Abstract
While the effects of TCR affinity and TGFβ on CD8+ T-cell function have been studied individually, the manner in which TCR affinity dictates susceptibility to TGFβ-mediated suppression remains unknown. To address this issue, we utilized OVA altered peptide ligands (APLs) of different affinities in the OT-I model. We demonstrate that while decreased TCR ligand affinity initially results in weakened responses, such interactions prime the resultant effector cells to respond more strongly to cognate antigen upon secondary exposure. Despite this, responses by CD8+ T cells primed with lower-affinity TCR ligands are more effectively regulated by TGFβ. Susceptibility to TGFβ-mediated suppression is associated with downregulation of RGS3, a recently recognized negative regulator of TGFβ signaling, but not expression of TGFβ receptors I/II. These results suggest a novel tolerance mechanism whereby CD8+ T cells are discriminately regulated by TGFβ according to the affinity of the ligand on which they were initially primed. In addition, because of the major role played by TGFβ in tumor-induced immune suppression, these results identify the affinity of the priming ligand as a primary concern in CD8+ T-cell-mediated cancer immunotherapeutic strategies.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-011-1043-1) contains supplementary material, which is available to authorized users.
Keywords: Tumor-induced suppression, TGFβ, CD8+ T cells, T-cell receptor affinity, RGS3
Introduction
CD8+ T cells emerge from the thymus bearing T-cell receptors (TCRs) with a wide range of affinities. Key to the understanding of TCR affinity and T-cell function has been the development of altered peptide ligands (APLs). Studies using APLs demonstrate high-affinity interactions between the TCR and peptide-MHC class I complexes (pMHC) result in greater induction of CD8+ T-cell responses [1, 2]. However, the manner in which APLs with differential TCR affinity dictate susceptibility to TGFβ-mediated suppression remains unknown.
Transforming growth factor beta (TGFβ) is an immunoregulatory cytokine with activity affecting T-cell proliferation, differentiation, survival, and self-tolerance [3–7]. TGFβ signals through a heterotetrameric complex of TGFβ receptor (TGFβR)-I and TGFβRII, which phosphorylates the receptor-regulated Smad signaling proteins (R-Smads), including Smad2 and Smad3. These R-Smads then complex with the co-Smad, Smad4, which together translocate to the nucleus to activate transcription of certain TGFβ-responsive genes [8, 9]. The inhibitory Smads, including Smad7, act by preventing phosphorylation of the R-Smads, while the noncanonical inhibitor of TGFβ signaling, the regulator of G-protein signaling (RGS)-3, acts by forming complexes with the R-Smads and co-Smad and prevents the activation of TGFβ-induced gene transcription [10]. Mice with T cells that lack the ability to respond to TGFβ rapidly experience multiorgan, multitarget T-cell-mediated autoimmunity without any prior modification of the T-cell repertoire [11–13]. Studies analyzing these mice have demonstrated that self-reactive T cells exist in the natural repertoire and that TGFβ signaling is required to prevent these responses in the normal physiological state. Additional studies have shown that TGFβ-insensitive polyclonal CD8+ T cells possess enhanced antitumor function and can prevent tumors from developing [14]. However, little is known about natural variations in CD8+ T-cell sensitivity to TGFβ signaling.
TCR affinity and TGFβ-mediated suppression have been individually shown to regulate CD8+ T-cell responses. However, the interplay between these variables remains unknown. In this study, we now demonstrate that while decreased TCR ligand affinity initially results in weakened responses, such interactions prime the resultant effector cells to respond more strongly to cognate antigen upon secondary exposure. In spite of this, responses by CD8+ T cells primed with lower-affinity TCR ligands are more effectively suppressed by TGFβ. These results highlight antigen affinity as an important concern in cancer immunotherapy that may not be addressed by vaccination or increasing the density of the presented antigen.
