Abstract
Treacher Collins syndrome (TCS) is a rare craniofacial disorder characterized by facial anomalies and ear defects. TCS is caused by mutations in the TCOF1 gene and follows autosomal dominant inheritance. Recently, mutations in the POLR1D and POLR1C genes have also been identified to cause TCS. However, in a subset of patients no causative mutation could be found yet. Inter- and intrafamilial phenotypic variability is high as is the variety of mainly family-specific mutations identified throughout TCOF1. No obvious correlation between pheno- and genotype could be observed. The majority of described point mutations, small insertions and deletions comprising only a few nucleotides within TCOF1 lead to a premature termination codon. We investigated a cohort of 112 patients with a tentative clinical diagnosis of TCS by multiplex ligation-dependent probe amplification (MLPA) to search for larger deletions not detectable with other methods used. All patients were selected after negative screening for mutations in TCOF1, POLR1D and POLR1C. In 1 patient with an unequivocal clinical diagnosis of TCS, we identified a 3.367 kb deletion. This deletion abolishes exon 3 and is the first described single exon deletion within TCOF1. On RNA level we observed loss of this exon which supposedly leads to haploinsufficiency of TREACLE, the nucleolar phosphoprotein encoded by TCOF1.
Key Words: Deletion, Multiplex ligation-dependent probe amplification, TCOF1, Treacher Collins syndrome
Introduction
Treacher Collins syndrome (TCS, OMIM 154500) is a rare craniofacial disorder with an incidence of about 1 in 50,000 live births [Posnick and Ruiz, 2000]. It is characterized by craniofacial anomalies like down-slanting palpebral fissures often accompanied by lower eyelid colobomas and a paucity of eyelashes medial to the defect, cleft palate, hypoplasia of mandible and zygomatic complex, as well as external and middle ear defects and conductive hearing loss [Dixon et al., 2007].
In about 80% of the patients TCS is caused by heterozygous mutations of the TCOF1 gene [Splendore et al., 2000; Teber et al., 2004; Horiuchi et al., 2005]. Recently, heterozygous mutations in POLR1D and compound heterozygous mutations in POLR1C have been identified as the cause of TCS subgroups 2 (OMIM 613717) and 3 (OMIM 248390), respectively [Yao et al., 1996; Dauwerse et al., 2011]. But there is still a subset of patients in whom no causative mutation has been found so far [Splendore et al., 2000; Teber et al., 2004; Horiuchi et al., 2005; Masotti et al., 2005].
TCOF1 encodes for the nucleolar phosphoprotein TREACLE which is involved in ribosomal biogenesis as are the proteins encoded by POLR1D and POLR1C [Larkin and Guilfoyle, 1997; Wise et al., 1997; Isaac et al., 2000; Valdez et al., 2004; Dixon et al., 2006]. Haploinsufficiency for Treacle in mice led to a reduced production of mature ribosomes thus causing restriction of cell cycle progression, reduced proliferation and high levels of apoptosis of the normally highly proliferative cranial neural crest cells from which facial bones and cartilages are derived [Dixon et al., 2006; Jones et al., 2008]. A reduction of the number of these cells at a critical time point early in embryogenesis leads to the cranial malformations typical for TCS [Dixon et al., 2007; Trainor, 2010]. In humans the phenotype shows a high inter- and intrafamilial variability ranging from mild, sometimes unaffected mutation carriers to severe forms leading to intrauterine death [Edwards et al., 1996; Splendore et al., 2002; Dixon et al., 2004; Teber et al., 2004; Masotti et al., 2005]. The spectrum of mutations within the TCOF1 gene that causes TCS is very heterogeneous [Gladwin et al., 1996; The Treacher Collins Syndrome Collaborative et al., 1996; Splendore et al., 2002, 2005; So et al., 2004; Teber et al., 2004; Horiuchi et al., 2005; Dixon et al., 2007; Masotti et al., 2009]. The majority of the mutations are insertions and deletions of up to 40 nucleotides that result in premature termination codons [Gladwin et al., 1996; Edwards et al., 1997; Ellis et al., 2002; Teber et al., 2004; Trainor et al., 2009]. Exon deletions or duplications in TCOF1 have not been published yet. Hitherto no obvious correlation between the type or location of the mutation and the phenotype could be observed [Trainor, 2010].
