SUMMARY
In Saccharomyces cerevisiae, the Ku heterodimer contributes to telomere maintenance as a component of telomeric chromatin and as an accessory subunit of telomerase. How Ku binding to double-stranded DNA (dsDNA) and to telomerase RNA (TLC1) promotes its telomeric functions is incompletely understood. We demonstrate that deletions designed to constrict the DNA-binding ring of Ku80 disrupt non-homologous end-joining (NHEJ), telomeric gene silencing and telomere length maintenance, suggesting that these functions require Ku's DNA end-binding activity. Contrary to the current model, a mutant Ku with low affinity for dsDNA also loses affinity for TLC1 both in vitro and in vivo. Competition experiments reveal that wild-type Ku binds dsDNA and TLC1 mutually exclusively. Cells expressing the mutant Ku are deficient in nuclear accumulation of TLC1, as expected from the RNA-binding defect. These findings force reconsideration of the mechanisms by which Ku assists in recruiting telomerase to natural telomeres and broken chromosome ends.
INTRODUCTION
Telomeres provide a shield at chromosome ends to maintain the integrity of the cell's genome (reviewed in (Jain and Cooper, 2010)). Telomeric DNA consists of multiple repeats of a short sequence; it is mainly dsDNA but terminates in a single-stranded 3'-overhang. The telomeric repeats are synthesized by the ribonucleoprotein enzyme telomerase (Greider and Blackburn, 1989). In S. cerevisiae, the telomerase reverse transcriptase (Est2 or TERT) and the RNA component (TLC1) are necessary and sufficient for enzyme catalysis (Lingner et al., 1997; Singer and Gottschling, 1994; Zappulla et al., 2005). In addition to providing the template for telomeric DNA synthesis, the 1.2 kb TLC1 RNA acts as a scaffold to provide binding sites for the accessory proteins Est1, the Sm complex, and Ku (Zappulla and Cech, 2004).
Ku is best known for its essential role in DNA break repair via NHEJ (Boulton and Jackson, 1996; Milne et al., 1996). The Ku heterodimer is composed of Ku70 and Ku80 (70.6 kDa and 71.2 kDa, respectively, in yeast). It binds dsDNA in a non- specific manner through a preformed ring, which limits Ku to sliding onto DNA from DNA breaks or termini (Walker et al., 2001). The Ku-DNA interaction is dynamic in vivo unless Ku is bound to other proteins, as observed in NHEJ studies (Chen and Tomkinson, 2011; Zhang et al., 2007). In the two-face model, the Ku80 side of the yeast heterodimer is responsible for Ku's telomeric functions and the Ku70 side is responsible for Ku's role in NHEJ (Ribes-Zamora et al., 2007).
In S. cerevisiae, Ku also contributes to telomerase function. If Ku is absent or unable to interact with telomerase, the native telomeres shorten; also, Ku-depleted cells have reduced capabilities in de novo telomere addition at broken chromosome ends, resulting in a lower frequency of gross chromosomal rearrangements (Boulton and Jackson, 1996; Gravel et al., 1998; Myung et al., 2001; Nugent et al., 1998; Polotnianka et al., 1998; Porter et al., 1996; Stellwagen et al., 2003). When binding between Ku and the TLC1 RNA is disrupted, nuclear localization and retention of the holoenzyme are impaired (Gallardo et al., 2008) and the steady-state level of TLC1 RNA decreases (Mozdy et al., 2008; Zappulla et al., 2011). Ku also contributes to telomeric chromatin structure through its interaction with proteins such as Sir4 (Roy et al., 2004; Ribes-Zamora et al., 2007). This interaction facilitates the formation of a cap that not only protects the yeast telomeres from DNA break repair machinery but also silences the expression of genes located near the telomere (Gottschling et al., 1990; Mishra and Shore, 1999).
In genetic screens for loss of this telomere silencing, Ku was found to interact with a 48-nucleotide stem-loop of TLC1 (Peterson et al., 2001). This region is strongly conserved among several budding yeast species, suggesting a sequence-specific interaction between Ku and the RNA (Dandjinou et al., 2004; Zappulla and Cech, 2004). In vitro binding experiments revealed that mutating three nucleotides was enough to disrupt this interaction, which indicates a specific RNA structure is required for Ku to bind (Peterson et al., 2001; Stellwagen et al., 2003). Mutagenesis screening of Ku subunits indicated that the RNA interaction is disrupted in the yku80-135i allele (Stellwagen et al., 2003), but since this five-amino-acid insertion is in the hydrophobic core of the Ku80 subunit, its effect on RNA binding is likely to be indirect.
How does telomerase use the interaction between its RNA component and Ku to contribute to the synthesis of telomeric DNA? Because it has been thought that Ku can simultaneously bind dsDNA and the TLC1 RNA, Ku was postulated to tether the telomere and TLC1 during the recruitment of telomerase to natural telomeres or broken chromosomal ends (Bertuch and Lundblad, 2003; Fisher et al., 2004; Fisher and Zakian, 2005; Pennaneach et al., 2006; Peterson et al., 2001). More specifically, it has been proposed that Ku helps recruit telomerase to telomeres in G1 phase, before telomerase is active, and then promotes telomerase action in late S phase (Fisher et al., 2004).
To test the importance of DNA binding to Ku's involvement in telomerase recruitment to telomeres and in other processes, we designed a series of deletion mutations to inhibit DNA binding. We found that a mutant with reduced DNA affinity was defective in NHEJ, telomeric gene silencing and telomere length maintenance. In contrast to expectation, the mutant that was defective in DNA binding was also defective in TLC1 RNA binding. Using competition experiments, we discovered that the binding of WT Ku to TLC1 RNA and to dsDNA is mutually exclusive. These findings contradict the recruitment model of Ku binding both DNA and RNA simultaneously and lead to a new model for the mechanism by which Ku's interaction with TLC1 RNA contributes to telomerase recruitment.
RESULTS
Deletions within Ku's DNA-binding Loop Decrease NHEJ and Silencing
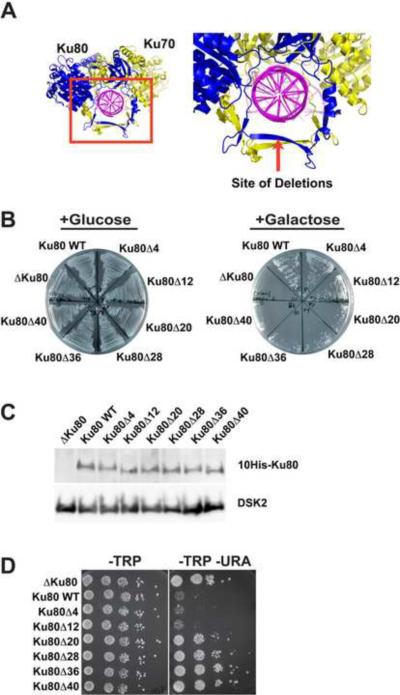
In an attempt to engineer separation-of-function mutants of Ku that would still bind TLC1 RNA but not DNA, a series of internal deletions was made in the primary DNA-binding site of Ku80 (Figure 1A). Larger deletions should prevent dsDNA from sliding through the ring of the heterodimer. Since yeast Ku binds dsDNA and recruits the repair machinery to DNA breaks (reviewed in (Daley et al., 2005; Riha et al., 2006)), an in vivo NHEJ assay was utilized to screen for defective Ku mutants that may have lost the ability to bind DNA. In this assay, an engineered HO endonuclease cut-site is cleaved by the HO endonuclease whose expression is under the control of a galactose promoter, and cell viability is monitored (Lee et al., 1999; Ribes-Zamora et al., 2007). It was observed that the DNA-binding loop of Ku80 could be truncated by twelve amino acids and still function like WT (Figure 1B and S1A). Deletions of 20 amino acids or more largely prevented the repair of the cleaved chromosome; these deletion mutants were almost as defective as Δku80 (the vector control strain). The NHEJ defect was not due to instability of the Ku mutant proteins, because protein levels for all mutants were similar to WT (Figure 1C); furthermore, heterodimerization was preserved (see below).
