Abstract
The evolution of novelty in tightly integrated biological systems, such as hormones and their receptors, seems to challenge the theory of natural selection: it has not been clear how a new function for any one part (such as a ligand) can be selected for unless the other members of the system (e.g., a receptor) are already present. Here I show—based on identification and phylogenetic analysis of steroid receptors in basal vertebrates and reconstruction of the sequences and functional attributes of ancestral proteins—that the first steroid receptor was an estrogen receptor, followed by a progesterone receptor. Genome mapping and phylogenetic analyses indicate that the full complement of mammalian steroid receptors evolved from these ancient receptors by two large-scale genome expansions, one before the advent of jawed vertebrates and one after. Specific regulation of physiological processes by androgens and corticoids are relatively recent innovations that emerged after these duplications. These findings support a model of ligand exploitation in which the terminal ligand in a biosynthetic pathway is the first for which a receptor evolves; selection for this hormone also selects for the synthesis of intermediates despite the absence of receptors, and duplicated receptors then evolve affinity for these substances. In this way, novel hormone-receptor pairs are created, and an integrated system of increasing complexity elaborated. This model suggests that ligands for some “orphan” receptors may be found among intermediates in the synthesis of ligands for phylogenetically related receptors.
According to the neodarwinian theory of evolution, novel functions arise as the phenotypic outcome of natural selection acting on random mutations. Complex organs and functions are thought to be the result of a gradual selective process of elaboration and optimization (1). Tightly integrated systems of interacting parts, such as those that characterize much of metazoan biology at the molecular level, pose an apparent challenge to this theory, because it is not clear how a new function for any protein can be selected for unless the other members of the complex are already present (2).
Vertebrate steroid hormones and the intracellular protein receptors that mediate their cellular effects elegantly illustrate this problem. In the absence of a ligand, what function does a new receptor serve? And without a receptor, what selection pressures guide the evolution of a new ligand? The six related steroid receptors in vertebrates—the estrogen receptors alpha and beta (ERα and ERβ), progesterone receptor (PR), androgen receptor (AR), glucocorticoid receptor (GR), and mineralocorticoid receptor (MR)—were created by a series of duplications from a common ancestral receptor gene (3). The classic model suggests that duplicated genes rapidly become pseudogenes unless they are subject to unique selection pressures (4). In theory, new receptors may evolve simultaneously with new ligands (5), or gene duplications may allow multifunctional proteins to take on greater specificity (6).
The history of steroid receptor diversification remains largely unknown. No steroid receptors have been found in any species outside the vertebrates, although an ortholog of the estrogen-related receptor (ERR), the nuclear receptor most closely related to the steroid receptors (3), is present in the Drosophila melanogaster genome (Genpept sequence 2891028). (Orthologs are related genes in different genomes, descended from a speciation event; paralogs are related genes in the same genome, descended from a gene duplication.) Orthologs of all steroid receptors present in tetrapods have been identified in teleosts (7, 8), indicating that all six types existed by the time ray-finned fish split from the lineage leading to tetrapods some 400 million years ago (9). PCR screens have identified short fragments of an ER, GR, and AR in shark and a single steroid receptor of indeterminate type in hagfish (8), but this approach cannot distinguish a failure to amplify a gene from its true absence in an organism. I have therefore identified steroid receptors in the sea lamprey Petromyzon marinus, which diverged from the jawed vertebrates (gnathostomes) about 450 million years ago (9). Because other gene families contain fewer members in the lamprey than in gnathostomes (10), I anticipated that lamprey would contain a relatively ancient subset of steroid receptors. I used an extensively parallel PCR screen to identify steroid receptor sequences and a phylogenetic approach to determine whether all steroid receptors orthologous to those in extant vertebrates had been obtained. The sequences and functional characteristics of ancestral receptor proteins were reconstructed to illuminate the timing and mechanisms by which the steroid receptor family achieved its current diversity.
Methods
Molecular Methods.
Total RNA was extracted from the liver of adult sea lampreys with RNA-zol (Tru-tetst) and reverse transcribed (Superscript from GIBCO). An EcoRI-digested cDNA library was prepared in lambda-ZAPII (Stratagene). Degenerate PCR was conducted with ramped temperature profiles (11). For each receptor, at least ten degenerate primers (five in each direction) were used in nested PCR in up to all possible combinations; primers and temperatures are available on request. Products were cloned into pCR2.1 (Invitrogen) and sequenced automatically in both directions. To obtain the entire DNA-binding domain, hinge, and ligand-binding domain, the RACE (rapid amplification of cDNA ends) technique (12) was modified for use on a cDNA library, using nested gene-specific primers that anneal to degenerate PCR products and universal primers that anneal to sequences in the phage.
Sequences, Alignment, and Phylogenetic Analysis.