Materials and methods
Cells and mice
All cells were cultured in RPMI supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA), 2 mM l-glutamine (Mediatech, Manassas, VA), and 1% penicillin/streptomycin (Mediatech, Manassas, VA), unless otherwise noted. Six-week-old, specific-pathogen-free C57BL/6-Tg(TcraTcrb)1100Mjb/J (OT-I) mice were purchased from Jackson Laboratories. All mice were housed at The University of Chicago animal facility under conventional conditions, and animal experimentation was conducted in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines.
Peptides
OVA257 (SIINFEKL) and, in order of decreasing reported affinity for the OT-I TCR, the APLs Y3 (SIYNFEKL), Q4 (SIIQFEKL), T4 (SIITFEKL), and V4 (SIIVFEKL) were purchased from New England Peptide (Gardner, MA).
In vitro activation and restimulation
Irradiated feeder EL-4 cells were loaded for 2 h with OVA257 peptide or OVA APL (1 μg/ml) and washed twice to remove unloaded peptide. OT-I splenocytes were co-cultured with feeder EL-4 cells for two days in media supplemented with 30 U/ml IL-2 (R&D Systems, Minneapolis, MN) prior to the addition of TGFβ1 (EMD Chemicals, Inc., Gibbstown, NJ) at a final concentration of 20 ng/ml. Some cells were washed in PBS and stained for analysis of extracellular and intracellular markers before the addition of TGFβ1. At day 5, cells were restimulated with OVA257 peptide (100 ng/ml) overnight in the presence of GolgiPlug (BD Biosciences, San Diego, CA) and TGFβ1 at a final concentration of 20 ng/ml as described. Cells were then washed in PBS and stained for surface and intracellular markers. To examine initial responses to the OVA APLs, OT-I splenocytes were cultured with various concentrations (ranging from 5 μM to 1 pM) of the peptide for a total of 8 h (6 h after the addition of GolgiPlug) without exogenous IL-2. Cells were washed in PBS and stained for analysis by flow cytometry.
Antibodies and flow cytometry
All mouse antibodies against cell surface and intracellular markers were purchased from Ebioscience (San Diego, CA), except APC-Cy7 anti-CD3 (BD Biosciences, San Diego, CA), Pacific Orange anti-CD8 (Invitrogen, Carlsbad, CA), PE anti-TGFβRII (R&D Systems, Minneapolis, MN), Pacific Blue anti-T-bet (BioLegend, San Diego, CA), FITC anti-KLRG1 (Southern Biotech, Birmingham, AL), anti-TGFβRI (Santa Cruz Biotechnology, Santa Cruz, CA), and anti-RGS3. The anti-RGS3 antibody has been described previously [15]. Extracellular and intracellular marker staining was performed as previously described [16].
Statistical analyses
Sigmoidal dose–response and exponential association curves were fit to data using GraphPad Prism (GraphPad Software, San Diego, CA). For the sigmoidal dose–response curves, separate curves for each peptide were accepted only if the extra sum-of-squares F test yielded a P value of less than 0.05. The goodness of fit for the exponential association curves is indicated by R 2 value. To compare cytokine suppression across multiple experiments, one-way ANOVA with a Tukey HSD post-test was used to calculate P values. P values below 0.05 were deemed significant.
