Abstract
Background:
Sharing digital pathology images for enterprise- wide use into a picture archiving and communication system (PACS) is not yet widely adopted. We share our solution and 3-year experience of transmitting such images to an enterprise image server (EIS).
Methods:
Gross pathology images acquired by prosectors were integrated with clinical cases into the laboratory information system's image management module, and stored in JPEG2000 format on a networked image server. Automated daily searches for cases with gross images were used to compile an ASCII text file that was forwarded to a separate institutional Enterprise Digital Imaging and Communications in Medicine (DICOM) Wrapper (EDW) server. Concurrently, an HL7-based image order for these cases was generated, containing the locations of images and patient data, and forwarded to the EDW, which combined data in these locations to generate images with patient data, as required by DICOM standards. The image and data were then “wrapped” according to DICOM standards, transferred to the PACS servers, and made accessible on an institution-wide basis.
Results:
In total, 26,966 gross images from 9,733 cases were transmitted over the 3-year period from the laboratory information system to the EIS. The average process time for cases with successful automatic uploads (n=9,688) to the EIS was 98 seconds. Only 45 cases (0.5%) failed requiring manual intervention. Uploaded images were immediately available to institution- wide PACS users. Since inception, user feedback has been positive.
Conclusions:
Enterprise- wide PACS- based sharing of pathology images is feasible, provides useful services to clinical staff, and utilizes existing information system and telecommunications infrastructure. PACS-shared pathology images, however, require a “DICOM wrapper” for multisystem compatibility.
Keywords: DICOM, digital image, LIS, PACS, pathology, wrapper
INTRODUCTION
An electronic picture archiving and communications system (PACS) has been successfully used in the field of radiology for storage, rapid retrieval, and widespread access to digital images.[1,2] Digital images can be acquired using multiple modalities and many PACS users at different sites can have simultaneous, remote access to images. Both electronic images and reports can be transmitted digitally via PACS. The major components of a PACS include the imaging devices, secure network for transmission of patient information, workstations for interpreting and reviewing images, and storage servers. PACS not only offers capabilities for off-site viewing and reporting, but also helps manage radiology workflow, and provides an electronic platform for digital images to be interfaced with other information systems such as the electronic medical record (EMR). Early PACS developments focused mainly on the radiologist's requirements for diagnosis and image interpretation. However, current imaging informatics at the enterprise level has shifted toward the development of new multimedia and communication tools geared toward the needs of other users such as pathologists, surgeons, cardiologists, and even patients themselves.[3,4] The universal format for PACS image storage and transfer is Digital Imaging and Communications in Medicine (DICOM). Recent initiatives have helped create a DICOM imaging standard so that pathology images too can be incorporated into PACS.[5,6] For example, the DICOM supplements 122 and 145 provide flexible object information definitions dedicated respectively to pathology specimen description and WSI acquisition, storage, and display.[7]
The potential benefits of a PACS specifically for pathology has been addressed by very few authors.[4,8,9] Moreover, the actual integration of digital pathology images into an enterprise-wide PACS has not yet been widely adopted. Previously described PACS-systems in pathology were limited in scope, involving restricted access to only the pathology department or a select group of medical subspecialties. Integration of digital pathology images into an enterprise-wide PACS requires that images from a pathology-based laboratory information system (LIS) are compatible with the DICOM format. Adhering to this format ensures that images from multiple sources can be integrated within one universally accessible EMR, by all clinicians of all medical specialties. It has been suggested that the benefits of integrating digital pathology images into pre-existing electronic health records include greater accessibility to these images by all clinicians, and amalgamation of clinical findings for patient care, education and research. Traditionally, sharing of pathology images has been achieved by paper (printed photos) and/or e-mail-based (e-mail with attached image files) systems. However, such actions require extra effort by the pathologist, leaving less time available for clinical service duties. Additional limitations of these systems include minimal documentation, patient traceability and lack of institution-wide access. To overcome these limitations, we share our solution and 3 year experience of transmitting digital gross pathology images to an enterprise image server (EIS) or PACS that provides all our clinicians access to them.