Amino acid sequences of the DNA- and ligand-binding domains of lamprey receptors were inferred and aligned to those of 70 other publicly available steroid and related receptors (see Table 2, which is published as supplemental data on the PNAS web site, www.pnas.org). An elision alignment (13) was prepared in clustalx (14) with gap:change costs in the series 1, 2, 4, . . . , 32 by using the Gonnett weight model. Results that failed to align the AF-2 activation function, which is conserved among all nuclear receptors, were discarded; the remaining alignments (costs 2 to 32) were assembled into a master data matrix. Phylogenetic analyses using parsimony were conducted in PAUP* (15) by using heuristic strategies of multiple random addition and tree bisection–reconnection. A stepmatrix was prepared from the Gonnett model of amino acid transformation (16) by setting diagonal elements to zero and all other elements to the probability of each replacement type if a replacement occurred, then correcting for triangle inequalities. To find the most parsimonious gene family phylogeny, tree lengths were calculated as L = A + wD, where A is the number of amino acid replacements, D the number of gene duplications and losses in the reconciled tree (17), and w the weight of a gene duplication/loss relative to a replacement (18). An initial and conservative value of 10 was chosen for w, because duplications and losses of entire genes are expected to occur much less frequently than amino acid changes; the impact of higher and lower values of w was explored analytically. Analyses were conducted without constraint and with multiple topological constraints that limited searches to all possible trees that require fewer gene duplications/losses than the most parsimonious unconstrained tree. Tree lengths, branch lengths, and branch supports were normalized by the average cost of an amino acid change, calculated by dividing the length of the most parsimonious tree with the stepmatrix in effect to the length of that same tree when characters were treated as unordered. Trees were rooted on three nuclear receptor subfamilies closely related to the steroid receptors (3). Confidence in individual nodes was calculated as branch support values—the number of extra steps required in the most parsimonious tree in which that clade does not appear (19)—by using auto-decay software (20).
Reconstruction of Ancestral Sequences, Functions, and Branch Lengths.
Maximum likelihood sequences of ancestral receptors and branch lengths were reconstructed on the most parsimonious phylogeny with paml software (21)—using a single alignment of 45 steroid receptor sequences for computational efficiency—the Jones amino acid transformation model, and an iteratively estimated gamma distribution of rates (α = 0.74652, four categories). Aspects of the ligand specificity of extant receptors were coded as characters and reconstructed for ancestral receptors on the same tree by the parsimony method (22). Ratios of relative rates of sequence divergence were calculated from Poisson-corrected amino acid distances, based on the mean distances of all pairs of ingroup and outgroup sequences (23). The departure of the ratio of means from unity was evaluated by a two-sample t test assuming unequal variances. Teleost receptors were excluded from relative rate tests because of possible rate anomalies after an additional genome-wide duplication (24).
Identification of Paralogous Groups Syntenous with Steroid Receptors.
From the OMIM (On-line Mendelian Inheritance in Man) database (http://www.ncbi.nlm.nih.gov), the chromosomal locations of human AR, PR, GR, and MR were ascertained, and the list of genes mapped to the same chromosomes was evaluated for other potential groups of tetralogous genes (25), based on similarity of name among genes shared on two or more of the same chromosomes. Paralogy was verified by blast searches of all available human protein sequences (critical value E < 0.001) (26), and families with greater than eight members in the human genome were excluded.
Results and Discussion
Lampreys Contain an Ancient Subset of Steroid Receptors.
An extensively parallel PCR strategy yielded lamprey steroid receptors when primers designed from gnathostome PR, ERα, and GR were used. The first receptor was most similar to the gnathostome PR and was named lamprey PR; the second was similar to gnathostome ERα and ERβ and was named lamprey ER; the third was similar to both of the vertebrate corticoid receptors MR and GR and was named lamprey CR (Table 1). For those receptors that were not amplified, reactions were repeated under varying conditions, and additional primers were designed when possible. Despite more than 200 unique degenerate PCR reactions, no authentic steroid receptor sequences were amplified for the ERβ, AR, or MR.
Table 1.
Pairwise similarities among extant and reconstructed steroid receptor sequences
| humERα | humERβ | humPR | humGR | humMR | humAR | |
|---|---|---|---|---|---|---|
| Lamprey ER | 63 | 60 | 30 | 32 | 30 | 30 |
| Lamprey PR | 29 | 30 | 66 | 58 | 65 | 59 |
| Lamprey CR | 33 | 32 | 60 | 58 | 60 | 58 |
| AncSR1 | 71 | 68 | 35 | 36 | 33 | 33 |
| AncSR2 | 32 | 30 | 77 | 68 | 72 | 67 |
The percent of identical amino acid sites in the combined DNA- and ligand-binding domains is shown for each pair.
