan-Caridad et al., 2007), and Zic3 (Lim et al., 2007). Most of these have been shown to be expressed in the ICM of blastocysts and are down-regulated or post-translationally modified upon differentiation; however, many of these are not exclusively expressed by pluripotent ES cells and can be found in other cell types in the soma. In the context of pluripotency network control, transcription factors like fgf4, utf1, rex1 are all direct targets of POU5F1 regulation; whereas, essrb and rif1 are primary targets of both POU5F1 and NANOG (Loh et al., 2006). Multiple others, including zic3 appear to be controlled either directly or indirectly by the regulatory core of POU5F1, NANOG and SOX2. Thus, POU5F1, SOX2, and NANOG collaborate or function independently to form a regulatory circuitry consisting of auto-regulatory and feed-forward loops that promote pluripotency and inhibit differentiation (Figure 1A).
The generation of induced pluripotent stem (iPS) cells using POU5F1/SOX2/MYC/KLF4 (in mouse) or POU5F1/SOX2/NANOG/LIN28 (in human) to reprogram adult cells into pluripotent cells has further demonstrated the role of these transcription factors in maintaining pluripotentiality; and recent studies have shown how POU5F1 and SOX2 alone with the histone deactylase inhibitor valproic acid can generate pluripotent cells (Huangfu et al., 2008). The reprogramming process is time dependent, and recent work from Brambrink et al and Stadtfeld et al have shown the sequence of events that occurs during this reprogramming process (Brambrink et al., 2008; Stadtfeld et al., 2008). Using inducible constructs and mouse embryonic fibroblasts, the authors found that during mouse iPS cell derivation, endogenous alkaline phosphatase in activated, followed by the expression of stage specific embryonic antigens and a loss of Thy1 expression. Expression of endogenous nanog and pou5f1 genes is observed only late in the process, marking the full reprogramming of these cells. At least in these experiments, the transduced cDNAs (Pou5f1, Sox2, Klf4 and c-Myc) must be expressed for at least 12 days to generate iPS cells. Subsequently, silencing of these exogenous genes is necessary for normal cell differentiation.
Epigenetics and Chromatin
Dolly the cloned sheep (Wilmut et al., 1997) and the establishment of iPS cells (Okita et al., 2007; Takahashi et al., 2007) have challenged the dogma that only totipotent (fertilized egg up to the 8 cell stage) or ES cells are capable of generating a viable organism. Since these clones were generated from adult somatic cell nuclei or cells, the results establish that cell reprogramming through altered epigenetic control is intimately related to pluripotentiality.
The term epigenetics specifically refers to heritable changes in a genotypic change that is caused by mechanisms other than changes in the primary DNA sequence; consequently, non-genetic factors cause the organism’s genes to behave differently (Bibikova et al., 2008; Surani et al., 2007). A more rigorous definition of epigenetics refers specifically to traits that can be passed on as a heritable trait; however, modifications to chromatin that normally occur during cellular differentiation (e.g., ES cells differentiating to mature cardiomyocytes or neurons) may also be an apt use of this term (Ptashne, 2007). During morphogenesis, for example, totipotent stem cells evolve into various pluripotent cell types that can fully differentiate into almost 200 different cell types including adult stem cells, neurons, muscle cells, liver cells, blood cells, epithelium, endothelium, and others. Since the primary DNA sequence does not change during this process, epigenetic modifications represent key regulators of developmental events that include X-inactivation, genomic imprinting, patterning by Hox genes and cellular differentiation and development (Surani et al., 2007). Importantly, epigenetic modifications may be maintained long-term, including through cell divisions, and multiple generations of cellular progeny.
Central to epigenetic control is chromatin, a complex of DNA, histones, and other proteins that make up chromosomes (Meshorer, 2007; Misteli, 2004; Misteli, 2007). The fundamental unit of chromatin is the nucleosome, which is comprised of two copies each of four core histones (H2A, H2B, H3 and H4) that form a histone octamer wrapped inside of 147 bp of genomic DNA (Figure 1B). The DNA bridging two adjacent nucleosomes is termed linker DNA. In eukaryotes, chromatin is folded into dynamically regulated structures, and changes to chromatin state directly influence genome activity and nuclear function (Gasser, 2002; Misteli, 2001). Its structure can be specifically influenced by histone modifications (e.g., methylation, acetylation, phosphorylation and ubiquitination) and binding proteins that may only transiently associate with chromatin. Alternatively, genes like pou5f1 and nanog can be turned off through cytosine methylation modifications to the promoter regions (Hattori et al., 2007; Hattori et al., 2004).
The dynamics of histone modification and association with chromatin binding proteins are particularly evident in ES cells and other multipotent cells during differentiation (Meshorer, 2007). During neuronal differentiation, for example, Meshorer et al (2006) showed that the number, size and distribution of heterochromatin foci is altered (Meshorer and Misteli, 2006); and following retinoic acid stimulation, Park et al, (2004) found an increase in the amount of compact heterochromatin (Park et al., 2004). Although chromatin reorganization during differentiation is not restricted to ES cells (Arney and Fisher, 2004), the unique aspects of ES cell chromatin dynamics have led to suggestions that the chromatin state holds some of the secrets of pluripotency (Meshorer and Misteli, 2006).