METHODS
At our institution, comprising 20 geographically separated medical centers, the PACS landscape consists of a federation of 12 separate PACS which allow physicians at any location to view imaging studies regardless of the imaging source location and physical site location of the user. At three of our core facilities we deployed a solution to incorporate gross pathology images into this PACS. An overview of the process by which digital gross pathology images are properly formatted and eventually transmitted to the PACS enterprise image server (EIS) is summarized in Figure 1. Pathology residents, assistants, and surgical pathology technicians acquire gross pathology images at our institution using Nikon DS-U2 cameras. The cameras are associated with a PicsPlus interface of our LIS (Cerner CoPath v.3.2, Cerner Corporation, Kansas City, MO), which automatically integrates these images into an image gallery (LIS-integrated image management system) associated with all accessioned and clinical data for corresponding cases directly into the LIS. The images are stored in JPEG2000 format on a networked image server (PicsPlus Server), and can be accessed via the LIS through the PicsPlus interface. To enable integration into the EIS, the following sequence takes place: each day close to 5 pm the LIS is programmed to automatically conduct a search for cases containing digital gross images that were taken within the past 24 hours, excluding gross images from autopsy cases. The list of cases generated in an ASCII text file is forwarded to an institutional EIS (Philips iSite v.3.5, Andover, MA), on which the Enterprise DICOM Wrapper (EDW) software resides. EDW (v.15) was developed within our institution. Although the majority of the workflow logic for the EDW was coded in the Microsoft .Net framework utilizing C# (Microsoft Corporation, Redmond, WA), the DICOM wrapping component utilized C++. The EIS is the same PACS that clinicians use to access radiology reports and DICOM-compliant radiology images at our institution.
Figure 1.
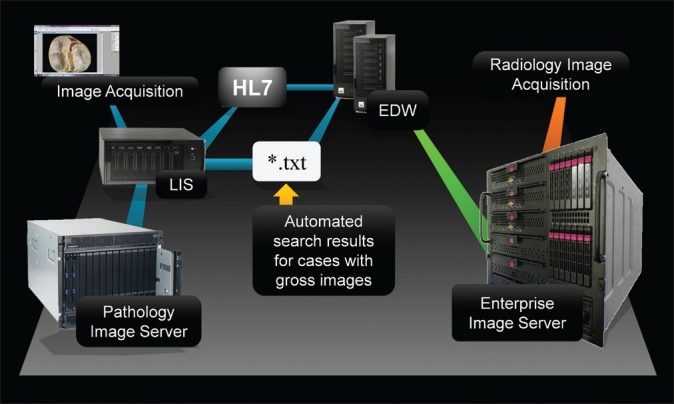
Overview of the electronic transmission pathway for digital gross pathology images from the laboratory information system (LIS) via an enterprise DICOM wrapper (EDW) to an enterprise image server, enabling enterprise- wide access to PACS users where the also access radiology images. Both HL7- based image order data with concomitant text file (*.txt) patient data are forwarded to the EDW to generate DICOM- compliant image files
Concurrently, the LIS generates HL7-based orders for each patient, identical to orders generated when radiologic studies are ordered for patients. However, in this case the HL7-based image orders contain the locations of both digital pathology images and patient data, and an order for each patient with a digital pathology image is then forwarded to the EDW. The HL7 Message Routing interface engine that is responsible for field mapping, field replacement, and HL7 routing utilizes the Microsoft .NET framework and C#. Both text files and HL7-based orders are analyzed to verify that patient data are accurate, and that the images are correctly matched with each patient. After verification, each image along with the correct patient data are then “wrapped” according to DICOM standards-specifically, new JPEG images are generated for each patient, containing embedded patient fields (such as patient name, medical record number, etc.) in Extensible Markup Language (XML) format. These fields are populated with patient metadata from the HL7-based image order [Figure 2]. A second verification process then occurs to ensure that all patient fields are completed, after which the EDW then transfers each image to the PACS server, at which point the images are immediately accessible to all clinicians with access to this PACS on an institution-wide basis.
Figure 2.