A PCR screen cannot exclude the possibility that orthologs to these receptors exist in the lamprey genome but were not recovered because of a lack of expression or extreme sequence divergence. Gene family phylogenetic analysis, however, can determine the timing of gene duplication events relative to speciation events and thereby offers a powerful method to distinguish a false negative PCR result from a real lack of these receptors in any taxon (27). The most parsimonious phylogeny of the steroid receptor gene family was inferred based on the sequences of the three lamprey receptors and 70 other publicly available sequences. Any gene family tree implies a certain number of gene duplications and losses, and genealogical inference should take account of both sequence evolution and changes in the presence/absence of a gene. In a phylogenetic context, the most parsimonious and therefore best supported tree is the one that minimizes the sum of weighted amino acid replacements and gene duplications/losses (18). When using this approach and a conservative cost ratio for duplications/losses to amino acid replacements, the single most parsimonious tree of the steroid receptors (Fig. 1) implies no extra duplications and losses beyond the minimum required to explain the distribution of receptors in lamprey and gnathostomes. This phylogeny is the most parsimonious for any and all cost ratios greater than 3, an implausibly low value, because amino acid replacements are almost certainly more than three times as likely as duplication or loss of an entire gene. The tree is well supported, with 3 to 24 extra amino acid changes required to impose alternative relationships at any of the nodes relevant to this analysis, indicating that the sequence data's support for this phylogeny is unlikely to be due to chance effects or phylogenetic noise.
Figure 1.
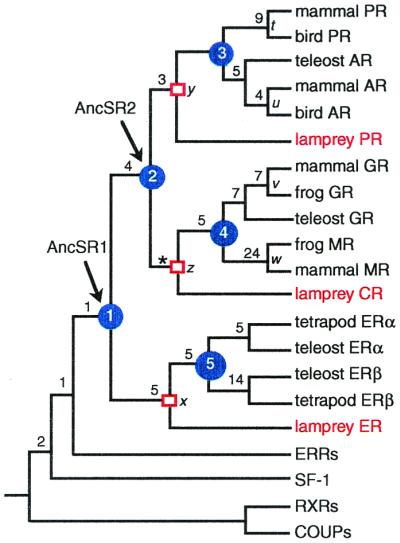
Phylogeny of the steroid receptor gene family. A reduced version is shown of the single most parsimonious phylogeny of 73 receptor sequences when the relative weight of gene duplications/losses to amino acids w > 3. (Length = 3,209 aa changes + 8 duplications + 0 losses. For unreduced phylogeny, see Fig. 7, which is published as supplemental data.) Support for each clade is shown as the number of extra steps required for the labeled node not to appear in the most parsimonious tree (19); all support values are insensitive to w except *, shown for w = 10. Blue circles indicate gene duplications within the steroid receptor family; red squares mark the lamprey–gnathostome divergence; and unmarked nodes represent other speciation events. Ancestral steroid receptors are indicated. Italicized node labels correspond to Fig. 3. Tree length = 3,209 aa changes, eight duplications, zero losses; consistency index = 0.628; retention index = 0.870.
The steroid receptor phylogeny indicates that unique orthologs to the AR, MR, and ERβ were not recovered from the lamprey because these receptors were created by gene duplication in the jawed vertebrate lineage, after the lamprey–gnathostome divergence. If the AR gene, for example, had appeared by duplication before this cladogenetic event, then the lamprey PR would form a clade with the gnathostome PRs to the exclusion of the gnathostome ARs; the phylogeny obtained here, however, shows that the lamprey–gnathostome divergence occurred before the gene duplication that created the AR. By similar reasoning, lampreys must possess one estrogen receptor ancestral to the gnathostome ERα and ERβ, and one corticoid receptor ancestral to the GR and MR. This analysis does not rule out the possibility of independent gene duplications in the lamprey lineage that may have created other novel receptor paralogs, but it does indicate that the three sequences recovered represent the entire complement of steroid receptors orthologous to the six found in jawed vertebrates.
Steroid Receptors Diversified in Two Serial Genome Expansions.
The receptor phylogeny suggests that two serial duplications of an ancestral steroid receptor occurred before the divergence of lamprey and jawed vertebrates. The first created an estrogen receptor and a 3-ketosteroid receptor, whereas the second duplicated the latter gene to produce a corticoid receptor and a receptor for 3-ketogonadal steroids (androgens, progestins, or both). The ancestral vertebrate therefore had three steroid receptors—an estrogen receptor, a receptor for corticoids, and a receptor that bound androgens, progestins, or both. At some later time within the gnathostome lineage, each of these three receptors duplicated yet again to yield the six steroid receptors currently found in jawed vertebrates: the ER to create ERα and ERβ, the corticosteroid receptor to yield the GR and the MR, and the 3-ketogonadal steroid receptor to create the PR and the AR.
This finding is consistent with the hypothesis that the genome of “higher” vertebrates is the result of two genome duplication events that occurred early in chordate evolution (24, 28). The timing of these events has remained unclear, however, because the number of members of other gene families in the lamprey has been consistent with two competing hypotheses: two duplications before the lamprey–gnathostome divergence, or one duplication before and one after (29). The number of steroid receptors in lamprey and their phylogenetic relations to gnathostome sequences clearly support the latter hypothesis.