Figure 1.
Truncations within the Ring of Ku80 Impair NHEJ and Silencing (Telomere Position Effect)
(A) Internal deletions within the loop that slides over the dsDNA were designed on the basis of the crystal structure of human Ku (Walker et al., 2001).
(B) Using a strain with an engineered HO endonuclease cut-site, the Ku mutants, along with the WT strain and Δku80, were streaked onto plates containing either glucose or galactose. Galactose induces the production of HO endonuclease, triggering dsDNA break repair in cells containing functional Ku. The cells were plated at 100 generations.
(C) Western blot of the mutant yeast protein content after 20 generations. The loading control is DSK2, an endogenous protein. The Ku80 mutant to WT protein ratios (n = 4) were: Ku80Δ4 0.9 ± 0.1; Ku80Δ12 0.7 ± 0.3; Ku80Δ20 0.8 ± 0.3; Ku80Δ28 1.2 ± 0.6; Ku80Δ36 1.0 ± 0.3; Ku80Δ40 1.3 ± 0.4.
(D) Silencing assay for the URA3 gene at 20 generations. This strain is described in Experimental. See also Figure S1.
Ku contributes to subnuclear localization of telomeres and to higher order chromatin structure, causing transcriptional silencing of genes near the telomere (Gottschling et al., 1990; Laroche et al., 1998; Mishra and Shore, 1999). In a yeast strain with a telomere-proximal URA3 gene (Ribes-Zamora et al., 2007), Ku mutants with deletions of up to twelve amino acids within the DNA-binding loop behaved like WT and were unable to express the URA3 gene (Figure 1D and S1B). Deletions of 20 amino acids or greater allowed URA3 expression, like the Δku80 control. In a strain with a telomere-proximal ADE2 gene, the WT strain and ku mutants with deletions of twelve amino acids or less were not able to express ADE2 and thus appeared red in color (Figure S1C). The vector control and mutants with deletions of 20 amino acids or more were able to express ADE2 and appeared white in color (Figure S1C). These results are in agreement with the silencing results seen for URA3.
Both the NHEJ assay and the silencing assays were carried out over multiple generations. No differences were observed between 20 or 40, 100, and 200 generations (Figure 1B, 1D and S1) and the yeast did not senesce.
Deletions within the Ku DNA-binding Loop Decrease Telomere Length
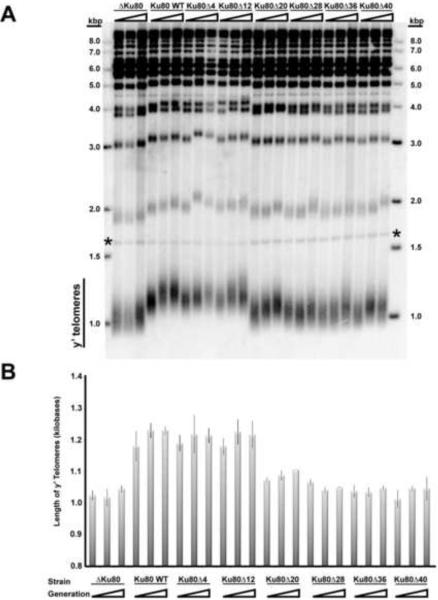
The Ku deletion mutants were assayed for telomere length maintenance. In Figure 2A, Southern blots demonstrate that the mutants with deletions of up to 12 amino acids remained capable of maintaining telomeres at WT length. Deletion of twenty or more amino acids resulted in shorter telomeres similar to the Δku80 control. This is the same trend observed in the NHEJ and silencing assays.
Figure 2.
Mutations within Ku's DNA-binding Loop Reduce Telomere Length
(A) Southern blot of Xho1-linearized yeast genomic DNA shows the length of Y' telomeres (heterogeneous distribution) and non-Y' telomeres (discrete bands at 2 kbp and above) over the course of generations 20, 100, and 200. The * symbol denotes the loading control, which is a restriction fragment of Chromosome IV.
(B) Measurement of average telomere length. The triangle represents 20, 100, and 200 generations. See also Figure S2.
The telomere length of the mutants was monitored over time up to generation 200. In the experiment shown in Figure 2A, it appeared that telomere length increased in WT and ku80Δ20 cells between 20 and 100 generations. However, measurements in independent experiments showed no convincing evidence of time-dependent telomere length change (Figure 2B). The main conclusion is Ku80 can tolerate the removal of several amino acids from the ring and still maintain WT telomeric length, whereas deletions of 20 amino acids or more are defective.
One explanation for the decrease in telomere length for the defective Ku mutants could be decreased levels of telomerase, since Ku has been implicated in the cellular abundance of TLC1 RNA (Mozdy et al., 2008; Zappulla et al., 2011). When analyzed by Northern hybridization, the TLC1 RNAs in the mutant strains all had the correct size (Figure S2). While we did see a modest decrease in the TLC1 RNA levels for Δku80 by quantitative real-time RT-PCR, the other mutant strains all showed TLC1 RNA levels similar to wild type (Figure S2). The variability in TLC1 RNA level between experiments, presumably due to variability in level of the plasmid expressing TLC1, precluded the observation of any small changes in TLC1 abundance due to Ku80 mutations. These data did not support the idea that lower telomerase levels could explain the shorter telomeres.
Ku Mutant Defective in DNA Binding is Defective in Binding RNA
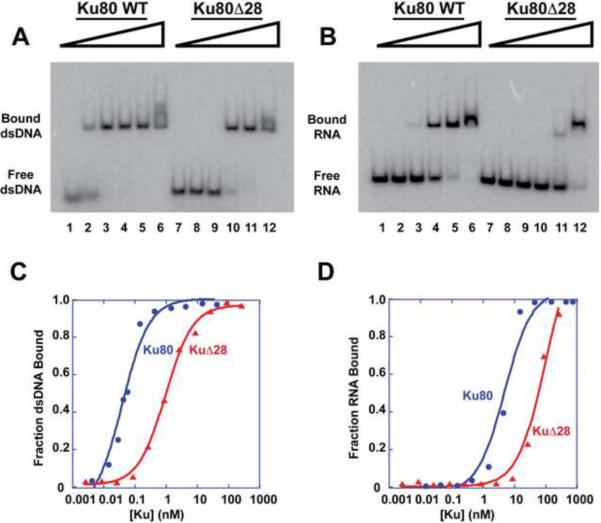
To confirm that the defect in Ku function for the truncated mutants was due to decreased DNA binding, the Ku80Δ28 heterodimer was purified to homogeneity from yeast and tested for DNA binding (Figure S3A). Because our measurements showed a very slow off-rate of DNA from Ku, overnight incubations were required to reach equilibrium (see Extended Experimental Procedures). The dsDNA contained a telomeric 3' overhang (Figure S3B); a double-stranded region of 22 base pairs was chosen because it was long enough to permit the binding of one Ku per DNA (Ma and Lieber, 2001). Figures 3A and 3C demonstrate that Ku80Δ28 protein has an affinity for the DNA (active Kd = 0.78 nM; active Kd refers to the Kd being corrected with respect to amount of active protein) that is about ten-fold weaker than that of WT (active Kd = 0.08 nM); this correlates with Ku80Δ28 being defective in vivo. At sufficiently high concentrations of WT Ku, a second shifted band of dsDNA can be seen, which presumably corresponds to two Ku per DNA molecule.