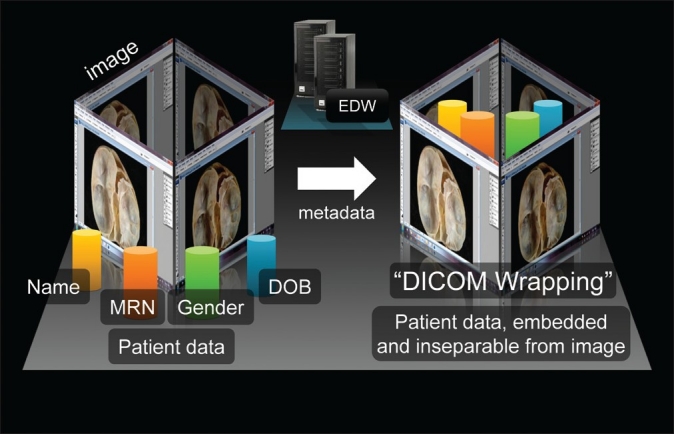
A simplified view of “DICOM” wrapping, as used by the Enterprise Data Wrapper (EDW) software, to combine a digital gross pathology image (JPEG2000 format) with patient metadata (part of the inbound HL7 message), resulting in a new DICOM-compliant image with embedded patient metadata. This format is required in order for the image to be uploaded to the enterprise image server. (MRN = medical record number; DOB = date of birth; DICOM = Digital Imaging and Communications in Medicine.)
An outline of data flow, specifically through the EDW, is provided in Figure 3. We utilized a combination of available third-party DICOM and imaging libraries to open JPEG images and “wrap DICOM.” Many years ago, when the EDW was written, libraries operated at a much lower level. For current programmers, more advanced and now commercially available software libraries (i.e. LeadTools, Lead Technologies, Charlotte, NC, http://www.leadtools.com/sdk/medical-imaging.htm) abstracts away much of the imaging and DICOM details, allowing programmers to focus on how to obtain patient demographics and image data, and away from the technicalities of the metadata and wrapping. For some users, “DICOM wrapping” can be a manual process though a web page, where the user uploads images and ties in HL7-based demographics (name, age, accession, etc.). In our scenario, the EDW is analogous to a web- based XML form, except that the EDW “fills out the form” automatically with JPEG file names and patient demographics.
Figure 3.
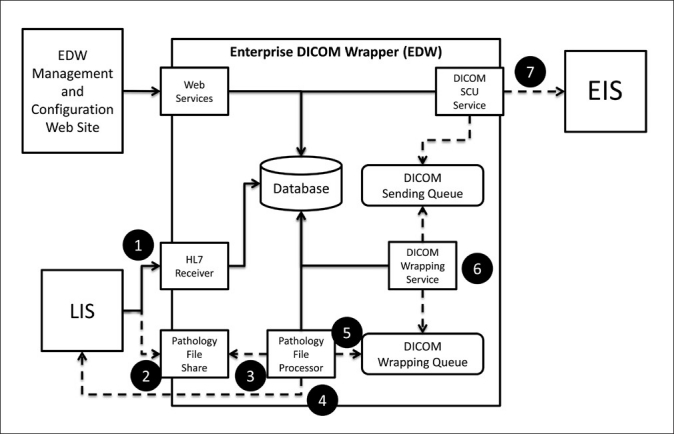
Software flow diagram for the Enterprise DICOM Wrapper (EDW). The steps outlined in the diagram are as follows: 1. The Laboratory Information System (LIS) sends the HL7 order. 2. The LIS sends the text file. 3. A “Pathology File Processor” receives the text file and registers the patient and exam in an EDW database. The HL7 order received is similar to orders generated for radiology images, so HL7 order modification by the pathology file processor is not necessary. 4. The Pathology File Processor retrieves JPEG images sent from the LIS. 5. The Pathology File Processor places JPEG images into the DICOM Wrapping Queue. 6. The DICOM Wrapping Service retrieves the next wrapping job, wraps the jpegs, and places the newly created DICOM in the DICOM Sending Queue. 7. DICOM Service Class User (SCU) retrieves the next send job and sends the wrapped DICOM to the Enterprise Image Server (EIS)
In regard to hardware, the pathology image server runs on an IBM VM server with a dual-core Intel Xeon processor (2.27 GHz), 3 GB of ram and Windows XP Enterprise (32-bit). The server on which the EDW resides is an IBM blade physical server with a dual-core Intel Xeon processor (3.6 GHz), 6 GB of RAM and Windows Server 2003 R2 (32-bit). More powerful servers were not chosen due to an overall low volume of daily cases - only hundreds of images per day, as opposed to tens of thousands of images.