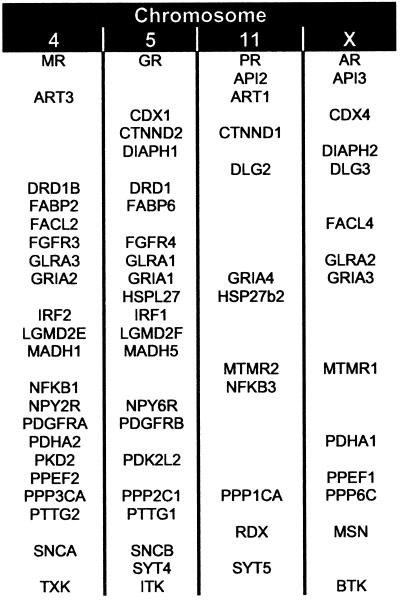
Gene mapping data also support serial genome duplications as the mechanism by which steroid receptors diversified. When a gene family is created by large-scale duplications rather than by local processes like tandem duplication or transposition, its members will be syntenous—mapped to the same chromosome—with members of other gene families that proliferated in the same events. Redundant genes will often be lost after gene duplications; therefore, many of the paralogous groups that result from two rounds of gene duplication are expected to contain two or three rather than four members (25). The complete human genome sequence suggests that large-scale block duplications have occurred, but mapping data from a single species are not enough to distinguish whole-genome duplications from regional copying of chromosomes or their parts (30). Mapping data from the human genome (Fig. 2) indicate that the AR, PR, GR, and MR on chromosomes X, 4, 5, and 11 are syntenous with members of at least 30 other gene families. The actual number of syntenous gene families is likely to be considerably higher, because the identification criteria used were conservative. The large number of linked paralogous groups indicates that the steroid receptors diversified as the result of two rounds of large-scale genome expansion rather than by gene-specific mechanisms like transposition or tandem duplication. Complete genome sequences from lamprey and other gnathostomes, along with development of numerical models of the chromosomal distribution of genes, are required for statistical testing of this hypothesis.
Figure 2.
Steroid receptors diversified by large-scale genome expansions. Paralogous members of gene families with two or more members on the same chromosomes as the human 3-ketosteroid receptors are shown, without regard to map order.
The Ancestral Steroid Receptor Was an Estrogen Receptor.
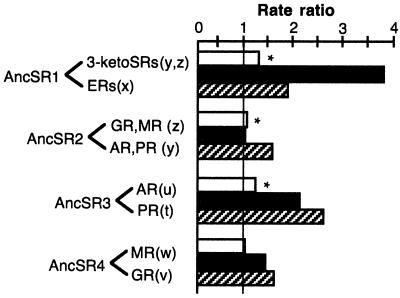
The classical model of gene duplication suggests that redundancy will free one member of the resulting pair of genes from the constraints imposed by natural selection, and its sequence will diverge rapidly, yielding either a pseudogene or, less commonly, a gene with novel functions, the sequence of which will then be constrained again by selection (4). An ancestral protein is likely to have been most similar in sequence and therefore in function to the descendant gene that diverged more slowly after the duplication event than the one with a more rapid evolutionary rate. Relative rate tests based on amino acid distances and reconstruction of branch lengths by parsimony and maximum likelihood all indicate that the rate of amino acid replacement after duplication of the ancestral steroid receptor (AncSR1) was up to four times greater in the lineage leading to the 3-ketosteroid receptors than in that leading to the estrogen receptors, a result that is statistically significant at a P = 0.001 level in the relative rate test (Fig. 3). The gross difference in sequence divergence rates suggests that that the ancestral steroid receptor was a functional estrogen receptor, the sequence of which was conserved among descendant ERs.
Figure 3.
Divergence rates of steroid receptor sequences after gene duplications indicate that the first steroid receptor was an estrogen receptor. Grouped bars show the ratio of the rate of amino acid replacement on the upper branch to that in the lower. White bars, rate ratio based on the relative rate test; black bars, ratio of parsimony branch lengths; hatched bars, ratio of maximum likelihood branch lengths. Outgroups for relative rate tests, from top to bottom, are estrogen-related receptors (ERRs), tetrapod ERs, lamprey PR, and lamprey GR. *, statistically significant departure from unity, P < 0.001. Parsimony and likelihood branch lengths are proportional to the number of weighted amino acid changes on paralogous branches that descend from duplication of an ancestral steroid receptor to an equivalent speciation event, with labels corresponding to nodes in Fig. 1.