Results
Priming with low-affinity peptide ligands gives rise to effectors with enhanced function
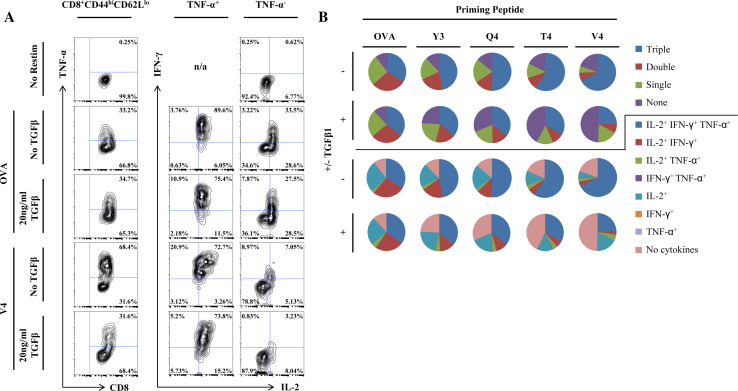
In order to investigate the effect of TCR signaling strength at priming on effector function, OT-I CD8+ T cells were primed with EL4 cells loaded with wild-type (wt) OVA257 or one of four single residue-substituted versions of the peptide, referred to collectively as APLs [1]. These APLs bear substitutions only at TCR-facing residues and not anchor residues and accordingly have been shown to affect OT-I TCR-binding affinity without affecting affinity of binding to the class I MHC molecule, H-2Kb. Restimulation with 100 ng/ml wt OVA257 was performed at day 5 after priming. While single cytokines are often used as measures of effector CD8+ T-cell function, a number of studies have shown that the simultaneous expression of several cytokines correlates far better with protective immunity than the magnitude of any one single cytokine [17–20]. Therefore, antigen-specific production of the cytokines IFN-γ, TNF-α and IL-2 were used as measures of effector function. In contrast to our expectations, a greater proportion of those cells primed with the lower-affinity APLs produced all three cytokines than those primed with higher-affinity ligands, including wt OVA257 (Fig. 1a, b, Table 1). Previous studies have reported that decreased antigen density or availability can lead to enhanced responses at restimulation by maintaining increased levels of TCR and the coreceptor CD8 at the cell surface [21, 22]. However, in response to decreased ligand affinity at the same concentration in our system, no differences were observed with respect to CD3ε or CD8 expression at the cell surface (Supplemental Fig. 1). Furthermore, the effect of lower-affinity ligands was not mediated by activating fewer cells during the 5-day priming period, as the expression of CD44 and CD62L in CD8+ T cells remain unchanged between stimulatory cultures regardless of ligand affinity (Supplemental Fig. 2).
Fig. 1.
Priming with lower-affinity peptide ligands leads to enhanced secondary responses but also increased sensitivity to TGFβ-mediated suppression. OT-I splenocytes were primed on peptide-loaded, irradiated EL4 cells for 5 days in vitro prior to restimulation with 100 ng/ml OVA257 for all groups. Half of all samples were incubated in 20 ng/ml TGFβ starting at day 2 and during restimulation. Cells were analyzed by flow cytometry for polycytokine