RESULTS
In total, 26,966 gross images from 9,733 cases were transmitted from the laboratory information system to the EIS. The average process time for cases with successful automatic uploads (n=9,688) to the EIS was 98 seconds; these times represent the processing time for the patient data and images to run though the EDW system into the EIS, but does not include any processing time in the LIS. Only 45 cases (0.5%) failed, requiring a manual transmission by the information technology support staff; for these cases, the average process time was 1 day, 8 minutes and 10 seconds. More specifically, these 45 cases failed due to a reported “time synchronization” error between the batch process creating the text file and availability of the jpeg images. Moving the batch an hour or so past the end of the working day resolved these errors.
Uploaded images were immediately available to institution-wide PACS users. The graphic user interface used by clinicians to view these images is shown in Figure 4. Since inception, user feedback has been positive (informally evaluated via correspondence and at tumor boards) in several ways. First, digital gross pathology images were readily available to surgeons to help determine follow-up treatment protocols for their patients (e.g., transplant trials based on the amount and type of resected tissue). Second, surgeons no longer had to repeatedly contact pathologists to request gross pathology images of the specimens they removed. Third, these images were available for immediate review in subspecialty-based surgical oncology conferences and for counseling specific patients during clinic appointments. Access to these images has also allowed surgeons to more easily interpret the text-based gross description, including the locations of lesions and the orientation of the specimen, in the pathology reports they receive.
Figure 4.
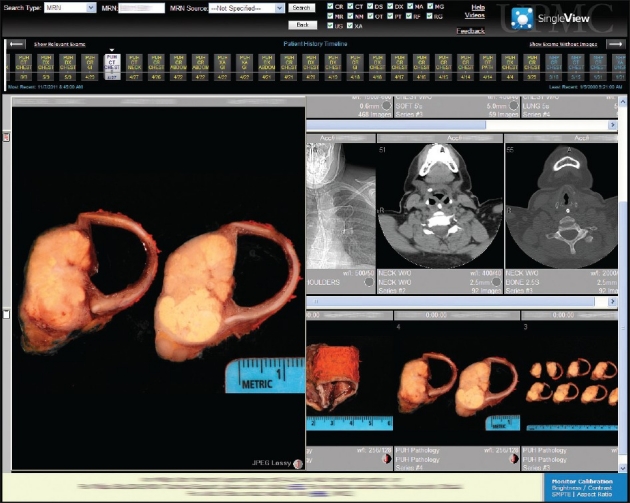
Screen capture showing the PACS graphic user interface used by clinicians to view radiology images. In this case of tracheal carcinoma, simultaneous access to both radiology and pathology images is possible via the same enterprise image server
DISCUSSION
We demonstrate that enterprise-wide PACS-based sharing of pathology images can be achieved. Digital gross pathology images in JPEG2000 format stored on our networked image server were accessible via the LIS only to the pathology department. In this format, which did not conform to the DICOM standard, the image files did not contain embedded patient metadata and thus could not be integrated into the PACS. However, employing EDW software to convert these files to DICOM format permitted digital gross pathology images to be made available outside the LIS to many PACS users. Pathology images were often available to institution-wide PACS users before many of the pathology cases were signed out. Nonetheless, the same holds true for any radiographic image in the PACS which is made immediately available to clinicians for viewing after acquisition, before the radiology report is even complete.
Any user with privileged access to the PACS can copy or save any image, such as for a presentation or publication, without obtaining permission from the pathology department. Once again, the same holds true for any radiology image stored in the PACS. Given this similarity, if dissemination of gross pathology images to the non-pathology community occurs, the expected consequences would hence be similar to the dissemination of radiology images to the non-radiology community. Yet, the availability of radiology images to non-radiology staff through PACS remains vast, in many hospitals nationwide. Given this widespread availability, the theoretical benefits of a radiology PACS appear to outweigh the risks of this dissemination. Also, there is no direct user interface that allows PACS users (including pathologists) to remove an image from the EIS. Once an image is permanently archived it cannot be removed from the system. If contacted, the information services department can “remove” an unwanted image from the user interface, but they cannot truly delete any images once they have been permanently stored as the image and associated information is still fully auditable and available on the EIS. Since the system relies on an automated process, if the resident or prosector who captured the image decides to post-process and re-upload an image the next day, both the original and edited image will appear in the medical record, and a manual request must be obtained to remove the original image. Despite these limitations, no reports of unauthorized use, security breaches or patient-image-data mismatches have been reported since the initiation of our enterprise-wide sharing of digital gross pathology images.