The amino acid sequence of the ancestral steroid receptor was reconstructed with a maximum likelihood approach; this ancestral sequence, interpreted in light of structure-function data on extant vertebrate receptors, strongly support the inference that AncSR1 was an estrogen receptor. The reconstructed ancestral receptor is 71% identical to the human ERα, but radically less similar to the PR, AR, GR, and MR (Table 1). In its DNA-binding domain, the ancestral receptor shares 61 of 66 residues with the human ERα, but no more than 41 with any of the nonestrogenic receptors (see Fig. 6, which is published as supplemental data). The ancestral sequence's P-box, the sequence of which determines the distinct specificity of the ER and the GR for their respective target sequences (31), has the exact sequence of estrogen receptors rather than that found in the AR, PR, GR, and MR (Fig. 6). This result indicates that the ancestral steroid receptor activated genes with estrogen-response elements (a palindrome of AGGTCA) rather than those with the response elements recognized by the other steroid receptors (a palindrome of AGAACA). These data strongly suggest that ancestral steroid receptor bound estrogens and activated genes regulated by classic estrogen-response elements.
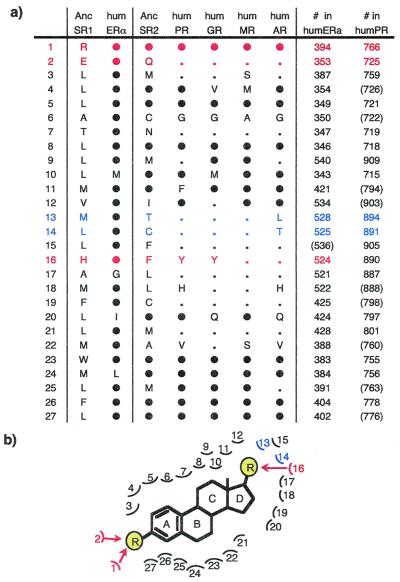
Reconstructions for specific amino acid positions that contact hormone in the crystallized human ERα provide more specific support (Fig. 4). Of 20 positions that vary among steroid receptors, 16 contain the residue characteristic of the estrogen receptor, and all four exceptions are conservative replacements. Most tellingly, twelve of the residues that AncSR1 shares with the ER are diagnostic of estrogen receptors in that all other steroid receptors have different amino acids at these positions. The reconstructed ancestral receptor has the ER-characteristic residue at the critical position that forms hydrogen bonds with and discriminates between the 3-hydroxyl group of estrogens and the 3-keto group of all other steroid hormones, as well as at the position that bonds with and discriminates the 17-hydroxyl group of estrogens from the methylketo moiety of progesterone and corticoids (32, 33). At seven of the eight positions that surround the steroid D-ring, which varies among estrogen, progesterone, and corticoids, the ancestral receptor's amino acid is identical to the ER but different from the other receptors. If the estrogen receptor is the most ancient of all steroid receptors, it is likely also to have the widest taxonomic distribution, suggesting that a broad array of taxa may be potentially sensitive to synthetic environment pollutants that interact with the estrogen receptor (34).
Figure 4.
Maximum likelihood reconstructions of ancestral sequences indicate that the first steroid receptor was an estrogen receptor and the first 3-ketosteroid receptor was not an androgen receptor. (a) Aligned amino acids forming the ligand-binding pocket of the ancestral steroid receptor (AncSR1), the ancestral 3-ketosteroid receptor (AncSR2), and five human steroid receptors, based on homology to human steroid receptors with solved structures. Colors and residue numbers refer to the positions shown in b. Filled circles (●) indicate amino acids identical to AncSR1; small dots are identical to AncSR2 but not AncSR1. Red, amino acids making direct hydrogen bonds with ligands (32, 33); blue, residues critical to discriminate androgens from C21 steroids in the androgen receptor (35). Amino acid numbers of homologous positions in the crystallized human receptors are at right; parentheses indicate positions that do not contact ligand in the indicated receptor. (b) Schematic of the ligand-binding pocket of ancestral steroid receptors with generic steroid hormone, based on homology to the crystal structures of human ERα and human PR (32, 33). Red and blue residues as in a. Yellow circles marked “R” indicate substituents that vary among steroid hormones.
The Second Receptor to Evolve Was a Progesterone Receptor.
Maximum likelihood sequence reconstructions indicate strongly that the ancestral 3-ketosteroid receptor (AncSR2) bound C21 steroids—progestins or corticoids—and not C19 androgens, and they suggest that this protein was a progesterone receptor. The combined DNA- and ligand-binding domains are 81% identical to the human PR, but considerably less similar to the AR, GR, and MR (Table 1). Of the amino acids that form the ligand-binding pocket in the human PR and are not shared with the AR, GR, and MR, the ancestor shares more residues with the PR—and fewer with the AR—than with any other extant receptor (Fig. 4). Positions that align with C891 and T894 in the human PR are critical, because crystallographic studies show that these amino acids contact the C20 keto group unique to progesterone and corticoids (33), and mutation of the former residue in the human AR to that found in the other receptors causes the AR to bind and trans-activate in the presence of progesterone and corticoids (35). At both of these positions, the reconstructed ancestor contains residues characteristic of the PR, GR, and MR, but not AR, indicating that it bound C21 steroids rather than C19 androgens like testosterone or dihydrotesterone.