production in the CD3+CD8+CD44hi antigen-experienced CD8+ T-cell gate. a Contour plots demonstrating differential cytokine output by cells primed with OVA257 and the V4 APL in the presence and absence of TGFβ. Cytokine output for cells primed for 5 days with OVA257 but not restimulated is also shown. b Pie charts representing the proportion of effector CD8+ T cells producing all three cytokines (triple), a set of only two cytokines (double), only a single cytokine (single), or no cytokines (none), or the proportion of cells producing a precise combination of the three cytokines. Data shown are representative of at least three individual experiments with similar results
Table 1.
Cytokine profile of cells primed on OVA or APLs in the presence and absence of TGFβ
| Priming peptide | ± TGFβ1 | Triple | Double | IL-2+, IFN-γ+ | IL-2+, TNF-α+ | IFN-γ+, TNF-α+ | Single | IL-2+ | IFN-γ+ | TNF-α+ | No cytokines | Cumulative | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL-2+ | IFN-γ+ | TNF-α+ | ||||||||||||
| OVA | − | 33.98 | 29.29 | 27.02 | 2.27 | 0.00 | 26.58 | 26.34 | 0.18 | 0.06 | 10.24 | 89.61 | 36.31 | 61.18 |
| + | 35.28 | 28.59 | 24.12 | 4.35 | 0.13 | 24.51 | 24.25 | 0.26 | 0.00 | 11.60 | 88.00 | 39.76 | 59.79 | |
| Y3 | − | 46.99 | 21.14 | 17.91 | 3.14 | 0.09 | 19.73 | 19.46 | 0.27 | 0.00 | 12.10 | 87.50 | 50.22 | 65.26 |
| + | 36.38 | 17.63 | 12.92 | 4.47 | 0.24 | 22.01 | 21.83 | 0.12 | 0.06 | 24.05 | 75.60 | 41.15 | 49.66 | |
| Q4 | − | 51.03 | 14.45 | 11.97 | 2.48 | 0.00 | 20.74 | 20.52 | 0.18 | 0.05 | 13.76 | 86.00 | 53.56 | 63.18 |
| + | 35.66 | 14.11 | 9.94 | 3.90 | 0.26 | 19.00 | 18.80 | 0.17 | 0.03 | 31.27 | 68.30 | 39.86 | 46.03 | |
| T4 | − | 59.10 | 10.52 | 6.17 | 4.07 | 0.28 | 12.45 | 12.03 | 0.28 | 0.14 | 17.86 | 81.37 | 63.59 | 65.83 |
| + | 33.56 | 9.94 | 5.24 | 4.38 | 0.32 | 14.22 | 13.72 | 0.40 | 0.11 | 42.27 | 56.90 | 38.37 | 39.52 | |
| V4 | − | 67.47 | 7.26 | 3.93 | 2.62 | 0.71 | 5.95 | 5.55 | 0.20 | 0.20 | 19.31 | 79.58 | 71.01 | 72.32 |
| + | 24.67 | 8.19 | 2.54 | 5.22 | 0.44 | 17.69 | 17.58 | 0.07 | 0.04 | 49.45 | 50.01 | 30.37 | 27.72 | |
OT-I splenocytes were primed and restimulated as shown in Fig. 1. The percentages of the resulting effector CD8+ T cells producing a particular set of cytokines or all of those producing an individual cytokine (Cumulative) are shown. Data shown are representative of at least three individual experiments with similar results
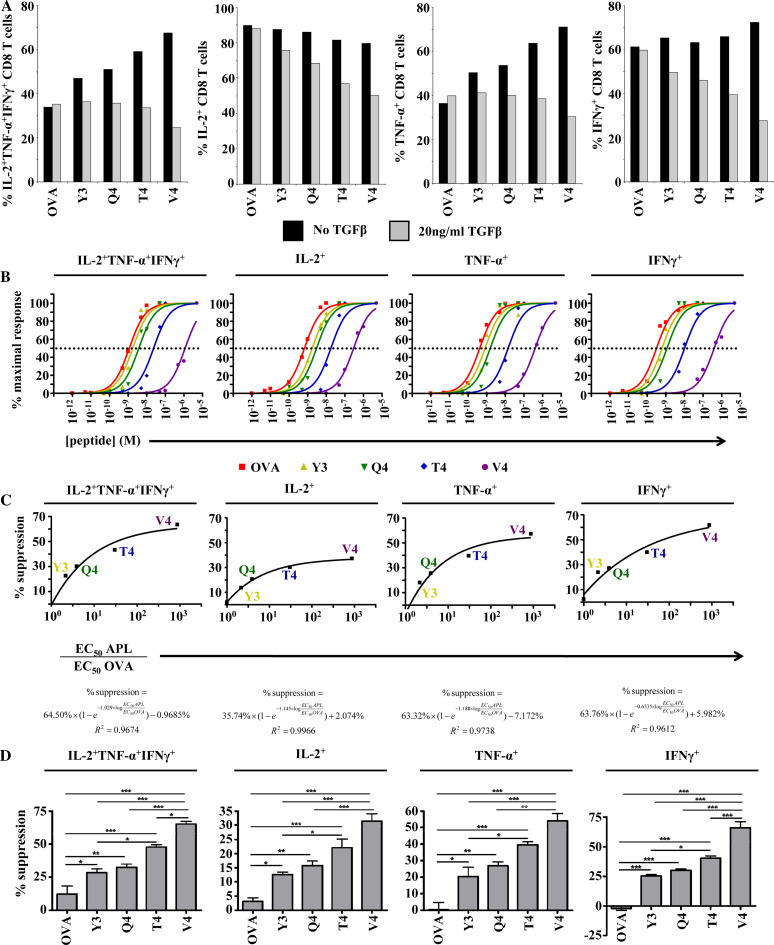
Fig. 2.