We here report on a 3.367 kb deletion affecting exon 3 and neighboring sequences within TCOF1 in a woman with unequivocal clinical diagnosis of TCS.
Material and Methods
Material
Genomic DNA was isolated from EDTA whole blood using the Flexi Gene DNA Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Total RNA was isolated from blood using the PAXgene Blood RNA Kit (PreAnalitiX, Hombrechtikon, Switzerland) following the manufacturer's instructions. An additional DNAse treatment was conducted with RQ1 DNAse (Promega, Wisc., USA) according to the manufacturer's recommendations to remove remaining traces of genomic DNA. A written informed consent approved by the ethics committee of the Universitätsklinikum Essen, Germany (07–3321), was obtained from the patient and the father. No material was available from the deceased mother.
Mutation Analysis
Sequencing of the TCOF1 gene was performed as described previously [Teber et al., 2004]. In addition, mutation analysis of the POLR1C and POLR1D genes, the 2 recently identified genes causative for TCS, was also performed as published before [Dauwerse et al., 2011].
Microarray Analysis
A (genome-wide) copy number analysis was conducted using the Affymetrix genome-wide human SNP array 6.0 (Affymetrix, Santa Clara, Calif., USA). The array was processed according to the manufacturer's instructions and data analysis was performed using the Genotyping Console software (Affymetrix).
Multiplex Ligation-Dependent Probe Amplification
Gene dosage of 25 exons of the TCOF1 locus was further analyzed by multiplex ligation-dependent probe amplification (MLPA) using the SALSA MLPA KIT P310-B1 TCOF1 (MRC Holland, Amsterdam, The Netherlands) (missing exons 9, 21 and 22; see manual). Hybridization, ligation and polymerase chain reactions (PCR) were carried out according to the manufacturer's instructions. Amplification products were analyzed by capillary electrophoresis using the ABI3100 capillary sequencer. Data analysis was carried out using the Gene Marker Software (Softgenetics, State College, Pa., USA).
Identification of the Breakpoints via Long-Range PCR (and Sequencing)
To confirm the deletion and to identify its breakpoints, a long-range PCR using the Expand long-range dNTPack (Roche, Mannheim, Germany) was performed using the primers F: 5′-GTGCTGGGGATTAATTGTGG-3′ and R: 5′-CCCATAGGCAATAGCTTGGA-3′ which generate a 6,840 bp product if a wild-type allele is present. The PCR protocol was used as stated in the manufacturer's manual using a magnesium-chloride (MgCl2)-containing buffer, a final primer concentration of 0.6 μM, no DMSO and about 500 ng of genomic DNA. The PCR was carried out on a Veriti cycler (Applied Biosystems, Foster City, Calif., USA) with the PCR conditions: first denaturation at 95°C for 2 min, then 10 cycles with 95°C for 10 s, 60°C for 15 s and 68°C for 7 min followed by 20 cycles of 95°C for 10 s, 60°C for 15 s and 68°C for 7 min plus 20 s in each cycle and a final elongation step of 7 min.
Sequencing of the breakpoints was performed using an additional primer (F_Seq: 5′-AGTGGGTAGAGAGTGGCAGG-3′) after ExoSap-It purification (USB, Ohio, USA, see manual) with the BigDye Terminator v1.1 Cycle Sequencing Kit on an ABI-3100 automatic capillary genetic analyzer (Applied Biosystems). For sequence analysis we used the software Sequencing Analysis (Applied Biosystems) and Geneious (Biomatters, Auckland, New Zealand).