Branch length comparisons support this inference. By all methods of calculation, the rate of sequence divergence after the duplication of the common ancestor of the androgen and progesterone receptors (AncSR3) is considerably greater in the lineage leading to the AR than in that leading to the PR, and the difference is statistically significant in the relative rate tests (Fig. 3). This result suggests that the androgen receptor is a more recent evolutionary novelty than the progesterone receptor.
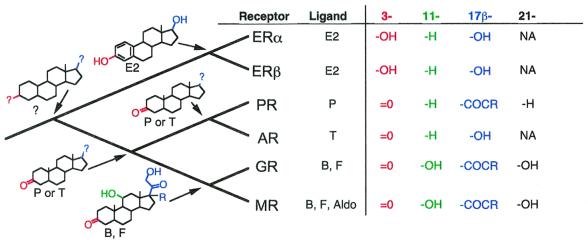
Reconstruction of the ability of ancestral receptors to bind specific ligands by using a parsimony-based algorithm, which explains shared states as due to descent from a common ancestor, indicates that the ancestral 3-ketosteroid receptor was a PR (Fig. 5). This reconstruction shows that AncSR2 did not bind corticoids; the capacity to bind 11- and 21-hydroxylated steroids evolved later on the branch leading to the GR and the MR. If the AR was a late evolutionary novelty—as both branch lengths and sequence reconstructions show—and if the ancestral 3-ketosteroid receptor bound progestins or androgens but not corticoids, then the second receptor to evolve would have been a PR.
Figure 5.
Reconstruction of ligand-binding characteristics of ancestral steroid receptors indicates that the ancestral 3-ketosteroid receptor did not bind corticoids. Substituents at critical positions of the ligands that each vertebrate receptor binds were coded as characters (Right). Character states at ancestral nodes were reconstructed on the reduced phylogeny of steroid receptor paralogs with a parsimony-based algorithm, and the inferred structures of the ligands bound by each ancestral receptor are shown (Left); colored groups correspond to characters in the matrix. ?, substituent groups that could not be unambiguously reconstructed; NA, not applicable. P, progesterone; E2, estradiol; T, testosterone; B, corticosterone; F, cortisol; Aldo, aldosterone.
Ligand Exploitation: A Mechanism for the Evolution of Endocrine Complexity.
These findings support a model for hormone/receptor evolution in the steroid receptor family, the dynamics of which may also apply to other kinds of receptors and their ligands. If, as I have shown, the first receptor to evolve was an estrogen receptor, then the terminal hormone in the pathway for steroid biosynthesis was the first to function as a hormone acting through an intracellular receptor. In the synthesis of estradiol and other estrogens, progesterone and testosterone are synthesized as intermediates. These steroids (and the enzymes that produce them) would therefore have been present during the period when only a receptor for estrogen existed. After one to three duplications of the estrogen receptor gene, followed by considerable sequence divergence, receptors emerged that gave these intermediate compounds novel signaling functions. The advent of corticoid signaling would have required enzymes for 21- and 18-hydroxylation to be added to the pathway.
This evolutionary history provides one solution to the problem posed by the classical model—how can a ligand or a receptor be maintained without the other in a system governed by natural selection? Once an organism depended on estrogen/ER signaling for physiological or developmental functions that contribute to fitness, then the same constraints that selected for the synthesis of estrogen and its receptor would by necessity have selected for the synthesis of other steroids in the pathway, although none of them yet signaled through nuclear receptors. Redundant receptors created by gene duplication could then diverge in sequence from their ancestors and evolve affinity for these steroids, creating signaling functions for what were once intermediates. I call this process ligand exploitation, because it involves the cooption of existing metabolites to serve as novel hormones by duplicated receptors; this model reverses the evolutionary dynamics previously proposed for hormone-receptor evolution (36). Ligand exploitation can occur whether ancestral receptors regulated cellular processes through direct transcriptional activation, via signal transduction pathways in the cytosol or membrane, or both, as extant steroid receptors do (37, 38). If ligand exploitation is a general mechanism for the evolution of new receptor-hormone pairs, then ligands for some “orphan” receptors may be found among intermediates formed in the synthesis of ligands for phylogenetically related receptors.
Evolution of Endocrine Specificity.
The elaboration of the steroid receptor family by gene duplication and ligand exploitation allowed increasingly specific hormonal control over physiological functions. Estrogen regulation, presumably of reproductive maturation and function, appears to be the most ancient of all modes of steroid/receptor control, a conclusion supported by the apparent role of estrogen in branchiostome and echinoderm reproduction (39, 40). Progesterone control over ovulation, oviposition, or other aspects of reproduction also appears to be quite ancient, as indicated by the presence of corpora atretica and lutea in hagfish ovaries (41), but not as old as estrogen signaling. Hormonal control over sexual dimorphism appears to be a relatively recent evolutionary novelty: if the androgen receptor was created by a gene duplication after the lamprey lineage diverged from other vertebrates, then androgen-mediated masculinization and estrogen-mediated feminization must be unique to the gnathostomes. Supporting this view, the lamprey testis binds estradiol but not androgens with high affinity (42); in both male and female lamprey, estradiol regulates reproductive maturation and behavior, but androgens do not appear to play any role, and plasma levels of neither hormone are sexually dimorphic (43, 44).