Degree of TGFβ-mediated suppression is dependent on the affinity of the ligand on which CD8+ T cells are primed. a CD8+ T cells were primed and restimulated as shown in Fig. 1. Bar graphs indicate the percentages of CD3+CD8+CD44hi cells producing all three cytokines or producing each cytokine, regardless of whether these cells produced any other cytokines. b OT-I splenocytes were stimulated in vitro for 8 h with decreasing concentrations of OVA257 or the APLs. Following stimulation, cells were analyzed for polycytokine production. For each peptide, responses were normalized to the peak response. The data were fit with sigmoidal dose–response curves. Separate curves were accepted for each peptide as the extra sum-of-squares F test yielded P values less than 0.0001 for each plot. A dotted line denotes 50% maximal stimulation. EC50 values were derived from the intersection between the sigmoidal dose–response curves and the line denoting 50% maximal stimulation. c The degree to which effector responses were suppressed by TGFβ was plotted against the derived EC50 values for each peptide relative to the derived EC50 value for OVA257. The equation defining the best-fit curve and its R 2 value is shown for each plot. d The degree of suppression of cytokine production by TGFβ was measured across four individual experiments. Columns represent mean suppression. Error bars represent standard deviation. *P < 0.05, **P < 0.01, ***P < 0.001. Suppression was defined by (% cytokine+noTGFβ − % cytokine+TGFβ)/% cytokine+noTGFβ. Data shown are representative of at least three individual experiments with similar results
Priming with low-affinity peptide ligands gives rise to effectors with increased susceptibility to TGFβ signaling
As peptide ligand affinity was found to control the quality and magnitude of the secondary response, we endeavored to also investigate the role of ligand affinity during priming on later sensitivity to immune regulation by TGFβ. This was accomplished by priming the OT-I CD8+ T cells in the same manner as before but incubating half of the cells in media containing physiological concentrations (20 ng/ml) of TGFβ1 [23] beginning at day 2 after priming and during restimulation at day 5. As the magnitude of the secondary effector response was enhanced by lower-affinity ligands, we expected to find that these cells were also more resistant to TGFβ-mediated immune regulation. However, while there was little apparent effect of TGFβ on cells primed with higher-affinity ligands, there were large apparent suppressive effects on those primed with the lower-affinity ligands (Fig. 1a, b, Table 1). Furthermore, this effect was observed with respect to production of all three cytokines (Fig. 2a, Table 1).
Ligand affinity at priming correlates with susceptibility to suppression
To further gain insight into the functional effects of APLs with lower affinities, we stimulated naive OT-I CD8+ T cells with various concentrations of OVA257 or the APLs and measured cytokine output. As expected, higher concentrations of the lower-affinity APLs were necessary to reach half-maximal stimulation (EC50) and the maximum proportion of cells producing cytokines with stimulation from lower-affinity ligands was also lower than those stimulated with higher-affinity ligands (Fig. 2b and Supplemental Fig. 3). Plotting these experimentally determined relative EC50 values, which are similar to previously published values for these peptides [1], against the degree of suppression with each peptide ligand reveals a clear relationship between peptide ligand affinity during priming and later susceptibility to TGFβ-mediated suppression during the effector phase (Fig. 2c). The curves that best correlate with the data are defined by exponential association equations, suggesting that as the affinity of the TCR for the priming ligand decreases, the degree of suppression will eventually approach an asymptotic maximum. In addition, all of these equations include y-intercepts that fall near zero (some are negative), which corresponds to the suppressive effect of 20 ng/ml TGFβ on cells originally primed with wt OVA257. Hence, lower-affinity peptide priming leads to increased susceptibility to TGFβ-mediated suppression. Statistically significant differences in the degree of TGFβ-mediated suppression were evident with respect to each cytokine or concurrent production of all three cytokines (Fig. 2d).