mRNA Analysis
Reverse transcriptase (RT)-PCR was conducted according to standard protocols, followed by a PCR using primers spanning exons 1–4 of TCOF1 (F_Ex1: 5′-CTTCCCCTGATCTACCACCA-3′ and R_Ex4: 5′-CCAGTCTTGCCAGCTTTCTC-3′). The PCR was carried out using AmpliTaq Gold (Applied Biosystems) and a touchdown PCR program [Zeschnigk et al., 2009] modified as follows: first denaturation at 95°C for 5 min, 14 cycles with 95°C for 20 s, annealing for 30 s with temperature decreasing 0.5°C each cycle starting from 66°C and elongation for 30 s at 72°C followed by 35 cycles with 95°C for 20 s, 59°C for 30 s and 72°C for 30 s and a final elongation for 5 min at 72°C. An amplification of an RNA-specific product for the ACTB gene was carried out to exclude a contamination with genomic DNA [Kanber et al., 2009]. The 2 obtained cDNA amplicons were gel purified using the QIAquick gel extraction Kit (Qiagen) according to the manufacturer's instructions. The PCR primers F_Ex1 and R_Ex4 were used for sequencing.
Results and Discussion
Clinical Report
We investigated a cohort of 112 patients with a tentative clinical diagnosis of TCS. All patients were selected after negative screenings for mutations in the TCOF1, POLR1D and POLR1C genes.
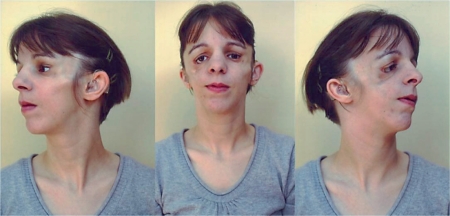
The patient was born at term with normal birth measurements to nonconsanguineous, healthy parents. Family history is unremarkable. She reached her developmental milestones in a normal range: Sitting with 6 months, walking without support at the age of 15 months and speaking first words at the age of 12 months. She presented with a classical TCS phenotype showing down-slanting palpebral fissures, a right-sided lower eyelid coloboma with absence of eyelashes medial to the defect, hypoplasia of the zygomatic complex as well as conductive deafness and median cleft palate (see fig. 1 and Teber et al. [2004] patient M18662). She had normal external ears, but the middle ear lacked the ambos. Cleft palate was surgically corrected at the age of 6 years, reconstructive surgery of lower eyelid coloboma, middle ear anomalies and zygomatic hypoplasia were performed at the age of 7, 11 and 16 years, respectively.
Fig. 1.
The patient at the age of 26 years, showing the characteristic facial anomalies of TCS: down-slanting palpebral fissures, a lower eyelid coloboma, hypoplasia of the zygomatic complex and low-set, small ears. Earlier photographs were published previously [Teber et al., 2004].
According to the severity score based on clinical findings introduced by Teber et al. [2004], the patient is severely affected with 12 out of 20 points.
Molecular Analyses
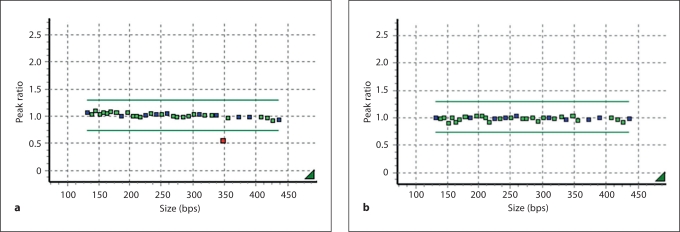
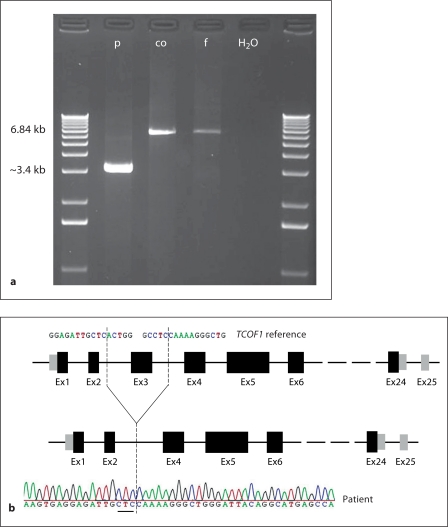
MLPA analyses of TCOF1 in a cohort of 112 patients with a tentative clinical diagnosis of TCS showed an aberrant result in 1 patient (M18662) only. Here a reduced dosage for the probe representing exon 3 (fig. 2) was observed in the patient but not in her healthy father. No material of the deceased mother was available for analysis. The nature of the aberrant MLPA result was further analyzed by long-range PCR and sequencing of the deletion junction fragment led to the identification of the breakpoints and revealed a 3.367 kb deletion abolishing the entire exon 3 and neighboring sequences of introns 2 and 3 (chr5: 149,741,531–149,741,534: 149,744,897– 149,744,900; GRCh37/hg19; 3 bp flanking the breakpoints cannot exactly be assigned either to the proximal or distal deletion breakpoint, see fig. 3). A previously conducted Affymetrix genome-wide SNP array 6.0 showed no aberrant results in this area due to scarcity of probes (in this region) (results not shown).