Figure 3.
Ku80Δ28 Protein Loses Affinity for Both dsDNA and TLC1-KBS RNA
(A) Binding of 22-bp dsDNA with a 14-nt telomeric 3' overhang by purified WT Ku and Ku80Δ28 proteins assayed by EMSA. (Throughout this paper, Ku80Δ28 protein refers to a heterodimer of Ku70 and Ku80Δ28.) Lanes 1 – 6 contain the following amounts of active WT Ku protein: 0; 0.02 nM; 0.2 nM; 1.7 nM; 17.2 nM; 172 nM. Lanes 7 – 12 contain active Ku80Δ28 protein in the same amounts as in lanes 1 – 6.
(B) Binding of TLC1-KBS RNA to WT Ku and Ku80Δ28 assayed via EMSA. The amounts of active protein are the same as in (A).
(C) Graphical representation of the DNA binding seen in (A). The active Kd for WT Ku is 0.08 nM. The active Kd for Ku80Δ28 is 0.78 nM.
(D) Graphical representation of (B). The fitted data yielded active Kd = 4.9 nM for WT Ku and active Kd = 87.1 nM for Ku80Δ28 binding to TLC1-KBS RNA. See also Figure S3.
The binding of Ku80Δ28 and WT Ku to a 95-nt RNA comprising the Ku-binding site (KBS) of TLC1 RNA was also measured (Figure S3C, Figure 3B and 3D). Ku80Δ28 had a greatly diminished affinity for the RNA (active Kd = 87.1 nM) compared to WT (active Kd = 4.9 nM). This finding was unexpected; we had hypothesized that a smaller ring would inhibit DNA binding but not affect TLC1 RNA binding, based on data suggesting that RNA binding occurred at a different site on Ku80 (Stellwagen et al., 2003).
Mutation of the Ku ring inhibits TLC1 RNA binding in vivo
Because of the profound effect of the Ku80Δ28 mutation on TLC1 RNA binding in vitro, it seemed likely that this mutant Ku would also be defective in TLC1 RNA binding in vivo. This was tested by a co-immunoprecipitation (co-IP) experiment.
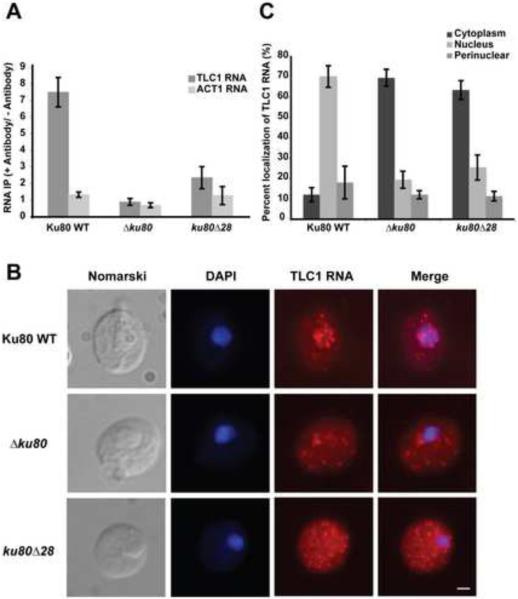
Yeast were formaldehyde cross-linked, Myc-tagged Ku was IP'd on beads coated with anti-Myc antibodies, and the co-IP'd TLC1 RNA was quantified by real-time RT-PCR. The data showed substantial and specific association of TLC1 RNA with Ku in the WT strain but about 75% reduced binding in the ku80Δ28 mutant strain (Fig. 4A). Control experiments showed that the co-IP was dependent on anti-Myc antibody, was eliminated in the Δku80 strain, and was specific due to the lack of substantial pull-down of ACT1 RNA (Fig. 4A). The yeast strains contained similar amounts of WT and Ku80Δ28 protein prior to the IP (Fig. 1C), and the amount of Myc-tagged protein recovered by IP was similar for WT Ku and Ku80Δ28 proteins as assessed by Western blot (Figure S4). Thus, our experiments provide no evidence that other factors in vivo are able to compensate for the loss of the primary binding interaction between Ku and TLC1 RNA; the greatly reduced RNA binding by the mutant Ku in vivo correlates with the substantial destabilization of mutant Ku-RNA interaction measured in vitro.
Figure 4.
Ku80Δ28 Loses Association with TLC1 RNA in vivo and the RNA Accumulates in the Cytoplasm
(A) TLC1 RNA immunoprecipitation with Myc-tagged Ku proteins analyzed by real-time RT-PCR. Cells were subjected to formaldehyde crosslinking to preserve RNA-protein interactions prior to immunoprecipitation on anti-Myc beads. The highest levels of pull-down (around 15-fold enrichment) corresponded to 2% of the input TLC1 RNA. ACT1 mRNA, which is not known to associate with Ku, served as a control for nonspecific binding. Bars indicate average of five biologic replicates performed on four different weeks, and error bars give SEM.
(B) Localization of endogenous TLC1 RNA in WT, Δku80 and ku80Δ28 strains was detected using fluorescent in situ hybridization. DAPI: DNA staining. Scale bar: 1 μm.
(C) Quantification of TLC1 RNA distribution in WT, Δku80 and ku80Δ28 strains. For each strain, a total of 300 cells were randomly scored, in three independent experiments. See also Figure S4.
Mutation of the Ku ring reduces nuclear localization of TLC1 RNA
Telomerase RNA biogenesis involves export of the TLC1 RNA to the cytoplasm and subsequent importation (Teixeria et al., 2002; Gallardo et al. 2008). Previous work showed that Ku70 is important for the nuclear retention of TLC1 RNA (Gallardo et al. 2008). Although the mutations studied here are in the Ku80 subunit, Ku functions primarily as a heterodimer, so we expected that an RNA-binding-defective Ku80 would also be defective in nuclear retention of TLC1 RNA. Reduced nuclear retention of telomerase would in turn help explain the observed short-telomere phenotype.
The cellular localization of endogenous TLC1 RNA in the ku80Δ28 strain was analyzed by fluorescent in situ hybridization. In contrast to the strain containing WT Ku, where TLC1 RNA was predominantly nuclear, the ku80Δ28 strain showed most of the TLC1 RNA in the cytoplasm (Figure 4B). The redistribution to the cytoplasm was not as complete as observed with the Ku80-knockout strain (Figure 4C). This result was in complete accord with the in vivo Ku-TLC1 RNA binding experiments (Figure 4A), which showed that the mutant Ku retained ~25% RNA-binding function.