Additional epigenetic complexity comes from lysine methylation that in histones may take one of several forms, including mono-, di-, or trimethyl. This array of possible modifications, among others, increases the potential for functional responses. For example, the methylation of Histone 3, Lysine 4 (H3K4) residues is generally associated with trithorax group proteins; while, the methylation of Histone 3, Lysine 27 (H3K27) is associated with polycomb group genes (Ringrose and Paro, 2004) (Figure 1B). The former is associated with gene activation, while the latter is generally associated with gene repression. The simultaneous methylation of both H3K4 and HeK27 is associated with a bipolar status, whereby a gene may be “poised” to either be fully activated or repressed (Bernstein et al., 2006; Lee et al., 2006). More specifically in ES cells, Polycomb group (PcG) proteins bind promoter regions of numerous lineage specific genes (Boyer et al., 2006; Lee et al., 2006). Activation of these genes would normally promote ES cell differentiation, but the PcG proteins keep them repressed, but in a “poised state” in undifferentiated cells. Thus, the dynamic regulation of developmental pathways by Polycomb repression complexes may be required for maintaining mouse and human ES cell pluripotency and plasticity (Boyer et al., 2006; Lee et al., 2006).
Based on what is currently known about epigenetic control during normal embryonic development (Surani et al., 2007), it is likely that these factors, among others, interact with chromatin remodeling proteins and histone modifying enzymes to modulate the chromatin state. Similarly and if chromatin remodeling is indeed part of the mechanism underlying pluripotency, it remains unclear whether reprogramming associated with the generation of iPS cells is associated with a partial or complete loss of the epigenetic status or whether reprogrammed cells retain “memory” of their former state, which may have beneficial or adverse effects on cellular pluripotency.
Unique Cell Cycle
Relative to most somatic cells, ES cells divide very rapidly. Undifferentiated mouse ES cells transit the cell cycle once every 8-12 hours, depending on the cell line and cultivation conditions (Figure 1C). Human ES cells take approximately 15-30 hours to transit this cycle [rev. in (Becker et al., 2006; Wobus and Boheler, 2005)]. Irrespective of the timing differences, both mouse and human ES cells have a very short G1 phase. In mouse it is roughly 1-2 hours (Savatier et al., 1994) and in primate and human cells it takes about 2.5-3.0 hours (Becker et al., 2006; Fluckiger et al., 2006). In contrast, mouse embryonic fibroblasts transit the G1 phase in 6-12 hours and many adult cells take even longer. In somatic cells, passage from G1 into the S phase normally requires mitogen activated cyclin dependent kinases (Cdk) 4 and 6, cyclins (D and E), and members of the retinoblastoma (Rb) tumor suppressor protein family, including pRb, p107 and p130. In general, Cdk activity is opposed by a number of cell cycle inhibitory proteins, including p15INK4B, p16INK4A, p18INK4C, p19INK4D, p19INK4D, p21CIP, p27KIP1 and p57Kip2. In ES cells, cyclin D is mostly inactive and cell division is driven by high and constitutively active Cdk2-cyclin E activity (Stead et al., 2002), low levels of p21Cip1 and p27Kip1, and hyperphosphorylation of members of the retinoblastoma (Rb) tumor suppressor protein family, including p107 (Savatier et al., 1994). The presence of hyperphosphorylated, inactive Rb proteins allows E2F-responsive genes to be transcribed independently of cell cycle progression (Stead et al., 2002; White et al., 2005).
The only cell cycle regulators that show cell cycle-dependent expression in undifferentiated mouse ES cells are Cdk1 and cyclin B1, both of which show regulation during the G2 phase of the cell cycle (Stead et al., 2002). An uncoupled G2 mitotic-spindle checkpoint, which normally helps maintain chromosomal integrity during cell divisions, does not however initiate apoptosis in mouse and human ES cells as it does in somatic cells; consequently, ES cells fail to undergo mitosis and develop polyploidy. The absence of a robust checkpoint at this phase of the cell cycle is a likely source of karyotypic abnormalities in ES cells and their derivatives (Mantel et al., 2007).
Differentiation of pluripotent ES cells leads to progressive up-regulation of D cyclins (Savatier et al., 1996), decreased Cdk activity and regulation by the Rb pathway (White et al., 2005). Moreover, proliferating mouse ES cells contain very little of the truncated form of cyclin A2, which binds to and activates cyclin-dependent kinase 2 (CDK2); but as cells differentiate the amount of truncated cyclin A2 increases (Anger et al., 2003). This increase may be associated with the activation of CDK2, which is essential for the cell cycle transition from G1 to S. Cdc6, an activator of S phase kinase (AKT), cyclin A2, cyclin B1 and B-Myb also appear to be highly abundant in ES cells, but are down-regulated upon differentiation (Fujii-Yamamoto et al., 2005; Tarasov et al., 2008a; Tarasov et al., 2008b). Importantly, changes in these cell cycle regulators with differentiation occur prior to loss in POU5F1, NANOG or SOX2 protein abundance, suggesting that the absence of robust cell cycle checkpoints may be critical to the maintenance of pluripotency. Concomitant with the establishment of robust cell cycle checkpoints is the enhanced potential for apoptosis and other forms of cell death (Mantel et al., 2007).
In contrast to mouse, a number of essential regulators show cell cycle dependent levels of expression in human ES cells. More specifically, Cdc25a, a phosphatase required for progression from G1 to the S phase, is most abundant in the G1 phase of the cell cycle, cyclin E is highest in G1 and S phase cells, Cdk2 shows higher expression in the S phase, cyclin A and c-Myc are highest in S and G2, and cyclin B1 in the G2 phase. (Neganova and Lako, 2008a; Neganova and Lako, 2008b). NANOG also regulates S-phase entry in hESCs via transcriptional regulation of cell cycle regulatory components (Zhang et al., 2009). Irrespective of any species differences, cell cycle regulation in ES cells is fundamentally different from that of other somatic cell types, and an understanding of the processes that maintain the unique features of pluripotent stem (including iPS cells) cell cycle regulation appear critical to understanding pluripotency.