Fig. 2.
MLPA analysis. a Result for the patient. b Result for the father. Each square represents 1 MLPA probe. The bars enclose the range for a normal dosage. The probe for exon 3 shows a reduced dosage in the patient.
Fig. 3.
a Result of the long-range PCR. PCR from the patient (p), a normal person (co) as control, the healthy father (f) of the patient and the negative control (H2O). Product length of the wild-type allele was 6,840 bp. For the patient only the preferentially amplified product for the deleted allele of 3.367 kb was visible, confirming the deletion detected by MLPA. b Sequencing of the breakpoint and scheme of TCOF1. The figure shows the result of the sequence analysis of the patient's long-range PCR product under a scheme of TCOF1 (upper part: wild type with exon 3, lower part: with deletion of exon 3). The dashed lines indicate the position of the deletion. The 3 underlined nucleotides in the sequence can be aligned to both sides of the breakpoint (chr5: 149,741,531–149,741,534: 149,744,897–149,744,900, GRCh37/hg19). For comparison, part of the wild-type sequence is shown above.
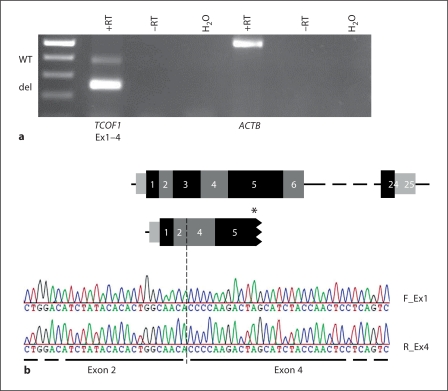
To further investigate the effect of the deletion on RNA level expression analysis was performed. Both expression of a transcript lacking exon 3 and the wild-type transcript were observed in RNA from peripheral blood of the patient (fig. 4a). Sequence analysis confirmed that exon 2 was directly spliced onto exon 4 (fig. 4b).
Fig. 4.
Results on RNA level. a Expression analysis of TCOF1 in peripheral blood RNA from the patient (M18662). The integrity of the RNA samples was demonstrated by amplification of an RNA-specific fragment of the ACTB locus. The upper TCOF1 product was derived from the wild-type allele (WT), while the lower product was derived from the allele harboring the deletion of exon 3 (del). +RT = RT-PCR with reverse transcriptase, –RT = RT-PCR without reverse transcriptase, H2O = RT-PCR without RNA. b Sequencing of the breakpoint on RNA level and scheme of TCOF1. The result of the sequencing of the patient's transcripts with the forward (F_Ex1) and reverse (R_Ex4) primer is shown. The schemes above depict the WT mRNA of TCOF1 and the transcript lacking exon 3. The breakpoint is indicated by a dashed line. The thus introduced premature stop codon is indicated by an asterisk in exon 5 together with the truncation. The results confirm that exon 2 is spliced directly onto exon 4. Black and grey squares = exons, small grey squares = UTR, horizontal dashed line indicates that not all exons are shown.