Separate control over osmolarity and response to stress must also have arisen after the lamprey–gnathostome divergence. In many jawed vertebrates, including mammals, osmolarity is regulated by the MR, whereas the GR controls long-term stress response (45). Lampreys, with a single CR, would be expected to have no ability to use steroids for independent regulation of these functions. Corticoids are found in plasma and appear to regulate osmolarity in lamprey and hagfish, but no research on their glucocorticoid effects is available (46, 47). The spawning behavior of most lamprey species, however, involves a migration from marine to freshwater environments accompanied by extreme and fatal changes in carbohydrate and protein metabolism, consistent with coordinate control of these functions. Independent regulation of the many physiological functions controlled by steroids in jawed vertebrates therefore appears to have been gradually elaborated from an ancient mechanism for estrogen regulation, as receptor genes duplicated, diverged, and exploited the middle steps of a biosynthetic pathway that was stabilized by natural selection acting on its endpoint.
Supplementary Material
Acknowledgments
I thank Darcy Kelley and Rob DeSalle for support and guidance, Darcy Kelley for generously hosting this research, Stacia Sower for lamprey tissues and comments. I also thank Matt Scherer, Emily Glick, Kwok Wu, and Bill Hahn for technical assistance and advice. This work was supported by National Science Foundation Grant DEB-98-70055.
Abbreviations
- ER
estrogen receptor
- AR
androgen receptor
- PR
progesterone receptor
- GR
glucocorticoid receptor
- MR
mineralocorticoid receptor
- AncSR
ancestral steroid receptor
Footnotes
References
- 1.Dawkins R. The Blind Watchmaker. New York: W. W. Norton; 1986. [Google Scholar]
- 2.Kauffman S A. The Origins of Order: Self-Organization and Selection in Evolution. Oxford: Oxford Univ. Press; 1993. [Google Scholar]
- 3.Thornton J W, DeSalle R. Syst Biol. 2000;49:183–201. [PubMed] [Google Scholar]
- 4.Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970. [Google Scholar]
- 5.Fryxell K J. Trends Genet. 1996;12:364–369. doi: 10.1016/s0168-9525(96)80020-5. [DOI] [PubMed] [Google Scholar]
- 6.Force A, Lynch M, Pickett F B, Amores A, Yan Y-L, Postlethwaite J H. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colombe L, Fostier A, Bury N, Pakdel F, Guiguen Y. Steroids. 2000;65:319–328. doi: 10.1016/s0039-128x(00)00090-8. [DOI] [PubMed] [Google Scholar]
- 8.Escriva H, Safi R, Hanni C, Langlois M C, Saumitou-Laprade P, Stehelin D, Capron A, Pierce R, Laudet V. Proc Natl Acad Sci USA. 1997;94:6803–6808. doi: 10.1073/pnas.94.13.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colbert E H, Morales M. Evolution of the Vertebrates: A History of the Backboned Animals Through Time. 4th Ed. New York: Wiley-Liss; 1991. [Google Scholar]
- 10.Sharman A C, Holland P W. Int J Dev Biol. 1998;42:617–620. [PubMed] [Google Scholar]
- 11.Compton T. In: PCR Protocols: A Guide to Methods and Applications. Innis M A, Gelfand D H, Sninsky J J, White T, editors. London: Academic; 1990. pp. 39–45. [Google Scholar]
- 12.Frohman M A. In: PCR Protocolos: A Guide to Methods and Applications. Innis M A, Gelfand D H, Sninsky J J, White T, editors. London: Academic; 1991. pp. 28–38. [Google Scholar]
- 13.Wheeler W C, Gatesy J, DeSalle R. Mol Phylogenet Evol. 1995;4:1–9. doi: 10.1006/mpev.1995.1001. [DOI] [PubMed] [Google Scholar]
- 14.Thompson J D, Higgins D G, Gibson T J. CLUSTALX, multiple sequence alignment program. Hamburg, Germany: European Molecular Biology Organization; 1997. , Version 1.63b. [Google Scholar]
- 15.Swofford D. PAUP* (Phylogenetic Analysis Using Parsimony and Other Methods) Sunderland, MA: Sinauer; 2000. , Version 4.0 beta. [Google Scholar]
- 16.Gonnett G H, Cohen M A, Benner B A. Science. 1992;256:1443–1445. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- 17.Page R D, Charleston M A. Mol Phylogenet Evol. 1997;7:231–240. doi: 10.1006/mpev.1996.0390. [DOI] [PubMed] [Google Scholar]