Binding of Ku to RNA or DNA is Mutually Exclusive
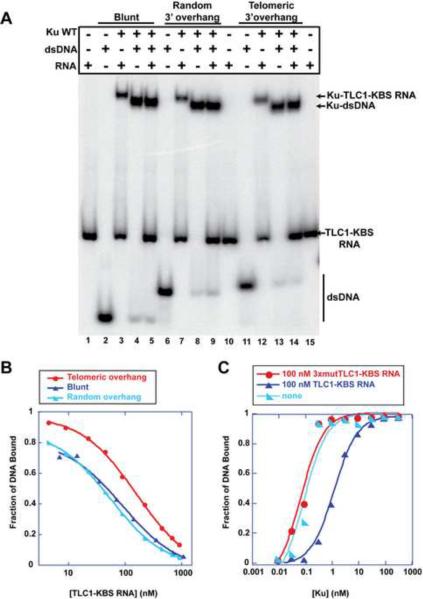
Since the Ku80 mutation interfered with both TLC1 RNA and DNA binding, we hypothesized that WT Ku would not be able to bind dsDNA and TLC1 RNA simultaneously. Three different forms of dsDNA were tested: DNA with blunt ends, a 3' overhang with a random sequence, and a 3' overhang with a telomere-like sequence (Figure S3B). Mammalian Ku binds all of these forms of DNA (Bianchi and de Lange, 1999; Dynan and Yoo, 1998). Similarly, we found that all three dsDNA forms were able to bind yeast WT Ku (Figure 5A). The shifted DNA-Ku band migrated slightly faster than the Ku bound TLC1-KBS RNA. In the lanes containing equal amounts of both dsDNA and RNA, the resulting shifted band migrated as the Ku-DNA complex. The amount of radiolabeled free RNA in lanes 5, 9 and 14 provides independent evidence that very little if any RNA was present in the shifted band. Under no circumstances did we detect a shifted band that could correspond to both DNA and RNA being simultaneously bound. Because of Ku's higher affinity for DNA than for RNA, the DNA-Ku complex dominates. Thus, we conclude either that RNA and DNA bind to the same site on Ku or that mutually exclusive binding sites for RNA and DNA exist on Ku. The DNA 3'end does not affect the preference of Ku to bind only to dsDNA versus forming the complex containing Ku, DNA and RNA.
Figure 5.
Mutually Exclusive Binding of RNA and DNA to WT Ku
(A) A mixing experiment shows the complexes that Ku forms with dsDNA containing different 3' overhangs and with TLC1-KBS RNA. Each arrow denotes the position in the gel for a particular complex.
(B) TLC1-KBS RNA and DNA compete against one another to bind Ku. Increasing amounts of TLC1-KBS RNA were added to samples containing Ku and radiolabeled dsDNA with different 3' overhangs. The fraction bound of each sample was calculated and fitted to a single binding site competition formula to calculate the Ki of the RNA.
(C) 100 nM of TLC1-KBS RNA and 3xmutTLC1-KBS RNA were added in equilibrium binding experiments to ascertain their effects on Ku's affinity for radiolabeled dsDNA with telomere-like 3'overhang. The fraction bound of the DNA was calculated and fitted to the Langmuir isotherm. See also Figures S5 and S6.
Because the ability of DNA to inhibit the Ku-RNA interaction was contrary to expectation, we conducted additional tests. To ensure that equilibrium was being achieved in our studies, the off-rates were measured for all dsDNAs and TLC1-KBS RNA (Figure S5). We observed that the RNA had a faster off-rate than the dsDNA (Table 1). For example, the blunt dsDNA had a koff = 0.007 ± 0.005 min−1 and t1/2 = 99 min, which was a substantially longer half-life than that reported for human Ku (Ma and Lieber, 2001). As a consequence of the slow off-rate, the incubation times for the binding experiments went overnight to ensure that equilibrium was achieved.
Table 1.
Summary of binding and competition data.
| 3xmutTLC1-KBS RNA | TLC1-KBS RNA | Telomeric 3'overhang | Random 3'overhang | Blunt end | |
|---|---|---|---|---|---|
| Apparent Kd [nM] | 353 ± 49 | 10.5 ± 0.7 | 0.17 ± 0.10 | 0.30 ± 0.22 | 0.18 ± 0.06 |
| koff [min−1] | 2.6 ± 1.2 | 0.005 ± 0.001 | 0.011 ± 0.002 | 0.007 ± 0.005 | |
| t1/2 [min] | 0.27 | 140 | 63 | 99 | |
| kon [M−1S−1] | 3.4×106 | 4.8×105 | 5.9×105 | 6.3×105 | |
| Ki for TLC1-KBS RNA [nM] | 51 ± 32 | 13 ± 5 | 19.5 ± 8.3 | ||
| IC50 for DNA [nM] | 40 ± 3 | 42 ± 9 | 38 ± 6 |
In an independent test for mutually exclusive binding, increasing amounts of TLC1-KBS RNA were able to prevent dsDNA from binding Ku (Figure 5B and S6A). The inhibition was specific: 3xmutTLC1-KBS RNA and Tetrahymena P4P6 RNA did not hinder DNA binding even at 1 μM (Figure S6B and S6C). TLC1-KBS RNA prevented the three different DNAs from interacting with Ku but at different concentrations (Figure 5B and Table 1), which reflect the differences in the affinity of Ku for these DNAs (Table 1). The Ki of TLC1-KBS RNA was calculated for each DNA tested using equations for a single-site binding model (Table 1). The calculated Ki's are equal within experimental error to the Kd of the TLC1-KBS RNA, as expected. The 22-bp blunt-ended DNA bound to yeast Ku with an apparent Kd of 180 pM, similar to the Kd of 160 pM reported for human Ku (Ma and Lieber, 2001), 340 pM reported for human Ku (Roberts and Ramsden, 2007) and 300 pM reported for yeast Ku using a 600-bp blunt-ended DNA (Chen and Tomkinson, 2011).
The reverse experiment was performed to determine if dsDNA could inhibit TLC1-KBS RNA from binding Ku (Figure S6D–E). All three forms of DNA tested hindered the RNA-Ku interaction equally well with similar IC50 values of approximately 40 nM (Table 1). Because these experiments were conducted at Ku and DNA concentrations far above Kd, they exhibit titration behavior (Shoichet, 2006); i.e., the binding of RNA is limited by the amount of free (non-DNA-bound) protein rather than the binding constant. The titration behavior is also reflected in the steepness of the curves. Thus, IC50 is not expected to equal the Kd for DNA binding.
RNA Competitively Inhibits DNA Binding to Ku
To determine if the inhibition was competitive or noncompetitive, an experiment was performed to see how the RNA affected the Kd of Ku binding to the dsDNA with the telomere-like sequence. In Figure 5C, the presence of 100 nM RNA shifted the curve to the right, an increase of the apparent Kd of DNA-binding to Ku by 17-fold, but did not change the maximum of the fraction bound. In parallel to enzyme kinetics, this behavior is indicative of a competitive inhibitor.
To confirm the competitive inhibition observed was a direct result of Ku binding to the RNA and not just a result of RNA being present, we performed the same experiment using an RNA nearly defective in Ku binding, 3xmutTLC1-KBS, which contains the three point mutations described previously (Peterson et al., 2001). We confirmed that the affinity of this mutant was substantially weaker than that of the WT RNA (Table 1); due to the ability to obtain more Ku with our modified protein purification protocol, we were able to use higher Ku concentrations to observe this weak binding. Even though the 3xmutTLC1-KBS RNA had a weak affinity for Ku, it was not able to competitively inhibit the DNA binding at 100 nM (Figure 5C).