The deletion leads to a frameshift on protein level and a premature stop codon in exon 5 (p.Gln55HisfsX73, according to reference AAR87774, fig. 4b and online suppl. fig. 1; for all online supplementary material, see www.karger.com/doi/10.1159/000335545). This is due to the fact that the last 2 nucleotides of exon 2 and the first nucleotide of exon 3 contribute to 1 codon (CAA: p.Gln55) and, in addition, the last nucleotide of exon 3 forms a codon with the first 2 nucleotides of exon 4 (ACC: p.Thr102). Introduction of a premature stop codon is the most frequent outcome of mutations in TCOF1 and probably leads to haploinsufficiency of TREACLE [Dixon et al., 2006, 2007; Gladwin et al., 1996]. Premature stop codons can lead to the translation of a truncated protein or to nonsense-mediated decay (NMD) of the mRNA [Cartegni et al., 2002]. Isaac et al. [2000] could not detect truncated proteins in fibroblasts of TCS patients, thus suggesting NMD. In contrast, Marsh et al. [1998] could show mislocalization of human TREACLE after introduction of 3 different truncating mutations into TCOF1 using the (African green monkey) cell line COS-1. Only for one of the inserted mutations, which terminates TREACLE at amino acid position 36, no protein could be detected [Marsh et al., 1998]. Expression studies on RNA level also showed ambiguous results. Masotti et al. [2009] investigated samples from 4 TCS patients with different mutations on RNA level. They detected aberrant transcripts in 2 of these samples, one of them carrying a mutation within exon 3 (4 bp insertion). They argued that NMD might not be the only mechanism regulating transcription. It is also possible that the kind of mechanism is somehow linked to the position of the mutation. However, in the case of our patient the detection of an aberrant transcript argues against complete NMD.
Most of the mutations identified in TCOF1 so far are point mutations or very small deletions which do not exceed a length of a few base pairs [Gladwin et al., 1996; Splendore et al., 2000, 2002; Teber et al., 2004; Horiuchi et al., 2005; Masotti et al., 2005]. The largest deletion yet identified spanned 40 nucleotides and was located in exon 20 [Ellis et al., 2002], whereas the large deletion reported here spans about 3 kb. Its size seems to have no influence on the phenotype as the patient is not more severely affected than other TCS patients [Teber et al., 2004] and shows no additional symptoms. This is in concordance with the previously described lack of correlation between the phenotype and the type and location of the mutation [Trainor, 2010].
Although the patients in the investigated cohort are clinically not characterized in much detail, our results suggest that the frequency of such large deletions within TCOF1 seems to be rather low (1 in 112 patients ≈ 0.9%) despite covering 25 of the known 28 exons of TCOF1 with the MLPA Kit used. More studies will be needed to estimate a more exact frequency. Nevertheless, analyses like MLPA that are capable of detecting larger deletions should be considered for TCS diagnostic in patients without point mutations or very small deletions and insertions in TCOF1, POLR1D and POLR1C.
Conclusions
This is the first report about a deletion within the TCOF1 gene identified by MLPA. The deletion is the largest one described so far in TCS patients. It spans 3.367 kb, abolishes the entire exon 3 and leads to a premature stop codon in exon 5 due to a frameshift. As expected from previous genotype/phenotype correlations, there seems to be no correlation between the size of the intragenic deletion, the phenotype and the severity of the phenotype. The frequency of large deletions seems to be rather low, although more studies are needed to estimate a more exact frequency. Analyses capable of detecting larger deletions should be considered for TCS diagnostic in patients without point mutations in TCOF1, POLR1D and POLR1C.
Supplementary Material
Amino acid sequence of TREACLE with and without deletion of exon 3. The upper part shows the first part of the amino acid (aa) sequence of the WT TREACLE. In the second part the aa sequence of TREACLE after deletion of exon 3 is depicted. The site of the deletion and the premature stop codon in exon 5 are highlighted by arrows.
Acknowledgements
We are grateful to the family for their participation in the study. This work was supported by the German Federal Ministry of Education and Research (BMBF) by grant number 01GM0802 to D.R.L. and D.W.