B-Myb and ES cell pluripotency
The transcription factor B-MYB (avian myeloblastosis viral oncogene homolog-2 (Mybl2)) has been implicated in somatic cell differentiation, cell cycle progression and apoptosis (Joaquin and Watson, 2003; Sala and Watson, 1999), and was recently implicated in the maintenance of pluripotency, chromatin stability and normal cell cycle progression (Figure 2) (Tarasov et al., 2008a). This protein contains an N-terminal DNA binding domain made up of three tandem repeats containing a helix-turn-helix motif and a C-terminal domain containing regulatory and transactivation domains that exhibit sequence-specific DNA-binding activity (Ness, 1996; Ness, 2003). Consistent with its function as a transcription factor, consensus myb bindings sites have been identified that are directly modulated by B-MYB, including apolipoprotein J and Cyclin B (Cervellera et al., 2000; Zhu et al., 2004) (Figure 2), but most genes regulated by B-MYB do not involve binding to consensus myb binding sites (e.gs., cyclin A, cyclin D1, B-Myb, Fgf4) (Bartusel et al., 2005; Johnson et al., 2002; Sala, 2005; Sala et al., 1999). The gene encoding B-Myb is regulated by E2F transcription factors, and its transcription in somatic cells is induced maximally at the G1/S boundary of the cell cycle (Joaquin and Watson, 2003; Sala, 2005). Although B-MYB was initially thought to regulate the progression from G1 to S phase, it also has important cellular functions in transit through G2/M. In Drosophila, dMyb (the homolog of mammalian B-Myb) mutants display a variety of abnormalities including spindle defects and increased aneuploidy (Manak et al., 2002); and in zebrafish carrying a loss-of-function mutation in the mybl2 gene (crash&burn mutant), embryos show defects in spindle formation, mitotic progression, and genomic instability (Shepard et al., 2005).
Figure 2.
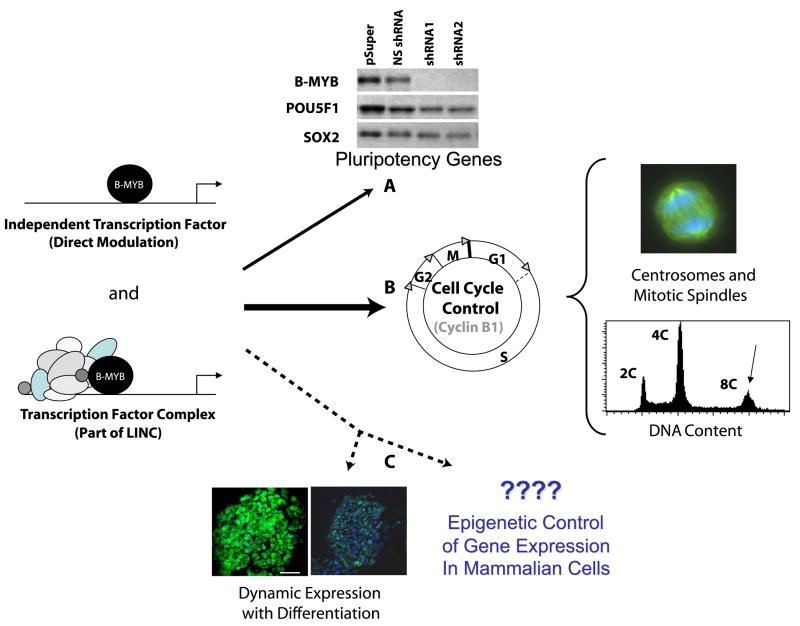
Probable roles of B-Myb in the regulation of the pluripotent state, either through function as a direct modulator of gene expression or part of a complex. A) B-MYB can modulate the expression of several pluripotency genes (e.g., Sox2 and Pou5F1). B) B-MYB is a direct modulator of cell cycle progression and gene (e.g., cyclin B1) expression. Reduced levels of B-Myb in pluripotent cells lead to polyploidy, centrosome and mitotic spindle abnormalities, and enhanced apoptosis. Differentiation of ES cells is also associated with a transient decrease in expression that may occur prior to the establishment of robust cell cycle controls. This transient decrease in expression is not associated with aneuploidy, but may be indicative of the establishment of cell cycle checkpoint controls. C) Differentiation of pluripotent cells promotes a rapid but transient loss of B-MYB. In this example, undifferentiated ES cells and ES cells following serum withdrawal are shown. In the latter, B-MYB expression is markedly reduced, but within another 12-18 hours, it returns to pre-differentiation levels of abundance. Because B-MYB can also modulate gene expression directly or as part of a protein complex (LIN complex), its effects are multifactorial, and recent evidence from Drosophila indicate that its expression together with that of either Lin-9 or E2F proteins can lead to epigenetic modifications (i.e., variegation) in gene expression. Currently, there is no direct evidence in pluripotent cells for epigenetic control regulated by B-MYB, but because its mechanisms of action are well conserved across species, it is a possibility that merits continued investigation.