An implicit yet crucial assumption with this solution is that every obtained gross pathology image will be automatically available, without restrictions, institution- wide. Accompanying this assumption are potential drawbacks. For instance, images with poor focus or composition that are not deleted from the pathology image server will automatically appear on the EIS, with the potential to elicit dissatisfaction of clinicians with pathology services. Given this potential, all residents and technicians are educated on the importance of taking effective gross images, and replacing any suboptimal images with more effective ones at the time of grossing.
Of particular importance is that all images, regardless of content, are open to misinterpretation by any non- pathologist. For instance, a resection margin that appears grossly free may be microscopically involved, or a tumor appearing grossly benign may indeed be malignant on histologic examination. To our knowledge, literature sufficiently investigating this specific pitfall in pathology is limited, but the issue has been explored in radiology with notable results. A cross-sectional study evaluating the interpretation of computed tomography (CT) scans for head trauma by “middle grade and consultant” emergency physicians recommended that these physicians not interpret these radiologic images without additional training.[10] A study involving 202 observers comparing the interpretations of lung nodule detection by CT scanning highlighted statistically significant differences in detection rates between radiologists and non-radiologists, in cases without computer-assisted detection technologies.[11] Conversely, retrospective review of 3,886 consecutive CT examinations by fellows versus general staff non-neuroradiologists revealed an overall relatively low discrepant rate between fellowship trainees and generalist staff, as well as negligible change in clinical management, though the discrepant rate was higher (greater than 5%) for those in the early months of fellowship training.[12] Over several years at our institution, we are not aware of any cases of inadequate patient care as a result of misinterpretation of gross pathology images. Nonetheless, the possibility of this occurring in the future cannot be entirely excluded. Excellent rapport between pathologists and clinicians, including discussion and reminders of referring to the final diagnostic pathology report for clinical decision making, is key to avoiding misinterpretation of digital gross images, and subsequent misguided patient care.
In addition to the aforementioned hardware and software requirements, two programmers were required to implement this service. This was a full time job for the senior programmer and a half time job for the junior, with costs amounting to 1.5 full time equivalents over a total of 15 months of project implementation time. Additional duties by programming staff included gathering application requirements, designing, manual coding, testing, documentation of progress, software deployment, and training of users. After implementation of this service, many of these duties were no longer needed. Ongoing maintenance of the service was accomplished by already employed members of our institutional information technology department, thereby reducing the need for additional costs after the implementation of this service.
CONCLUSIONS
Based on our experience, enterprise-wide PACS-based sharing of pathology digital images is feasible, at least in a large academic hospital setting. As we have demonstrated, this service can be cost effective if existing technology and communications infrastructure can be leveraged. The LIS-PACS partnership described herein, however, involved significant resource commitments, including 15 months of in-house programming time. Similar commercially available methods have since emerged.[13,14] While there is certainly much potential with digital pathology, one of the challenges facing many institutions remains the feasibility of permanently archiving large volumes of pathology images for long periods of time because of their size, compared to imaging from most other disciplines. Advantages of utilizing DICOM compliant digital pathology images include the ease of image distribution to a PACS, downstream multi-system compatibility with other electronic health records, and widespread sharing of images intended to improve client satisfaction and hopefully improve patient care. There is a concerted effort for institutions to begin leveraging their PACS beyond the radiology department for enterprise-wide initiatives such as integration of digital images into the EMR, building decision support tools, supporting quality assurance programs and as a research tool.[15] The LIS-PACS partnership at our institution continues to grow, with future plans for digital gross pathology images acquired at all of our centers to be sent to the PACS, to facilitate enterprise-wide sharing of digital photomicrographs, and perhaps even whole-slide images given the recent push to link these images with DICOM[16] and for PACS to handle whole-slide microscopic images.[17]