Ku70 separation-of-function mutants bind DNA and TLC1 RNA normally
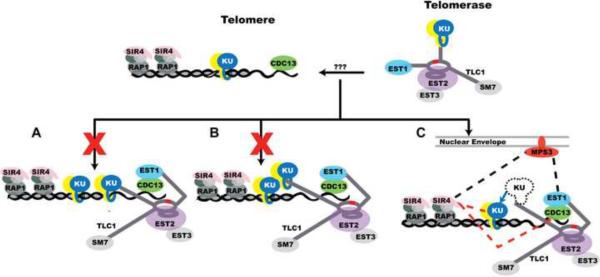
The inability of Ku to bind both DNA and TLC1 RNA simultaneously eliminates the current tethering model for Ku recruitment of telomerase (Figure 6A), but leaves open a revised model in which telomere-bound Ku would bind to telomerase-bound Ku (Figure 6B). This “synapse model” invokes the same Ku-Ku interactions that are thought to occur between Ku-bound DNA ends during NHEJ. Importantly, Ribes-Zamora et al. (2007) reported separation-of-function alleles of Ku70 with mutations distant from the DNA-binding ring that were defective in NHEJ but retained full telomeric functions. This observation would seem to provide a strong argument against Ku-Ku synapse formation being involved in telomerase recruitment, assuming that these Ku70 alleles in fact retained full DNA end-binding and TLC1 RNA-binding activities.
Figure 6.
Models of Ku's Role in Telomerase Recruitment
(A) Published model shows Ku binding simultaneously to TLC1 RNA and the dsDNA while recruiting telomerase to the chromosome end, which is not possible based on our work.
(B) Based on the action of Ku during NHEJ, telomere-bound Ku might bind to a telomerase-bound Ku to recruit telomerase to telomeres. However, the Ku70 separation-of-function mutants described by Ribes-Zamora et al. (2007) and data presented herein cause us to discount this model.
(C) In the new model, Ku recruitment of telomerase begins with its key role in nuclear import and retention. When telomerase-bound Ku encounters telomeric DNA, Ku may be handed off from TLC1 to the DNA (blue line). This hand-off may be necessary to prevent telomerase from being sequestered in telomeric heterochromatin by Ku-Sir4 binding. The Est1-Cdc13 protein-protein interaction then secures telomerase to the telomere. Other reported interactions include Est1 and Sir4 binding to the nuclear envelope protein Mps3 (black dashes) and Ku and Cdc13 binding to Sir4 (red dashes). See also Figure S7.
Therefore we used site-specific mutagenesis to introduce the D195A and D195R mutations into Ku70, purified the proteins, and performed quantitative dsDNA-binding and TLC1-KBS RNA-binding gel-shift experiments. As shown in Figure S7, both mutant proteins bound both nucleic acids with essentially WT affinity. Thus, we concur with the conclusion of Ribes-Zamora et al. (2007) that these are true separation-of-function mutants. The mutations do not perturb DNA or RNA binding, so they presumably interfere with protein-protein interactions required for NHEJ but not for telomerase function.
DISCUSSION
The prevailing paradigm in the telomerase field has been that Ku can bind DNA termini and TLC1 RNA independently, which led to a simple and elegant model by which Ku could help recruit telomerase to chromosome ends. Based on this paradigm, we set out to engineer separation-of-function alleles of yeast Ku that lost DNA end-binding and retained TLC1 RNA binding. Instead, our DNA-binding mutants of Ku showed substantially reduced binding to TLC1 RNA both in vitro and in vivo. While this genetic evidence was dramatic, it is important to remember that mutant phenotypes provide information about the mutant, from which the behavior of the WT is inferred. We therefore studied the binding of mixtures of TLC1 RNA and DNA to purified WT Ku protein, and we were able to confirm that Ku cannot bind both nucleic acids simultaneously. This finding leads to a new model of how Ku contributes to recruitment of telomerase to telomeres. In addition, the Ku mutants with reduced DNA binding are defective in NHEJ and telomeric gene silencing, providing direct evidence for the importance of DNA end-binding in these Ku functions.
Mutually Exclusive Binding of Ku to RNA and DNA
Our in vitro binding experiments showed that the binding of telomerase RNA or DNA to Ku is mutually exclusive. The TLC1-KBS RNA was able to competitively inhibit Ku from binding dsDNA and vice versa. The mixing experiment showed that a complex containing both TLC1-KBS RNA and dsDNA could not be formed, which further supports the conclusion that Ku will bind to either dsDNA or TLC1-KBS RNA, but not to both simultaneously. Finally, mutational analysis provided independent evidence for mutually exclusive binding, since the deletion designed to close the DNA-binding ring also inhibited RNA binding to a similar extent. The simplest way to achieve mutually exclusive binding is if RNA and DNA both bind to the ring of Ku or if the RNA-binding site physically overlaps with the DNA-binding site. The more accurate term “mutually exclusive binding” covers these possibilities and also the additional possibility of two non-overlapping binding sites where occupancy of either site causes a conformational change that precludes availability of the other site.
Competition between TLC1 and chromosome ends for yeast Ku has been observed in vivo. In silencing experiments, a plasmid was engineered to express an RNA containing three TLC1 Ku-binding sites in tandem and transformed into yeast. This RNA and similar constructs disrupted silencing (Peterson et al., 2001; Zappulla et al., 2011). These data are consistent with our in vitro observations that TLC1-KBS RNA and dsDNA compete against one another for Ku, although in vivo it is difficult to ascertain whether the Ku is bound to the very end of the chromosome through its DNA-binding ring or is bound at more internal telomeric sites.
Aptamer RNAs that bind human Ku have been identified by in vitro selection, and they were found to compete with dsDNA for Ku binding (Yoo and Dynan, 1998). These observations are very reminiscent of our findings for yeast Ku and TLC1-KBS. An intriguing possibility is that both TLC1-KBS and the aptamer RNAs are binding to the ring of Ku by mimicking the structure of B-form DNA, a known property of some RNAs (Bullock et al., 2010; Reiter et al., 2008). If the RNA were mimicking DNA structure, then the two substrates would naturally compete for the same binding site on Ku. The mutations in the 3× mutant RNA could then be disrupting its B-like helical structure, which might prevent the wider A-form RNA helix from even entering the Ku ring. One implication of this model is that there might be as-yet-unidentified Ku-binding RNA stem-loops that are completely unrelated in nucleotide sequence, but share a B-DNA-like helical structure.
Models for Yeast Telomerase Recruitment
The currently accepted model is that telomerase recruitment occurs via Ku simultaneously interacting with both dsDNA (telomeric DNA) and TLC1 RNA. Thus, Ku acts as a tether: Ku bound to telomerase could at the same time recognize and bind to telomeric DNA or to broken chromosome ends (Bertuch and Lundblad, 2003; Fisher et al., 2004; Fisher and Zakian, 2005; Pennaneach et al., 2006; Peterson et al., 2001). The present study challenges this model of Ku's contribution to telomerase recruitment, as we have demonstrated that DNA and RNA competitively bind Ku. Because Ku cannot bind DNA and RNA simultaneously, it cannot tether telomerase to the telomere or to a broken chromosome end (Figure 6A).