In a mouse knock-out model, the absence of B-MYB results in post-implantation embryonic lethality, and in vitro, explanted embryos fail to form an inner cell mass (Tanaka et al., 1999). Since the ICM is the source of ES cells, B-Myb must be critical for pluripotency. Consistent with this assumption, B-Myb is highly expressed in ES and pluripotent embryonic carcinoma (EC) cells. While over-expression of B-Myb in ES cells has marginal effects on the ES cell phenotype, short-hairpin RNA mediated knockdown of B-Myb causes a significant decrease in both Sox2 and Pou5F1 transcripts prior to an up-regulation in gene markers of differentiation (Figure 2A) (Tarasov et al., 2008a; Tarasov et al., 2008b). In chromosomal immunoprecipitation (ChIP) assays, B-Myb dynamically binds to the promoters of both sox2 and pou5f1, but not to nanog. Moreover, B-Myb modulates the activity of human pou5f1 promoter constructs in mouse ES cells, a finding that was supported by site specific mutagenesis of a putative myb-binding site. It was however a paradox when these same promoter constructs were introduced into 293T cells and only modest effects on promoter activity were observed with myb-binding site mutants. This led to speculation that other factors might bind to this site to regulate its activity in vitro. Binding to the sox2 promoter, but not the pou5f1 promoter, has been reconfirmed through ChIP-on-chip assays using an antibody with higher specificity to B-Myb than that employed in the original assays (unpublished data). This suggests that the major effect of B-Myb on pluripotency networks in mouse ES cells may depend on SOX2 and its control of the pluripotency transcriptional hierarchy in ES cells.
Most dramatically, however, knockdown of B-Myb alters cell cycle kinetics and causes ES cells polyploidy/aneuploidy (Figure 2B) (Tarasov et al., 2008a). The major cell cycle defects caused by the absence of B-Myb are a decrease in the number of cells in the S phase, and a significant accumulation of cells in G2/M. The changes in G2/M are not associated with an altered incidence of sister chromatin exchange, a change in γ-H2AX histone immunoreactivity (as an index of DNA strand breaks), or an increase in the number of DNA events associated with fragmentation, fusion, and breakage (abnormalities/chromosome); however, mitotic cells are characterized by an apparent broadening of metaphase plates, an accumulation of monopolar centrosome complexes, and pronounced mitotic spindle defects. The most dramatic change was the increase in DNA ploidy (8C, 16C and 32C). Because cyclin B1 is a known target for B-Myb and a decrease in cyclin B1 expression was observed following knockdown of B-Myb, the observed mitotic defects have been attributed, at least partially, to this protein. This would be consistent with prior results from megakaryocytes, where transient loss of cyclin B1 led to cellular polyploidy (Zhang et al., 1996). Unlike in megakaryocytes, however, the polyploid state in ES cells does not appear to be stable, and when cells are allowed to recover from the loss of B-Myb, polyploidy is lost. Many of the surviving “ES cells” are however aneuploid; consequently, these survivors can no longer be considered bona fide ES cells and are perhaps best described a possible cancer stem cells. Since knockdown of Sox2 induces polyploidy in mouse ES cells (Li et al., 2007), there is also a question as to whether these two factors lead to aneuploidy independently or if there is a direct interaction between the two.
Data from mammalian cells and from Drosophila have furthermore demonstrated that B-Myb is part of a dynamic DNA binding complex (Mammals:LINC; Drosophila:dREAM) that consists of at least five subunits (Korenjak et al., 2004; Schmit et al., 2007). In humans, LINC contains Lin-9, Lin-37, Lin-54, Lin-52 and RbAp48, and early in the cell cycle, this complex binds to E2F4 and either p130 or p107 to repress transcription of E2F target genes regulating the G1 to S cell cycle transition (Litovchick et al., 2007). In the S and G2 phase of non-pluripotent cells, this complex associates with B-MYB to either activate or repress the transcription of genes required for this cell cycle transition and mitosis. Moreover it was recently shown that a LIN-9-containing complex is recruited by B-MYB to activate transcription of G2/M genes in undifferentiated and pluripotent EC cells (Knight et al., 2009). Both B-MYB and LIN-9 were thus required for transcription of G2/M genes (e.g., cyclin B and survivin), but unlike what has been shown in somatic cells, members of the Rb family did not associate with the LIN complex until the EC cells were induced to differentiate. These findings demonstrated that B-MYB/LIN complexes are required for progression through mitosis in pluripotent cells lacking a robust G1/S cell cycle checkpoint.
B-MYB proteins are transiently and significantly reduced in abundance once differentiation begins, but this protein returns to pre-differentiation levels within a matter of hours (Figure 2C) (Tarasov et al., 2008a). This transition is correlated with the establishment of robust cell cycle checkpoint controls, a prolonged cell cycle transit time and enhanced potential for apoptosis (Yamanaka et al., 2008). During this transition, the amount of hyper-phosphorylated B-MYB is also reduced in the G2/M phase of the cell cycle. It is currently unclear whether there is a direct relationship between these events, but modulation of B-MYB function appears to be one of the earliest markers of pluripotent cell differentiation. While speculative, it is also possible that this transient loss of B-MYB permits more active control of cell cycle progression through Rb association with the LIN complex.
In Drosophila, Wen et al showed that dMYB (the Drosophila equivalent of B-Myb) is required for the in vivo expression of Polo kinase (Plk1 equivalent), a serine/threonine kinase regulates centrosome maturation and spindle assembly during mitosis. In Drosophila, E2F2 (mammalian equivalent of E2F4), RBF (Rb binding factors p107 or p130) and Mip130 (Lin-9) proteins, as part of the dREAM (Drosophila equivalent of the LIN) complex, also act in opposition to dMYB to repress the expression of Polo kinase (Wen et al., 2008). The absence of both dMyb and either Mip130 (Lin-9) or E2F2 however caused variegated expression of Polo kinase (i.e., and epigenetic modification). This resulted in either high or low levels of Polo kinase that were stably inherited through successive cell divisions in imaginal wing discs, demonstrating that this complex can and does exert epigenetic regulation of dREAM (LINC) regulated genes. Knight et al furthermore demonstrated that Lin-9 (as part of the complex) may regulate polo-like kinase 1 gene expression in mouse EC cells (Knight et al., 2009). Although some differences exist between the mammalian LIN and Drosophila (d)REAM complexes, we have confirmed that a number of MYB targets identified in Drosophila also bind (ChIP-on-chip, unpublished) B-MYB in mammalian ES cells (e.g. cyclin B, aurora kinases, polo-like kinase). Because the effects of B-Myb on mitosis, centrosome maturation and spindle formation are all relatively well conserved between Drosophila cell models and mammalian EC and ES cells, it is likely that this latter function of B-Myb will also be broadly conserved across species. Consequently, B-Myb should prove to be a critical regulator not only of cell cycle progression but also of epigenetic states in pluripotent ES cells (Figure 2C). When coupled with the findings that B-MYB regulates the expression of both pou5f1 and sox2, and is critical for cell cycle progression, it is reasonable to postulate that this transcription factor is intimately associated with and potentially a prime regulator of the three traits that define pluripotency.