- 18.Goodman M, Czelusniak J, Moore G W, Romero-Herrera A E, Matsuda G. Syst Biol. 1979;28:132–161. [Google Scholar]
- 19.Bremer K. Cladistics. 1995;10:295–304. [Google Scholar]
- 20.Eriksson T. AUTO-DECAY. Stockholm: Stockholm University; 1996. , Version 2.9.5. [Google Scholar]
- 21.Yang Z. PAML, Phylogenetic Analysis Using Maximum Likelihood. London: University College; 2000. , Version 2.0k. [Google Scholar]
- 22.Williams P L, Fitch W M. In: The Hierarchy of Life. Ferbholm B, Bremer K, Jurnvall H, editors. New York: Elsevier; 1989. pp. 453–470. [Google Scholar]
- 23.Li W-H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 24.Amores A, Force A, Yan Y L, Joly L, Amemiya C, Fritz A, Ho R K, Langeland J, Prince V, Wang Y L, et al. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 25.Spring J. FEBS Lett. 1997;400:2–8. doi: 10.1016/s0014-5793(96)01351-8. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Thornton J W, DeSalle R. Annu Rev Genom Hum Genet. 2000;1:41–73. doi: 10.1146/annurev.genom.1.1.41. [DOI] [PubMed] [Google Scholar]
- 28.Holland P W, Garcia-Fernandez J, Williams N A, Sidow A. Development (Cambridge, U.K.), Suppl. 1994. , 125–133. [PubMed] [Google Scholar]
- 29.Suga H, Hoshiyama D, Kuraku S, Katoh K, Kubokawa K, Miyata T. J Mol Evol. 1999;49:601–608. doi: 10.1007/pl00006581. [DOI] [PubMed] [Google Scholar]
- 30.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 31.Zilliacus J, Carlstedt-Duke J, Gustafsson J-A, Wright A P H. Proc Natl Acad Sci USA. 1994;91:4175–4179. doi: 10.1073/pnas.91.10.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brzozowski A M, Pike A C, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J A, Carlquist M. Nature (London) 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 33.Williams S P, Sigler P B. Nature (London) 1998;393:392–396. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- 34.Guillette L J, Crain D A. Environmental Endocrine Disruptors: An Evolutionary Approach. London: Taylor and Francis; 2000. [Google Scholar]
- 35.Veldscholte J, Ris-Stalpers C, Kuiper G G, Jenster G, Berrevoets C, Claassen E, van Rooij H C, Trapman J, Brinkmann A O, Mulder E. Biochem Biophys Res Commun. 1990;173:534–540. doi: 10.1016/s0006-291x(05)80067-1. [DOI] [PubMed] [Google Scholar]
- 36.Crews D, Willingham E, Skipper J K. Q Rev Biol. 2000;75:243–260. doi: 10.1086/393498. [DOI] [PubMed] [Google Scholar]
- 37.Watson C S, Campbell C H, Gametchu B. Exp Physiol. 1999;84:1013–1022. doi: 10.1111/j.1469-445x.1999.01903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian J, Kim S, Heilig E, Ruderman J V. Proc Natl Acad Sci USA. 2000;97:14358–14363. doi: 10.1073/pnas.250492197. . (First Published December 12, 2000; 10.1073/pnas.250492197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hines G A, Watts S A, Sower S A, Walker C W. Gen Comp Endocrinol. 1992;87:451–460. doi: 10.1016/0016-6480(92)90053-m. [DOI] [PubMed] [Google Scholar]
- 40.Fang Y Q, Zhao W X, Lin Q M. Sci China Ser B. 1994;37:842–850. [PubMed] [Google Scholar]
- 41.Jones R E, Baxter D C. In: Vertebrate Endocrinology: Fundamentals and Biomedical Implications. Pang P K T, Schreibman M P, editors. 4A. San Diego: Academic; 1991. pp. 205–302. [Google Scholar]
- 42.Ho S M, Press D, Liang L C, Sower S. Gen Comp Endocrinol. 1987;67:119–125. doi: 10.1016/0016-6480(87)90211-5. [DOI] [PubMed] [Google Scholar]
- 43.Sower S A, Plisetskaya E, Gorbman A. Gen Comp Endocrinol. 1985;58:259–269. doi: 10.1016/0016-6480(85)90342-9. [DOI] [PubMed] [Google Scholar]
- 44.Deragon K L, Sower S A. Gen Comp Endocrinol. 1994;95:363–367. doi: 10.1006/gcen.1994.1134. [DOI] [PubMed] [Google Scholar]
- 45.Tronche F, Kellendonk C, Reichardt H M, Schutz G. Curr Opin Genet Dev. 1998;8:532–538. doi: 10.1016/s0959-437x(98)80007-5. [DOI] [PubMed] [Google Scholar]
- 46.Norris D O. Vertebrate Endocrinology. Philadelphia: Lea and Feinger; 1980. [Google Scholar]
- 47.Weisbart M, Dickhoff W W, Gorbman A, Idler D R. Gen Comp Endocrinol. 1980;41:506–519. doi: 10.1016/0016-6480(80)90055-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.