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2012/3/1/10/93892
REFERENCES
- 1.Ralston MD, Coleman RM. Introduction to PACS. In: Branstetter BF, Rubin DL, Griffin S, Weiss DL, editors. Practical imaging informatics: Foundations and applications for PACS professionals. Vol. 3. New York: Springer; 2009. pp. 33–48. [Google Scholar]
- 2.Ratib O, Ligier Y, Scherrer JR. Digital image management and communication in medicine. Comput Med Imaging Graph. 1994;18:73–84. doi: 10.1016/0895-6111(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 3.Bergh B. Enterprise imaging and multi-departmental PACS. Eur Radiol. 2006;16:2775–91. doi: 10.1007/s00330-006-0352-9. [DOI] [PubMed] [Google Scholar]
- 4.Duncan LD, Gray K, Lewis JM, Bell JL, Bigge J, McKinney JM. Clinical integration of picture archiving and communication systems with pathology and hospital information system in oncology. Am Surg. 2010;76:982–6. [PubMed] [Google Scholar]
- 5.Le Bozec C, Henin D, Fabiani B, Schrader T, Garcia-Rojo M, Beckwith B. Refining DICOM for pathology-progress from the IHE and DICOM pathology working groups. Stud Health Technol Inform. 2007;129:434–8. [PubMed] [Google Scholar]
- 6.Daniel C, Macary F, Rojo MG, Klossa J, Laurinavičius A, Beckwith BA, et al. Recent advances in standards for Collaborative Digital Anatomic Pathology. Diagn Pathol. 2011;6(Suppl 1):S17. doi: 10.1186/1746-1596-6-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh R, Chubb L, Pantanowitz L, Parwani A. Standardization in digital pathology: Supplement 145 of the DICOM standards. J Pathol Inform. 2011;2:23. doi: 10.4103/2153-3539.80719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morelli S, Grigioni M, Giovagnoli MR, Balzano S, Giansanti D. Picture archiving and communication systems in digital cytology. Ann Ist Super Sanita. 2010;46:130–7. doi: 10.4415/ANN_10_02_05. [DOI] [PubMed] [Google Scholar]
- 9.Kalinski T, Hofmann H, Franke DS, Roessner A. Digital Imaging and electronic patient records in pathology using an integrated department information system with PACS. Pathol Res Pract. 2002;198:679–84. doi: 10.1078/0344-0338-00320. [DOI] [PubMed] [Google Scholar]
- 10.Boyle A, Staniciu D, Lewis S, Hugman A, Bauza-Rodriguez B, Kirby D, et al. Can middle grade and consultant emergency physicians accurately interpret computed tomography scans performed for head trauma.Cross-sectional study? Emerg Med J. 2009;26:583–5. doi: 10.1136/emj.2008.067074. [DOI] [PubMed] [Google Scholar]
- 11.Brown MS, Goldin JG, Rogers S, Kim HJ, Suh RD, McNitt-Gray MF, et al. Computer-aided lung nodule detection in CT: results of large-scale observer test. Acad Radiol. 2005;12:681–6. doi: 10.1016/j.acra.2005.02.041. [DOI] [PubMed] [Google Scholar]
- 12.Le AH, Licurse A, Catanzano TM. Interpretation of head CT scans in the emergency department by fellows versus general staff non- neuroradiologists: a closer look at the effectiveness of a quality control program. Emerg Radiol. 2007;14:311–6. doi: 10.1007/s10140-007-0645-6. [DOI] [PubMed] [Google Scholar]
- 13.Ladin M, Geertrui DS, Jacobs J. Agfa HealthCare unveils integrated digital pathology/PACS solution at RSNA. AGFA HealthCare. [Last accessed on 2011 Dec 9]. Available from: http://agfahealthcare.com/usa/en/main/news_events/news/archive/he20111129_lapitie_paris.jsp .
- 14.Apollo PACS. PathPACS: Apollo Enterprise Patient Media Manager. Apollo Enterprise Patient Media Services. [Last accessed on 2011 Dec 9]. Available from: http://apollopacs.com/products-solutions/pathpacs.php .
- 15.Stewart BK. Picture archiving and communication systems. In: Hendee WR, editor. Informatics in medical imaging. Vol. 16. United Kingdom: Taylor Andand Francis, MA; 2012. pp. 235–49. [Google Scholar]
- 16.Tuominen VJ, Isola J. Linking whole-slide microscope images with DICOM by using JPEG2000 interactive protocol. J Digit Imaging. 2010;23:454–62. doi: 10.1007/s10278-009-9200-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Punys V, Laurinavicius A, Puniene J. A data model for handling whole slide microscopy images in picture archiving and communications systems. Stud Health Technol Inform. 2009;150:856–60. [PubMed] [Google Scholar]