Conclusions
The three distinguishing traits of pluripotent cells include the presence of transcription factors (POU5F1, NANOG and SOX2) that regulate a transcriptional hierarchy to prevent differentiation and foster self-renewal; a poised chromatin state (H3K4me3 and H3K27me3) that permits rapid activation or repression of genes and microRNAs ; and a short cell cycle that lacks normal somatic cell cycle checkpoint controls in the G1 and G2 phases. The transcription factor B-MYB is required for expansion of the inner cell mass and derivation of embryonic stem cells. Although it can directly regulate the expression of some genes, recent results have demonstrated it to be part of a dynamic DNA binding complex that associates with E2F transcription factors and members of the pRb tumor suppressor family to activate or repress gene expression through direct transcriptional regulation or epigenetic controls. Since RB is a key cell cycle regulatory protein and senescence-inducing factor, the ability of B-MYB to associate with and possibly exclude RB from this DNA binding complex, may foster the escape of cells from normal cell cycle controls and cellular senescence. Because of its modulator effects on pou5f1 and sox2 gene expression, the reactivation of B-MYB may also be necessary for the establishment of pluripotent cells. Understanding its mechanisms of action and how it is involved with pluripotent traits may lead to novel methods of creating pluripotent cells and to differentiation paradigms capable of generating functional and chromosomally normal cells suitable for direct clinical applications. The next few years should therefore yield important insights into its function and continued research may lead to identification of other novel pathways regulated by Myb family members.
Acknowledgments
This research was supported entirely by the Intramural Research Program of the NIH, National Institute on Aging.
Abbreviations
- 2C, 4C, 8C
genome sizes (C- values), where 2C equals the genome size in either G0 or G1 and 4C equals the size in G2/M. 8C indicates a doubling of the normal genome size. Esrrb- estrogen related receptor, beta
- G1
Gap 1
- G2
Gap 2
- H3K4me1, 2, of 3
Histone 3, Lysine 4 monomethyl, dimethyl or trimethyl
- H3K27me3
Histone 3, Lysine 27, trimethyl
- LINC
LIN Complex; Human homologue of dREAM/Myb-MuvB complex whose composition is regulated at distinct phases of the cell cycle
- M
Mitosis
- Otx2
orthodenticle homolog 2 (Drosophila)
- Phc1
polyhomeotic-like 1 (Drosophila)
- S
S phase
- shRNA
short hairpin RNA
- Zfp219
zinc finger protein 219
References
- Ambrosetti DC, Basilico C, Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol Cell Biol. 1997;17(11):6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anger M, Bryja V, Jirmanova L, Hampl A, Carrington M, Motlik J, Dvorak P, Kubelka M. The appearance of truncated cyclin A2 correlates with differentiation of mouse embryonic stem cells. Biochem Biophys Res Commun. 2003;302(4):825–830. doi: 10.1016/s0006-291x(03)00270-5. [DOI] [PubMed] [Google Scholar]
- Arney KL, Fisher AG. Epigenetic aspects of differentiation. Journal of Cell Science. 2004;117(19):4355–4363. doi: 10.1242/jcs.01390. [DOI] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17(1):126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartusel T, Schubert S, Klempnauer KH. Regulation of the cyclin D1 and cyclin A1 promoters by B-Myb is mediated by Sp1 binding sites. Gene. 2005;351:171–180. doi: 10.1016/j.gene.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, Van Wijnen AJ, Stein GS. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol. 2006;209(3):883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie XH, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Laurent LC, Ren B, Loring JF, Fan JB. Unraveling epigenetic regulation in embryonic stem cells. Cell Stem Cell. 2008;2(2):123–134. doi: 10.1016/j.stem.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–353. doi: 10.1038/nature04733. Epub 2006 Apr 2019. [DOI] [PubMed] [Google Scholar]
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2(2):151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132(5):885–896. doi: 10.1242/dev.01670. Epub 2005 Jan 2026. [DOI] [PubMed] [Google Scholar]
- Catena R, Tiveron C, Ronchi A, Porta S, Ferri A, Tatangelo L, Cavallaro M, Favaro R, Ottolenghi S, Reinbold R, Scholer H, Nicolis SK. Conserved POU binding DNA sites in the Sox2 upstream enhancer regulate gene expression in embryonic and neural stem cells. J Biol Chem. 2004;279(40):41846–41857. doi: 10.1074/jbc.M405514200. Epub 42004 Jul 41815. [DOI] [PubMed] [Google Scholar]
- Cervellera M, Raschella G, Santilli G, Tanno B, Ventura A, Mancini C, Sevignani C, Calabretta B, Sala A. Direct transactivation of the anti-apoptotic gene apolipoprotein J (clusterin) by B-MYB. J Biol Chem. 2000;275(28):21055–21060. doi: 10.1074/jbc.M002055200. [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113(5):643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chapman G, Remiszewski JL, Webb GC, Schulz TC, Bottema CD, Rathjen PD. The mouse homeobox gene, Gbx2: genomic organization and expression in pluripotent cells in vitro and in vivo. Genomics. 1997;46(2):223–233. doi: 10.1006/geno.1997.4969. [DOI] [PubMed] [Google Scholar]