References
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Dauwerse JG, Dixon J, Seland S, Ruivenkamp CA, van Haeringen A, et al. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat Genet. 2011;43:20–22. doi: 10.1038/ng.724. [DOI] [PubMed] [Google Scholar]
- Dixon J, Ellis I, Bottani A, Temple K, Dixon MJ. Identification of mutations in TCOF1: use of molecular analysis in the pre- and postnatal diagnosis of Treacher Collins syndrome. Am J Med Genet A. 2004;127A:244–248. doi: 10.1002/ajmg.a.30010. [DOI] [PubMed] [Google Scholar]
- Dixon J, Jones NC, Sandell LL, Jayasinghe SM, Crane J, et al. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc Natl Acad Sci USA. 2006;103:13403–13408. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J, Trainor P, Dixon MJ. Treacher Collins syndrome. Orthod Craniofac Res. 2007;10:88–95. doi: 10.1111/j.1601-6343.2007.00388.x. [DOI] [PubMed] [Google Scholar]
- Edwards SJ, Fowlie A, Cust MP, Liu DT, Young ID, Dixon MJ. Prenatal diagnosis in Treacher Collins syndrome using combined linkage analysis and ultrasound imaging. J Med Genet. 1996;33:603–606. doi: 10.1136/jmg.33.7.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SJ, Gladwin AJ, Dixon MJ. The mutational spectrum in Treacher Collins syndrome reveals a predominance of mutations that create a premature-termination codon. Am J Hum Genet. 1997;60:515–524. [PMC free article] [PubMed] [Google Scholar]
- Ellis PE, Dawson M, Dixon MJ. Mutation testing in Treacher Collins Syndrome. J Orthod. 2002;29:293–297. doi: 10.1093/ortho/29.4.293. discussion 278. [DOI] [PubMed] [Google Scholar]
- Gladwin AJ, Dixon J, Loftus SK, Edwards S, Wasmuth JJ, et al. Treacher Collins syndrome may result from insertions, deletions or splicing mutations, which introduce a termination codon into the gene. Hum Mol Genet. 1996;5:1533–1538. doi: 10.1093/hmg/5.10.1533. [DOI] [PubMed] [Google Scholar]
- Horiuchi K, Ariga T, Fujioka H, Kawashima K, Yamamoto Y, et al. Mutational analysis of the TCOF1 gene in 11 Japanese patients with Treacher Collins syndrome and mechanism of mutagenesis. Am J Med Genet A. 2005;134:363–367. doi: 10.1002/ajmg.a.30357. [DOI] [PubMed] [Google Scholar]
- Isaac C, Marsh KL, Paznekas WA, Dixon J, Dixon MJ, et al. Characterization of the nucleolar gene product, treacle, in Treacher Collins syndrome. Mol Biol Cell. 2000;11:3061–3071. doi: 10.1091/mbc.11.9.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones NC, Lynn ML, Gaudenz K, Sakai D, Aoto K, et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med. 2008;14:125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanber D, Giltay J, Wieczorek D, Zogel C, Hochstenbach R, et al. A paternal deletion of MKRN3, MAGEL2 and NDN does not result in Prader-Willi syndrome. Eur J Hum Genet. 2009;17:582–590. doi: 10.1038/ejhg.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin RM, Guilfoyle TJ. Reconstitution of yeast and Arabidopsis RNA polymerase alpha-like subunit heterodimers. J Biol Chem. 1997;272:12824–12830. doi: 10.1074/jbc.272.19.12824. [DOI] [PubMed] [Google Scholar]
- Marsh KL, Dixon J, Dixon MJ. Mutations in the Treacher Collins syndrome gene lead to mislocalization of the nucleolar protein treacle. Hum Mol Genet. 1998;7:1795–1800. doi: 10.1093/hmg/7.11.1795. [DOI] [PubMed] [Google Scholar]
- Masotti C, Armelin-Correa LM, Splendore A, Lin CJ, Barbosa A, et al. A functional SNP in the promoter region of TCOF1 is associated with reduced gene expression and YY1 DNA-protein interaction. Gene. 2005;359:44–52. doi: 10.1016/j.gene.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Masotti C, Ornelas CC, Splendore-Gordonos A, Moura R, Felix TM, et al. Reduced transcription of TCOF1 in adult cells of Treacher Collins syndrome patients. BMC Med Genet. 2009;10:136. doi: 10.1186/1471-2350-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posnick JC, Ruiz RL. Treacher Collins syndrome: current evaluation, treatment, and future directions. Cleft Palate Craniofac J. 2000;37:434. doi: 10.1597/1545-1569(2000)037<0434:TCSCET>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- So RB, Gonzales B, Henning D, Dixon J, Dixon MJ, Valdez BC. Another face of the Treacher Collins syndrome (TCOF1) gene: identification of additional exons. Gene. 2004;328:49–57. doi: 10.1016/j.gene.2003.11.027. [DOI] [PubMed] [Google Scholar]