A second “synapsis” model is based on the idea that Ku binds to another Ku to promote the bridging of two DNA ends in NHEJ. In the nucleus, when the Ku-telomerase complex encounters a chromosome end, the Ku from the Ku-telomerase complex might bind to the Ku from the Ku-telomere complex in order to tether the telomere to telomerase (Figure 6B). A Ku-Ku joining event to facilitate the end-joining of DNA has been observed via atomic force microscopy with yeast Ku, although the authors of the paper noted that the end-bridging event happened less frequently than the one mediated by the Mre11/Rad50/Xrs2 complex (Chen et al., 2001). However, the Bertuch separation-of-function Ku70 mutants were defective in NHEJ (which is thought to require Ku-Ku interaction) but retained telomeric functions (Ribes-Zamora et al., 2007), and these mutants retain WT DNA binding and TLC1 RNA binding (Figure S7). This argues against the Ku-Ku synapse formation that mediates NHEJ being important for telomerase recruitment, causing us to disfavor the model of Figure 6B.
Our preferred model for Ku's role in telomerase recruitment, which integrates the results from this and other studies, is shown in Figure 6C. A primary function of Ku binding to telomerase is to promote its nuclear accumulation (Gallardo et al., 2008; this paper). The mutually exclusive binding of Ku to dsDNA and TLC1 RNA shown here is proposed to contribute to telomerase recruitment along with Cdc13-Est1 and nuclear envelope interactions. When telomerase encounters a dsDNA end, Ku is expected to become unbound from TLC1 RNA and engage the DNA, as it has higher affinity for DNA than for TLC1-KBS. This exchange of Ku between TLC1 and telomeric DNA could explain the Ku-dependent interaction between telomerase and telomeres observed by ChIP (chromatin immunoprecipitation) in the G1 phase of the cell cycle, prior to telomerase elongation of telomeres (Fisher et al., 2004). In late S-phase, telomerase then binds to the DNA end through two already well-established interactions: the base-pairing of the TLC1 template with the single-stranded DNA at the chromosome end, and the interaction of the Est1 subunit of telomerase with telomere-bound Cdc13 (Evans and Lundblad, 1999; Qi and Zakian, 2000; Chan et al., 2008).
Also during the S phase of the cell cycle, when telomeres are being elongated, the telomeres localize to the nuclear periphery via interactions between Sir4 and the nuclear envelope protein Mps3 (Bupp et al., 2007). Concurrently, Ku-bound telomerase is found to be associated with Mps3, which is dependent on Mps3 binding Est1 (Antoniacci et al., 2007; Schober et al., 2009). The interactions of telomerase-bound proteins, telomere-bound proteins and Mps3 may be important for preventing telomerase dissociation and fully elongating short telomeres (Teixeira et al., 2004; Chang et al., 2007), with regulation via Siz2 sumoylation of Sir4 and Ku (Ferreira et al., 2011).
DNA Binding is Necessary for Ku's Telomeric and DNA Repair Functions
The crystal structure of human Ku bound to dsDNA (Walker et al., 2001) revealed a preformed dsDNA-binding ring, providing a structural basis for understanding how Ku could recognize dsDNA breaks and facilitate repair via NHEJ. We found that the larger deletions in the DNA-binding beta strand of yeast Ku80 inhibited NHEJ, in agreement with our expectation. Our quantitative measurements of DNA binding by the Ku80Δ28 mutant protein indicate that a ten fold reduction in DNA affinity is sufficient for inhibition, although it is possible that this mutation could also interfere with protein-protein interactions required for NHEJ.
Our data on telomeric silencing show that the DNA-binding ring of Ku is also required for telomeric heterochromatin formation. Telomeric silencing relies on the interaction between Ku and Sir4 (Roy et al., 2004; see also Hediger et al., 2002), and Ku's functions at double-strand breaks and at telomeres have been separated by mutations that target potential protein binding sites on Ku (Bertuch and Lundblad, 2003; Ribes-Zamora et al., 2007). Our mutations in the ring of Ku are not near the 80α5 helix, so Ku's ability to bind Sir4 should not be affected (Ribes-Zamora et al., 2007). It is worth noting that the yKu80-135i mutant protein, a mutant defective in RNA binding, still retained an affinity for DNA, but it was reduced compared to wild type (Stellwagen et al., 2003). The reduced DNA affinity, however, was still strong enough to allow for a physiologically relevant interaction between the protein and dsDNA to permit the silencing of the reporter genes (Stellwagen et al., 2003). Because our DNA-binding-defective Ku mutant did not facilitate the silencing of either the ADE2 gene or the URA3 gene, we propose that Ku must bind to dsDNA for its gene-silencing function.
Shortening of telomeres is a more complex phenotype, in that inhibition of either Ku-TLC1 RNA interactions or Ku-telomeric DNA interactions could potentially contribute. Previous work using mutations or deletions in TLC1 RNA that prevent Ku binding showed redistribution of TLC1 from nucleus to cytoplasm, establishing one contribution of the Ku-RNA interaction to telomere length maintenance (Gallardo et al., 2008). The large reduction in nuclear localization of TLC1 RNA in the ku80Δ28 strain certainly contributes to the failure to maintain telomere length. Ku must also retain its ability to bind dsDNA in order to protect telomeres from nucleolytic degradation and recombination (Gravel et al., 1998; Polotnianka et al., 1998)
While this paper was under review, a publication appeared by Lopez et al. (2011) concluding that Ku must load directly onto chromosome ends to accomplish its telomeric functions. Our conclusion about the importance of Ku's DNA-binding activity for silencing of telomeric gene expression is consistent with that of Lopez et al., and the similar phenotypes observed with two very different sets of alleles make this conclusion even more robust. Concerning Ku's role in NHEJ, the different alleles in the two studies both reduced Ku's interaction with DNA but had opposite effects on NHEJ, so it appears that the two sets of mutations must perturb different properties of Ku as it engages the DNA breaks to facilitate repair. Finally, we remain circumspect regarding the conclusion that Ku's DNA-binding activity is required for telomerase function. The DNA-binding defective Ku alleles described by Lopez et al. (2011) did show some reduction of TLC1 RNA binding in vivo, and it would seem important to know whether TLC1 is still in the nucleus in these mutant Ku strains. Thus, while it remains entirely plausible that in the case of WT Ku and WT TLC1 RNA, Ku uses its ability to bind dsDNA to partake in the regulation of telomerase recruitment, our new findings that TLC1 RNA and dsDNA appear to bind in the same site on Ku make it challenging to use Ku mutants to distinguish between its telomerase-binding and DNA-binding functions.
EXPERIMENTAL PROCEDURES
Plasmids and Strains
NHEJ Assay
Assays used the YVL2236 (yku80-Δ = Δku80) strain containing a galactose-inducible HO endonuclease and an engineered HO cut-site (Ribes-Zamora et al., 2007). After transformation with the plasmid carrying a Ku80 construct and TRP gene, cells from generations 40, 100, and 200 were streaked onto -Trp -Ura plates containing either glucose or 2% galactose and grown at 30°C for 2–3 days.
Silencing Assays
Assays (Ribes-Zamora et al., 2007) used the YVL885 (Δku80) strain engineered to contain ADE2 and URA3 genes located near telomeres V-R and VII-L, respectively. Yeast were transformed with the plasmids carrying a Ku80 construct and TRP gene and streaked on plates. Cells from generations 20, 40, 100, or 200 were grown overnight. An equal number of cells for each construct was 5-fold serially diluted onto -Trp, -Trp -Ura, or -Trp - Ade plates. The yeast grew at 30°C for 2–3 days. To enhance the red pigmentation of cells grown on the -Trp -Ade plates, the cells were placed at 4°C for 4–7 days and then placed at 26°C for approximately 10–16 days or until a dark red pigment became apparent.