- Eisenstein M. Tracking the wily transcription factor. Nat Methods. 2006;3(5):341–341. doi: 10.1038/nmeth0506-341. [DOI] [PubMed] [Google Scholar]
- Fan Y, Melhem MF, Chaillet JR. Forced expression of the homeobox-containing gene Pem blocks differentiation of embryonic stem cells. Dev Biol. 1999;210(2):481–496. doi: 10.1006/dbio.1999.9279. [DOI] [PubMed] [Google Scholar]
- Fluckiger AC, Marcy G, Marchand M, Negre D, Cosset FL, Mitalipov S, Wolf D, Savatier P, Dehay C. Cell cycle features of primate embryonic stem cells. Stem Cells. 2006;24(3):547–556. doi: 10.1634/stemcells.2005-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii-Yamamoto H, Kim JM, Arai K, Masai H. Cell cycle and developmental regulations of replication factors in mouse embryonic stem cells. J Biol Chem. 2005;280(13):12976–12987. doi: 10.1074/jbc.M412224200. Epub 12005 Jan 12919. [DOI] [PubMed] [Google Scholar]
- Galan-Caridad JM, Harel S, Arenzana TL, Hou ZE, Doetsch FK, Mirny LA, Reizis B. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;129(2):345–357. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser SM. Nuclear architecture - Visualizing chromatin dynamics in interphase nuclei. Science. 2002;296(5572):1412–1416. doi: 10.1126/science.1067703. [DOI] [PubMed] [Google Scholar]
- Hattori N, Imao Y, Nishino K, Ohgane J, Yagi S, Tanaka S, Shiota K. Epigenetic regulation of Nanog gene in embryonic stem and trophoblast stem cells. Genes Cells. 2007;12(3):387–396. doi: 10.1111/j.1365-2443.2007.01058.x. [DOI] [PubMed] [Google Scholar]
- Hattori N, Nishino K, Ko YG, Hattori N, Ohgane J, Tanaka S, Shiota K. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J Biol Chem. 2004;279(17):17063–17069. doi: 10.1074/jbc.M309002200. [DOI] [PubMed] [Google Scholar]
- Hosler BA, Rogers MB, Kozak CA, Gudas LJ. An octamer motif contributes to the expression of the retinoic acid-regulated zinc finger gene Rex-1 (Zfp-42) in F9 teratocarcinoma cells. Mol Cell Biol. 1993;13(5):2919–2928. doi: 10.1128/mcb.13.5.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu DW, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26(11):1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Joaquin M, Watson RJ. Cell cycle regulation by the B-Myb transcription factor. Cell Mol Life Sci. 2003;60(11):2389–2401. doi: 10.1007/s00018-003-3037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LR, Johnson TK, Desler M, Luster TA, Nowling T, Lewis RE, Rizzino A. Effects of B-Myb on gene transcription: phosphorylation-dependent activity and acetylation by p300. J Biol Chem. 2002;277(6):4088–4097. doi: 10.1074/jbc.M105112200. [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132(6):1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight AS, Notaridou M, Watson RJ. A Lin-9 complex is recruited by B-Myb to activate transcription of G(2)/M genes in undifferentiated embryonal carcinoma cells. Oncogene. 2009;28(15):1737–1747. doi: 10.1038/onc.2009.22. [DOI] [PubMed] [Google Scholar]
- Korenjak M, Taylor-Harding B, Binne UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119(2):181–193. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Kuroda T, Tada M, Kubota H, Kimura H, Hatano SY, Suemori H, Nakatsuji N, Tada T. Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol Cell Biol. 2005;25(6):2475–2485. doi: 10.1128/MCB.25.6.2475-2485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125(2):301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Pan G, Cui K, Liu Y, Xu S, Pei D. A dominant negative form of mouse sox2 induces trophectoderm differentiation and progressive polyploidy in mouse es cells. J Biol Chem. 2007;15:15. doi: 10.1074/jbc.M702056200. [DOI] [PubMed] [Google Scholar]
- Lim LS, Loh YH, Zhang W, Li Y, Chen X, Wang Y, Bakre M, Ng HH, Stanton LW. Zic3 is required for maintenance of pluripotency in embryonic stem cells. Mol Biol Cell. 2007;18(4):1348–1358. doi: 10.1091/mbc.E06-07-0624. Epub 2007 Jan 1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7(2):165–171. doi: 10.1038/ncb1211. Epub 2004 Dec 2026. [DOI] [PubMed] [Google Scholar]
- Litovchick L, Sadasivam S, Florens L, Zhu XP, Swanson SK, Velmurugan S, Chen RS, Washburn MP, Liu XS, DeCaprio JA. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26(4):539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38(4):431–440. doi: 10.1038/ng1760. Epub 2006 Mar 2005. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, Plath K, Hochedlinger K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1(1):55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Manak JR, Mitiku N, Lipsick JS. Mutation of the Drosophila homologue of the Myb protooncogene causes genomic instability. Proc Natl Acad Sci U S A. 2002;99(11):7438–7443. doi: 10.1073/pnas.122231599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel C, Guo Y, Lee MR, Kim MK, Han MK, Shibayama H, Fukuda S, Yoder MC, Pelus LM, Kim KS, Broxmeyer HE. Checkpoint-apoptosis uncoupling in human and mouse embryonic stem cells: a source of karyotpic instability. Blood. 2007;109(10):4518–4527. doi: 10.1182/blood-2006-10-054247. Epub 2007 Feb 4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E. Chromatin in embryonic stem cell neuronal differentiation. Histol Histopathol. 