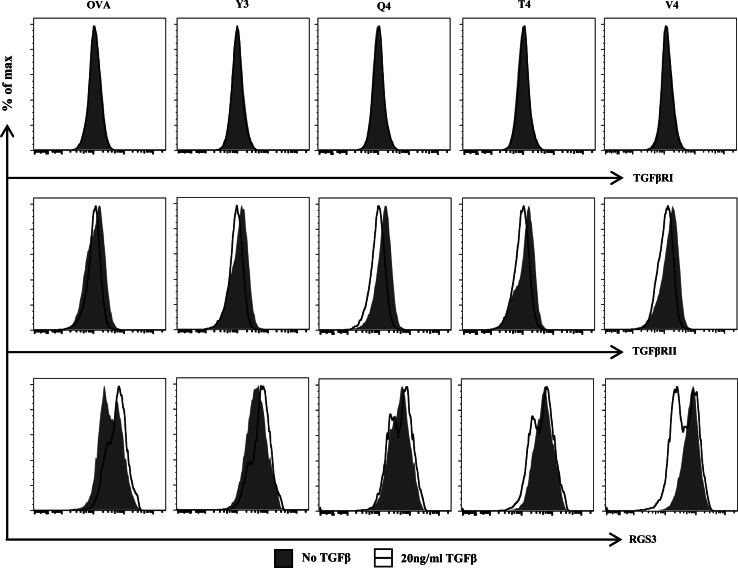
TCR affinity differentially regulates RGS3 expression in the presence of TGFβ
To determine the molecular basis for the increased susceptibility to TGFβ-mediated suppression, we similarly primed cells with wt OVA257 and each of the APLs and analyzed expression levels of proteins in the TGFβ signaling pathway, namely TGFβRI and RII. Yet for neither of these proteins did expression levels correlate with susceptibility to TGFβ-mediated suppression. There were no significant changes in expression for TGFβRI, TGFβRII, or the inhibitory Smad7 (Fig. 3 and data not shown). Interestingly, expression of RGS3, a recently defined noncanonical inhibitor of TGFβ signaling [10], was found to be higher in cells primed with lower-affinity peptide ligands (Fig. 3). In order to more completely recreate the suppressive conditions and thus accurately recreate the susceptible phenotype, the original priming scheme was followed. However, at day 5 after priming, the cells were analyzed for expression levels of each of the TGFβ pathway proteins. Once again, expression levels for the TGFβ receptors I and II were stable across the different peptide ligands (Fig. 3). However, while RGS3 once again was more highly expressed in cells that were primed with lower-affinity ligands in the absence of TGFβ, the presence of TGFβ reversed these phenotypes: RGS3 was upregulated in cells primed with high-affinity ligands and downregulated in cells primed with low-affinity ligands (Fig. 3). Thus, in the suppressive environment, cells primed with lower-affinity ligands adopted a more TGFβ-responsive phenotype. Interestingly, this change was not accompanied by any significant changes in activation status markers, such as CD44, CD62L, KLRG1, CD127, T-bet, or Eomes (Supplemental Fig. 2). In addition, no changes were observed with respect to CD3, CD44, CD62L, RGS3, TGFβRI, TGFβRII, KLRG1, CD127, or T-bet between priming cultures at day two, prior to the addition of TGFβ (Supplemental Fig. 4).
Fig. 3.
Ligand affinity during priming dictates later expression levels of RGS3. OT-I cells were primed by the various ligands and incubated as shown in Fig. 1. At day 5, the cells were analyzed for expression levels of TGFβRI, TGFβRII, and RGS3. Each histogram indicates the expression level of TGFβRI, TGFβRII, or RGS3 in CD3+CD8+CD44hi cells incubated with or without 20 ng/ml TGFβ. Data shown are representative of at least three individual experiments with similar results
Discussion
While previous studies have defined the role of TCR affinity in central tolerance, our study highlights the potential interplay between TCR affinity and TGFβ in peripheral tolerance. The outcome of low-affinity TCR ligand stimulation on CD8+ T-cell cytokine production during the primary response is known; however, the effects of such priming on secondary activation and effector function have not yet been defined. Through the use of OVA257 peptide and residue-substituted peptide OVA257 analogs for which the OT-I TCR has reduced affinity, we have shown that OT-I CD8+ T cells primed with lower-affinity ligands are better suited to respond to the cognate antigen upon restimulation. Furthermore, while it has been reported that functional avidity may be modulated by antigen dose through mechanisms involving altering surface expression levels of the TCR and the CD8 coreceptor [21, 22], we find that the levels of CD3ε and CD8 are unchanged at the surface regardless of the affinity of the priming ligand. This effect may partially underlie the importance of low-affinity self-reactivity in the periphery, where CD8+ T cells may be better suited to recognize foreign, high-affinity antigens and thus resolve infection because of earlier recognition of self-peptide ligands.