Western Blots
The protocol followed that of (Knop et al., 1999) with adjustments (see Supplemental Information). The membrane was cut below the 62 kDa SeeBlue Plus2 Prestained standard (Invitrogen LC5925). The blot portion containing the Ku mutants and other higher molecular weight proteins was probed either with Anti-His HRP Conjugate (Qiagen 34460) following the manufacturer's protocol or 6× His tag® from Abcam (ab9108). The other portion of the blot was probed with Dsk2 (Abcam ab4119). The blots that were probed with unconjugated antibodies went through a second blocking procedure using a goat anti-rabbit IgG-HRP (Santa Cruz SC-2054) secondary antibody. The proteins were then detected using Amersham ECL Plus Western Blotting Reagents (GE Healthcare RPN2132). PhosphoImager intensities of the DSK2 bands were measured using Imagequant, and normalization factors were generated and applied to the quantified Ku bands. The ratios of the Ku mutants to WT were calculated.
Southern and Northern Blots
20–50 ml cultures were grown in -Trp - Ura media until they reached an OD600 of 0.75 –1.0. The cells were then harvested, washed twice with sterile H2O, split in half after the second wash and the pellets were harvested. Northern and Southern Blots were carried out as described previously (Zappulla et al., 2005), except that the genomic DNA was isolated using the Gentra Puregene Yeast/Bact. Kit from Qiagen and the total RNA was probed for TLC1 and U1 RNAs.
Real-time PCR
TLC1 and ACT1 RNA levels were quantified using RT-PCR (Mozdy and Cech, 2006) as detailed in Extended Experimental Procedures.
RNA Preparation
TLC1-KBS and 3xmutTLC1-KBS were transcribed from PCR-amplified DNA that contained a 3'OMe on the last nucleotide and a T7 promoter. The transcribed RNA was purified as previously described (Kieft et al., 1999), but a 10% acrylamide gel was used. Contaminating DNA was removed using RQ1 DNase (Promega) according to the manufacturer's instructions. The purified RNA was dephosphorylated using Calf Intestinal Phosphatase (Roche) and 5'-end-labeled as previously described (Kieft et al., 1999).
Ku Expression and Purification
Yeast strain BJ2168 was co-transformed with the plasmid combinations pJP16 and pJP14, or pJP16 and pJP15. The plasmid pRS425TEF-YKU70 (Peterson et al., 2001) contained a point mutation or polymorphism (D473G), which was corrected. The transformed yeast were grown in - Trp - Leu media to an OD600 of 1.2. The cells were harvested, resuspended in lysis buffer (25 mM Hepes pH 7.5, 200 mM KCl, 2 mM MgCl2, 1 mM EDTA, 1 mM DTT), and stored at - 80°C. Thawed cells were treated with zymolase before being sonicated, and spun at 12, 000 rpm for 80 min. The resulting lysate was passed over nickel resin (Qiagen). The resin was washed with 5 column volumes (CV) of lysis buffer with 20 mM imidazole, 2.5 CV of lysis buffer with 50 mM imidazole, and 0.5 CV of lysis buffer with 250 mM imidazole. Ku eluted with lysis buffer containing 250 mM imidazole was concentrated and dialyzed in buffer A (50 mM Tris pH 8, 250 mM NaCl, 2 mM DTT, 1 mM EDTA). The protein was passed over a mono q column using buffer A and buffer B (50 mM Tris pH 8, 2 M NaCl, 1 mM EDTA). The fractions containing Ku were concentrated and passed over a Sup 200 column in buffer C (50 mM Tris pH 8, 500 mM NaCl, 2 mM DT, 1 mM EDTA). The fractions containing Ku were concentrated, dialyzed in storage buffer (25 mM Tris pH 8, 200 mM NaCl, 20% glycerol, 1 mM EDTA, 2 mM DTT), flash frozen and stored at - 80 °C. The percent active protein was measured using titration experiments and DNA as the substrate. The protein was consistently about 47% active, which is greater than the 15% reported for human Ku (Blier et al., 1993). For Figure 3, the Kd based on active protein concentration is reported, but in all other instances the apparent Kd is used.
Kinetics and Thermodynamics of Ku-RNA and Ku-DNA Interactions
Assays are described in detail in Supplemental Information. A typical binding buffer was 21 mM HEPES pH 7.5, 150 mM NaCl, 11% glycerol, 5 mM MgCl2, 1 mM EDTA, 25 μg/mL tRNA, 0.1 mg/ml BSA, 1 mM DTT.
Immunoprecipitation of TLC1-Ku Complexes
As detailed in Supplemental Information, yeast expressing myc-tagged Ku were treated with formaldehyde (1% final) and sonicated. Anti-myc antibody (Sigma M4439) was added to half of each sample. The other half comprised the “minus antibody” control. Complexes were pulled-down on Protein G Plus/Protein A Agarose (Calbiochem) beads and RNA was purified.
Fluorescent in situ hybridization
Yeast fixation and fluorescent in situ hybridization to detect endogenous TLC1 RNA were as described (Gallardo et al, 2008), except treatment with oxalyticase was between 15 and 23 min at 30°C. Hybridization with TLC1 specific probes was performed in 45% formamide.
Image acquisition and processing
All images were acquired using a Nikon Eclipse E800 epifluorescence upright microscope equipped with a 100× DIC H (1.4 NA) objective and with a Photometrics CoolSNAP fx CCD camera; 100 fields of yeast cells were acquired as Z stacks of 20 planes, with 0.5 μm between planes in the Z axis. Maximal projection of Z stacks was performed and merged with DAPI signal for quantification of localization. Images were acquired and processed with Metamorph software.
Other Methods
Supplementary Material
HIGHLIGHTS
Yeast Ku's telomeric functions require its DNA-binding ring
Ku mutation that inhibits DNA binding unexpectedly inhibits telomerase RNA binding
Competition experiments show mutually exclusive binding of DNA and RNA to Ku
Supports new model for recruitment of telomerase to telomeres by Ku
ACKNOWLEDGMENTS
We thank Alison Bertuch (Baylor College of Medicine) and Vicki Lundblad (Salk Institute) for generous gifts of yeast strains, Dan Gottschling (Fred Hutchinson Cancer Center) for providing plasmids and discussion, and Titia de Lange (Rockefeller University) for helpful ideas. We thank David B. McKay and Robert Batey (both Univ. of Colorado-Boulder) for assisting in the computational analysis of the competition data and Jacob Schwartz for help with the analysis of real-time RT-PCR data. We also thank Robert Batey, Jeffrey Kieft, Jayakrishnan Nandakumar, Jacob Schwartz, and Andrea Berman for comments on the manuscript. C.T. was an EXROP student of the Howard Hughes Medical Institute. This work was funded in part by Canadian Institutes of Health Research grant MOP89768 to P.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION Supplemental Information includes Extended Experimental Procedures, 2 tables and 7 figures, and can be found with this article online at XXXXXXXXXXX.
Author Contribution: JSP and TRC designed the experiments and wrote the paper with input from all authors. JSP performed in vivo based plating assays, Northern blots, purified yeast Ku and executed all in vitro based assays. KJG carried out Southern blots, Northern Blots, Westerns, plating assays, and quantitative RT-PCR. CT performed initial plating, Southern and Northern experiments. FO and PC designed and performed cytolocalization of TLC1.