2007;22(3):311–319. doi: 10.14670/HH-22.311. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nature Reviews Molecular Cell Biology. 2006;7(7):540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Misteli T. Nuclear structure - Protein dynamics: Implications for nuclear architecture and gene expression. Science. 2001;291(5505):843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- Misteli T. Spatial positioning: A new dimension in genome function. Cell. 2004;119(2):153–156. doi: 10.1016/j.cell.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: Cellular organization of genome function. Cell. 2007;128(4):787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113(5):631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Neganova I, Lako M. Cell cycle regulation in human embryonic stem cells. J Anat. 2008a;212(1):76–76. doi: 10.1111/j.1469-7580.2008.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neganova I, Lako M. G1 to S phase cell cycle transition in somatic and embryonic stem cells. J Anat. 2008b;213(1):30–44. doi: 10.1111/j.1469-7580.2008.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness SA. The Myb oncoprotein: regulating a regulator. Biochim Biophys Acta. 1996;1288(3):F123–139. doi: 10.1016/s0304-419x(96)00027-3. [DOI] [PubMed] [Google Scholar]
- Ness SA. Myb protein specificity: evidence of a context-specific transcription factor code. Blood Cells Mol Dis. 2003;31(2):192–200. doi: 10.1016/s1079-9796(03)00151-7. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Schoeler H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95(3):379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24(4):372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123(5):917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Ogawa K, Shimosato D, Takahashi K, Yagi R, Toyooka Y, Masui S, Matoba R, Ko MS. Forced expression of Tbx3 promotes LIF-independent self-renewal of mouse ES cells. Dev Biol. 2007;306(1):391–392. [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–317. doi: 10.1038/nature05934. Epub 2007 Jun 2006. [DOI] [PubMed] [Google Scholar]
- Okuda A, Fukushima A, Nishimoto M, Orimo A, Yamagishi T, Nabeshima Y, Kuro-o M, Boon K, Keaveney M, Stunnenberg HG, Muramatsu M. UTF1, a novel transcriptional coactivator expressed in pluripotent embryonic stem cells and extra-embryonic cells. Embo J. 1998;17(7):2019–2032. doi: 10.1093/emboj/17.7.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura-Nakanishi S, Saito M, Niwa H, Ishikawa F. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J Biol Chem. 2005;280(7):5307–5317. doi: 10.1074/jbc.M410015200. Epub 2004 Nov 5322. [DOI] [PubMed] [Google Scholar]
- Park SH, Park SH, Kook MC, Kim EY, Park S, Lim JH. Ultrastructure of human embryonic stem cells and spontaneous and retinoic acid-induced differentiating cells. Ultrastruct Pathol. 2004;28(4):229–238. doi: 10.1080/01913120490515595. [DOI] [PubMed] [Google Scholar]
- Ptashne M. On the use of the word ‘epigenetic’. Current Biology. 2007;17(7):R233–R236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the polycomb and trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. 2005;280(26):24731–24737. doi: 10.1074/jbc.M502573200. Epub 22005 Apr 24727. [DOI] [PubMed] [Google Scholar]
- Sala A. B-MYB, a transcription factor implicated in regulating cell cycle, apoptosis and cancer. Eur J Cancer. 2005;41(16):2479–2484. doi: 10.1016/j.ejca.2005.08.004. Epub 2005 Sep 2429. [DOI] [PubMed] [Google Scholar]
- Sala A, Saitta B, De Luca P, Cervellera MN, Casella I, Lewis RE, Watson R, Peschle C. B-MYB transactivates its own promoter through SP1-binding sites. Oncogene. 1999;18(6):1333–1339. doi: 10.1038/sj.onc.1202421. [DOI] [PubMed] [Google Scholar]
- Sala A, Watson R. B-Myb protein in cellular proliferation, transcription control, and cancer: latest developments. J Cell Physiol. 1999;179(3):245–250. doi: 10.1002/(SICI)1097-4652(199906)179:3<245::AID-JCP1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Savatier P, Huang S, Szekely L, Wiman KG, Samarut J. Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene. 1994;9(3):809–818. [PubMed] [Google Scholar]
- Savatier P, Lapillonne H, van Grunsven LA, Rudkin BB, Samarut J. Withdrawal of differentiation inhibitory activity/leukemia inhibitory factor up-regulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene. 1996;12(2):309–322. [PubMed] [Google Scholar]
- Schmit F, Korenjak M, Mannefeld M, Schmitt K, Franke C, von Eyss B, Gagrica S, Hanel F, Brehm A, Gaubatz S. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G(2)/M genes. Cell Cycle. 2007;6(15):1903–1913. doi: 10.4161/cc.6.15.4512. [DOI] [PubMed] [Google Scholar]