One of the central issues in immunology is the way in which autoimmunity is prevented or controlled. This is especially significant for T-cell-mediated autoimmunity, as T cells are positively selected on the basis of self-antigen recognition during thymic development yet are not generally self-reactive in the periphery. The prevailing explanation for this phenomenon is that because T cells that emerge from thymic development have only a low-affinity interaction with self-antigen due to negative selection, such an interaction could not normally lead to a productive immune response [24, 25]. Our results demonstrate an additional mechanism to avert autoimmunity, whereby CD8+ T cells that have been primed with such a low-affinity interaction become more sensitive to regulation by TGFβ while those primed with high-affinity interactions are minimally affected. This mechanism allows the host to selectively suppress those CD8+ T-cell clones that may be deleterious while maintaining those that, by virtue of TCR affinity for pMHC, may be considered strictly foreign antigen-specific, even while both are responding to the same antigen in the same microenvironment. The same principle can be applied to tumor-reactive CD8+ T cells, as they generally recognize tumor-associated antigens (most of which are unaltered self-proteins) with very low affinity. This mechanism to avoid autoimmunity would then allow the tumor to more effectively suppress CD8+ T-cell-mediated immune responses, particularly against the tumor itself, via TGFβ.
The immune system has evolved the capacity to mount responses to foreign pathogens while avoiding responses to self-antigens. From studies showing that loss of TGFβ signaling in T cells leads rapidly to multitarget autoimmunity, it can be inferred that self-reactive conventional T cells exist in the natural repertoire and are kept in check through normal levels of TGFβ [11, 12]. In addition, as mice reconstituted with TGFβ-insensitive CD8+ T cells prevent EL4 thymoma or B16 melanoma tumors from developing without any therapeutic intervention, these tumors appear to require the suppressive function of TGFβ to evade immune destruction [14, 26]. However, very little is known about differences in TGFβ sensitivity in T-cell populations. Sanjabi et al. have shown that during the contraction phase, short-lived effector cells respond to TGFβ by undergoing apoptosis, whereas memory progenitor effector cells are preferentially maintained despite elevated TGFβ levels [27]. There is evidence that both antigen affinity and availability during priming control the size of the resulting memory population [1, 28, 29]. It can be hypothesized that in our model, those CD8+ T cells primed with high-affinity ligands are more predisposed to becoming memory cells than those primed with low-affinity ligands.
A noncanonical function of RGS3 in regulating TGFβ signaling has recently been defined [10]. RGS3 has been shown to bind Smad2, Smad3, and Smad4, thereby impeding heteromerization of R-Smads and Smad4 and preventing TGFβ-induced, Smad-mediated transcriptional activation [10]. While no differences were observed with respect to expression levels of the type I or II TGFβ receptors or the inhibitory Smad, Smad 7, RGS3 expression correlated with sensitivity to TGFβ-mediated suppression of effector function in the presence of TGFβ1. Previous studies have shown that RGS protein expression may be modulated by TLR signaling in DCs [30] and as a result of activation in B cells [31]. However, our data represent the first evidence that RGS3 expression can be modulated in response to both TGFβ signaling and a program resulting from TCR affinity during priming.