REFERENCES
- Antoniacci LM, Kenna MA, Skibbens RV. The nuclear envelope and spindle pole body-associated Mps3 protein bind telomere regulators and function in telomere clustering. Cell Cycle. 2007;6:75–79. doi: 10.4161/cc.6.1.3647. [DOI] [PubMed] [Google Scholar]
- Bertuch AA, Lundblad V. The Ku heterodimer performs separable activities at double-strand breaks and chromosome termini. Mol Cell Biol. 2003;23:8202–8215. doi: 10.1128/MCB.23.22.8202-8215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, de Lange T. Ku binds telomeric DNA in vitro. J Biol Chem. 1999;274:21223–21227. doi: 10.1074/jbc.274.30.21223. [DOI] [PubMed] [Google Scholar]
- Blier PR, Griffith AJ, Craft J, Hardin JA. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem. 1993;268:7594–7601. [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock SL, Ringel I, Ish-Horowicz D, Lukavsky PJ. A'-form RNA helices are required for cytoplasmic mRNA transport in Drosophila. Nat Struct Mol Biol. 2010;17:703–709. doi: 10.1038/nsmb.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bupp JM, Martin AE, Stensrud ES, Jaspersen SL. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J Cell Biol. 2007;179:845–854. doi: 10.1083/jcb.200706040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Arneric M, Lingner J. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel 1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 2007;21:2485–2494. doi: 10.1101/gad.1588807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- Chen X, Tomkinson AE. Yeast Nej1 is a key participant in the initial end binding and final ligation steps of nonhomologous end joining. J Biol Chem. 2011;286:4931–4940. doi: 10.1074/jbc.M110.195024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- Dandjinou AT, Levesque N, Larose S, Lucier JF, Abou Elela S, Wellinger RJ. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr Biol. 2004;14:1148–1158. doi: 10.1016/j.cub.2004.05.054. [DOI] [PubMed] [Google Scholar]
- Dynan WS, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher TS, Taggart AK, Zakian VA. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat Struct Mol Biol. 2004;11:1198–1205. doi: 10.1038/nsmb854. [DOI] [PubMed] [Google Scholar]
- Fisher TS, Zakian VA. Ku: a multifunctional protein involved in telomere maintenance. DNA Repair (Amst) 2005;4:1215–1226. doi: 10.1016/j.dnarep.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Gallardo F, Olivier C, Dandjinou AT, Wellinger RJ, Chartrand P. TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J. 2008;27:748–757. doi: 10.1038/emboj.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Gravel S, Larrivee M, Labrecque P, Wellinger RJ. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- Jain D, Cooper JP. Telomeric strategies: means to an end. Annu Rev Genet. 2010;44:243–269. doi: 10.1146/annurev-genet-102108-134841. [DOI] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Laroche T, Martin SG, Gotta M, Gorham HC, Pryde FE, Louis EJ, Gasser SM. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr Biol. 1998;8:653–656. doi: 10.1016/s0960-9822(98)70252-0. [DOI] [PubMed] [Google Scholar]
- Lee SE, Paques F, Sylvan J, Haber JE. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr Biol. 1999;9:767–770. doi: 10.1016/s0960-9822(99)80339-x. [DOI] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Lopez CR, Ribes-Zamora A, Indiviglio SM, Williams CL, Haricharan S, Bertuch AA. Ku must load directly onto the chromosome end in order to mediate its telomeric functions. PLoS Genet. 2011;7:e1002233. doi: 10.1371/journal.pgen.1002233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Lieber MR. DNA length-dependent cooperative interactions in the binding of Ku to DNA. Biochemistry. 2001;40:9638–9646. doi: 10.1021/bi010932v. [DOI] [PubMed] [Google Scholar]
- Milne GT, Jin S, Shannon KB, Weaver DT. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4189–4198. doi: 10.1128/mcb.16.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra K, Shore D. Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by rif proteins. Curr Biol. 1999;9:1123–1126. doi: 10.1016/s0960-9822(99)80483-7. [DOI] [PubMed] [Google Scholar]
- Mozdy AD, Cech TR. Low abundance of telomerase in yeast: implications for telomerase haploinsufficiency. Rna. 2006;12:1721–1737. doi: 10.1261/rna.134706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy AD, Podell ER, Cech TR. Multiple yeast genes, including Paf1 complex genes, affect telomere length via telomerase RNA abundance. Mol Cell Biol. 2008;28:4152–4161. doi: 10.1128/MCB.00512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Chen C, Kolodner RD. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- Nugent CI, Bosco G, Ross LO, Evans SK, Salinger AP, Moore JK, Haber JE, Lundblad V. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- Pennaneach V, Putnam CD, Kolodner RD. Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae. Mol Microbiol. 2006;59:1357–1368. doi: 10.1111/j.1365-2958.2006.05026.x. [DOI] [PubMed] [Google Scholar]
- Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- Polotnianka RM, Li J, Lustig AJ. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol. 1998;8:831–834. doi: 10.1016/s0960-9822(98)70325-2. [DOI] [PubMed] [Google Scholar]
- Porter SE, Greenwell PW, Ritchie KB, Petes TD. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter NJ, Maher L.J.r., Butcher SE. DNA mimicry by a high-affinity anti-NF-kappaB RNA aptamer. Nucleic Acids Res. 2008;36:1227–1236. doi: 10.1093/nar/gkm1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes-Zamora A, Mihalek I, Lichtarge O, Bertuch AA. Distinct faces of the Ku heterodimer mediate DNA repair and telomeric functions. Nat Struct Mol Biol. 2007;14:301–307. doi: 10.1038/nsmb1214. [DOI] [PubMed] [Google Scholar]
- Riha K, Heacock ML, Shippen DE. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu Rev Genet. 2006;40:237–277. doi: 10.1146/annurev.genet.39.110304.095755. [DOI] [PubMed] [Google Scholar]
- Roberts SA, Ramsden DA. Loading of the nonhomologous end joining factor, Ku, on protein-occluded DNA ends. J Biol Chem. 2007;282:10605–10613. doi: 10.1074/jbc.M611125200. [DOI] [PubMed] [Google Scholar]
- Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 2009;23:928–938. doi: 10.1101/gad.1787509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet BK. Interpreting steep dose-response curves in early inhibitor discovery. J Med Chem. 2006;49:7274–7277. doi: 10.1021/jm061103g. [DOI] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 2003;17:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- Yoo S, Dynan WS. Characterization of the RNA binding properties of Ku protein. Biochemistry. 1998;37:1336–1343. doi: 10.1021/bi972100w. [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc Natl Acad Sci U S A. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Goodrich K, Cech TR. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat Struct Mol Biol. 2005;12:1072–1077. doi: 10.1038/nsmb1019. [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Goodrich KJ, Arthur JR, Gurski LA, Denham EM, Stellwagen AE, Cech TR. Ku can contribute to telomere lengthening in yeast at multiple positions in the telomerase RNP. RNA. 2011;17:298–311. doi: 10.1261/rna.2483611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hefferin ML, Chen L, Shim EY, Tseng HM, Kwon Y, Sung P, Lee SE, Tomkinson AE. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol. 2007;14:639–646. doi: 10.1038/nsmb1261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.