- Shepard JL, Amatruda JF, Stern HM, Subramanian A, Finkelstein D, Ziai J, Finley KR, Pfaff KL, Hersey C, Zhou Y, Barut B, Freedman M, Lee C, Spitsbergen J, Neuberg D, Weber G, Golub TR, Glickman JN, Kutok JL, Aster JC, Zon LI. A zebrafish bmyb mutation causes genome instability and increased cancer susceptibility. Proc Natl Acad Sci U S A. 2005;102(37):13194–13199. doi: 10.1073/pnas.0506583102. Epub 12005 Sep 13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2(3):230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead E, White J, Faast R, Conn S, Goldstone S, Rathjen J, Dhingra U, Rathjen P, Walker D, Dalton S. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene. 2002;21(54):8320–8333. doi: 10.1038/sj.onc.1206015. [DOI] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128(4):747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Sutton J, Costa R, Klug M, Field L, Xu D, Largaespada DA, Fletcher CF, Jenkins NA, Copeland NG, Klemsz M, Hromas R. Genesis, a winged helix transcriptional repressor with expression restricted to embryonic stem cells. J Biol Chem. 1996;271(38):23126–23133. doi: 10.1074/jbc.271.38.23126. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. Epub 2006 Aug 2010. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Patestos NP, Maekawa T, Ishii S. B-myb is required for inner cell mass formation at an early stage of development. J Biol Chem. 1999;274(40):28067–28070. doi: 10.1074/jbc.274.40.28067. [DOI] [PubMed] [Google Scholar]
- Tarasov KV, Tarasova YS, Tam WL, Riordon DR, Elliott ST, Kania G, Li J, Yamanaka S, Crider DG, Testa G, Li RA, Lim B, Stewart CL, Liu Y, Van Eyk JE, Wersto RP, Wobus AM, Boheler KR. B-MYB is Essential for Normal Cell Cycle Progression and Chromosomal Stability of Embryonic Stem Cells. PLoS One. 2008a;3(6):e2478. doi: 10.1371/journal.pone.0002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov KV, Testa G, Tarasova YS, Kania G, Riordon DR, Volkova M, Anisimov SV, Wobus AM, Boheler KR. Linkage of Pluripotent Stem Cell- Associated Transcripts to Regulatory Gene Networks. Cells Tissues Organs. 2008b;27 doi: 10.1159/000118787. [DOI] [PubMed] [Google Scholar]
- van den Berg DLC, Zhang WS, Yates A, Engelen E, Takacs K, Bezstarosti K, Demmers J, Chambers I, Poot RA. Estrogen-related receptor beta interacts with Oct4 to positively regulate Nanog gene expression. Mol Cell Biol. 2008;28(19):5986–5995. doi: 10.1128/MCB.00301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444(7117):364–368. doi: 10.1038/nature05284. Epub 2006 Nov 2008. [DOI] [PubMed] [Google Scholar]
- Wen H, Andrejka L, Ashton J, Karess R, Lipsick JS. Epigenetic regulation of gene expression by Drosophila Myb and E2F2-RBF via the Myb-MuvB/dREAM complex. Genes & Development. 2008;22(5):601–614. doi: 10.1101/gad.1626308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Stead E, Faast R, Conn S, Cartwright P, Dalton S. Developmental activation of the Rb-E2F pathway and establishment of cell cycle-regulated cyclin-dependent kinase activity during embryonic stem cell differentiation. Mol Biol Cell. 2005;16(4):2018–2027. doi: 10.1091/mbc.E04-12-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [see comments] [published erratum appears in Nature 1997 Mar 13; 386(6621):200] [DOI] [PubMed] [Google Scholar]
- Wobus AM, Boheler KR. Embryonic stem cells: prospects for developmental biology and cell therapy. Physiol Rev. 2005;85(2):635–678. doi: 10.1152/physrev.00054.2003. [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Li JL, Kania G, Elliott S, Wersto RP, Van Eyk J, Wobus AM, Boheler KR. Pluripotency of embryonic stem cells. Cell Tissue Res. 2008;331(1):5–22. doi: 10.1007/s00441-007-0520-5. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115(3):281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. Epub 2007 Nov 1920. [DOI] [PubMed] [Google Scholar]
- Zeineddine D, Papadimou E, Chebli K, Gineste M, Liu J, Grey C, Thurig S, Behfar A, Wallace VA, Skerjanc IS, Puceat M. Oct-3/4 dose dependently regulates specification of embryonic stem cells toward a cardiac lineage and early heart development. Dev Cell. 2006;11(4):535–546. doi: 10.1016/j.devcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, Lou Y, Yang J, Ma Y, Chai L, Ng HH, Lufkin T, Robson P, Lim B. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006;8(10):1114–1123. doi: 10.1038/ncb1481. Epub 2006 Sep 1117. [DOI] [PubMed] [Google Scholar]
- Zhang X, Neganova I, Przyborski S, Yang CB, Cooke M, Atkinson SP, Anyfantis G, Fenyk S, Keith WN, Hoare SF, Hughes O, Strachan T, Stojkovic M, Hinds PW, Armstrong L, Lako M. A role for NANOG in G1 to S transition in human embryonic stem cells through direct binding of CDK6 and CDC25A. J Cell Biol. 2009;184(1):67–82. doi: 10.1083/jcb.200801009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XF, Zhang J, Wang T, Esteban MA, Pei DQ. Esrrb Activates Oct4 Transcription and Sustains Self-renewal and Pluripotency in Embryonic Stem Cells. J Biol Chem. 2008;283(51):35825–35833. doi: 10.1074/jbc.M803481200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang ZG, Ravid K. The cell cycle in polyploid megakaryocytes is associated with reduced activity of cyclin B1-dependent Cdc2 kinase. J Biol Chem. 1996;271(8):4266–4272. doi: 10.1074/jbc.271.8.4266. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Chipperfield H, Melton DA, Wong WH. A gene regulatory network in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2007;104(42):16438–16443. doi: 10.1073/pnas.0701014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Giangrande PH, Nevins JR. E2Fs link the control of G1/S and G2/M transcription. Embo J. 2004;23(23):4615–4626. doi: 10.1038/sj.emboj.7600459. Epub 2004 Oct 4628. [DOI] [PMC free article] [PubMed] [Google Scholar]