- Splendore A, Silva EO, Alonso LG, Richieri-Costa A, Alonso N, et al. High mutation detection rate in TCOF1 among Treacher Collins syndrome patients reveals clustering of mutations and 16 novel pathogenic changes. Hum Mutat. 2000;16:315–322. doi: 10.1002/1098-1004(200010)16:4<315::AID-HUMU4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Splendore A, Jabs EW, Passos-Bueno MR. Screening of TCOF1 in patients from different populations: confirmation of mutational hot spots and identification of a novel missense mutation that suggests an important functional domain in the protein treacle. J Med Genet. 2002;39:493–495. doi: 10.1136/jmg.39.7.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splendore A, Fanganiello RD, Masotti C, Morganti LS, Passos-Bueno MR. TCOF1 mutation database: novel mutation in the alternatively spliced exon 6A and update in mutation nomenclature. Hum Mutat. 2005;25:429–434. doi: 10.1002/humu.20159. [DOI] [PubMed] [Google Scholar]
- Teber OA, Gillessen-Kaesbach G, Fischer S, Bohringer S, Albrecht B, et al. Genotyping in 46 patients with tentative diagnosis of Treacher Collins syndrome revealed unexpected phenotypic variation. Eur J Hum Genet. 2004;12:879–890. doi: 10.1038/sj.ejhg.5201260. [DOI] [PubMed] [Google Scholar]
- The Treacher Collins Syndrome Collaborative Group. Dixon J, Edwards SJ, Gladwin AJ, Dixon MJ, et al. Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. Nat Genet. 1996;12:130–136. doi: 10.1038/ng0296-130. [DOI] [PubMed] [Google Scholar]
- Trainor PA. Craniofacial birth defects: The role of neural crest cells in the etiology and pathogenesis of Treacher Collins syndrome and the potential for prevention. Am J Med Genet A. 2010;152A:2984–2994. doi: 10.1002/ajmg.a.33454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor PA, Dixon J, Dixon MJ. Treacher Collins syndrome: etiology, pathogenesis and prevention. Eur J Hum Genet. 2009;17:275–283. doi: 10.1038/ejhg.2008.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez BC, Henning D, So RB, Dixon J, Dixon MJ. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc Natl Acad Sci USA. 2004;101:10709–10714. doi: 10.1073/pnas.0402492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise CA, Chiang LC, Paznekas WA, Sharma M, Musy MM, et al. TCOF1 gene encodes a putative nucleolar phosphoprotein that exhibits mutations in Treacher Collins Syndrome throughout its coding region. Proc Natl Acad Sci USA. 1997;94:3110–3115. doi: 10.1073/pnas.94.7.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Yamamoto K, Nishi Y, Nogi Y, Muramatsu M. Mouse RNA polymerase I 16-kDa subunit able to associate with 40-kDa subunit is a homolog of yeast AC19 subunit of RNA polymerases I and III. J Biol Chem. 1996;271:32881–32885. doi: 10.1074/jbc.271.51.32881. [DOI] [PubMed] [Google Scholar]
- Zeschnigk M, Martin M, Betzl G, Kalbe A, Sirsch C, et al. Massive parallel bisulfite sequencing of CG-rich DNA fragments reveals that methylation of many X-chromosomal CpG islands in female blood DNA is incomplete. Hum Mol Genet. 2009;18:1439–1448. doi: 10.1093/hmg/ddp054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequence of TREACLE with and without deletion of exon 3. The upper part shows the first part of the amino acid (aa) sequence of the WT TREACLE. In the second part the aa sequence of TREACLE after deletion of exon 3 is depicted. The site of the deletion and the premature stop codon in exon 5 are highlighted by arrows.