Our data demonstrate that while lower TCR ligand affinity results in less intense initial responses, such interactions during priming lead effector cells to become better able to respond to the cognate antigen upon secondary antigen exposure. Despite this, these cells are also more effectively suppressed by TGFβ. Collectively, these results suggest a novel tolerance mechanism, whereby CD8+ T cells are discriminately regulated by TGFβ according to the affinity of the ligand on which they were initially primed. Furthermore, these findings suggest that the low-affinity TCR ligands expressed by tumors may render responding CD8+ T cells more sensitive to TGFβ-mediated suppression and that this programming may be avoided by initially priming CD8+ T cells with higher-affinity ligands.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Expression of CD3 and CD8 is not affected by priming ligand affinity. OT-I cells were primed with the various ligands and incubated as described in Fig. 1. At day 5 after priming, the cells were analyzed for surface expression of CD3 and CD8. Expression levels of these markers are shown in offset histograms. Data shown are representative of at least three individual experiments with similar results (TIFF 120 kb)
Neither ligand affinity nor TGFβ signaling affects CD8+ T-cell activation status markers. OT-I cells were primed with the various ligands and incubated as shown in Fig. 1. At day 5, the cells were analyzed for surface expression of CD44, CD62L, KLRG1, and CD127 and intracellular expression of the transcription factors T-bet and Eomes. Each histogram indicates the expression level of the marker in CD3+CD8+ cells incubated in the presence or absence of 20 ng/ml TGFβ. Data shown are representative of at least three individual experiments with similar results (TIFF 338 kb)
Initial responses to OVA APLs. OT-I splenocytes were stimulated in vitro for 8 h with decreasing concentrations of OVA257 or the APLs. Following stimulation, cells were analyzed for polycytokine production. The data were fit with sigmoidal dose–response curves. Separate curves were accepted for each peptide as the extra sum-of-squares F test yielded P values less than 0.0001 for each plot. Data shown are representative of at least three individual experiments with similar results (TIFF 130 kb)
No affinity-dependent phenotypic changes are observed before the addition of TGFβ. OT-I cells were primed with the various ligands and incubated as described for the first 2 days. The cells were then analyzed by flow cytometry. Expression levels of CD3, CD44, CD62L, RGS3, TGFβRI, TGFβRII, KLRG1, CD127, and T-bet are shown in offset histograms. Data shown are representative of at least three individual experiments with similar results (TIFF 390 kb)
Acknowledgments
We are grateful to Mary Jo Turk (Dartmouth Medical School, NH) and Anne Sperling (The University of Chicago, IL) for constructive discussions and the Flow Cytometry Facility at The University of Chicago for its invaluable support. This work was supported by the American Cancer Society (ACSLIB112496-RSG, to J.A.G.), American Cancer Society–Illinois Division (Young Investigator Award Grant #07-20, to J.A.G.), the National Institutes of Health (R21CA127037-01A1 to J.A.G. and R01GM85058 to N.O.D.), Cancer Research Foundation (Young Investigator Award, to J.A.G.), and the National Institutes of Health (T32 Immunology Training Grant, The University of Chicago, AI007090 to J.A.O., A.Z., and F.J.K.). The authors have no financial conflicts of interest to disclose.
References
- 1.Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458(7235):211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hommel M, Hodgkin PD. TCR affinity promotes CD8+ T cell expansion by regulating survival. J Immunol. 2007;179(4):2250–2260. doi: 10.4049/jimmunol.179.4.2250. [DOI] [PubMed] [Google Scholar]
- 3.Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor beta in human disease. N Engl J Med. 2000;342(18):1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 4.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198(12):1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulkarni AB, Huh CG, Becker D, Geiser A, Lyght M, Flanders KC, Roberts AB, Sporn MB, Ward JM, Karlsson S. Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proc Natl Acad Sci USA. 1993;90(2):770–774. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 7.Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441(7090):231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 8.Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1(3):169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 9.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/S0092-8674(03)00432-X. [DOI] [PubMed] [Google Scholar]
- 10.Yau DM, Sethakorn N, Taurin S, Kregel S, Sandbo N, Camoretti-Mercado B, Sperling AI, Dulin NO. Regulation of Smad-mediated gene transcription by RGS3. Mol Pharmacol. 2008;73(5):1356–1361. doi: 10.1124/mol.108.044990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorelik L, Flavell RA. Abrogation of TGFbeta signaling in T cells leads to spontaneous T cell differentiation and autoimmune disease. Immunity. 2000;12(2):171–181. doi: 10.1016/S1074-7613(00)80170-3. [DOI] [PubMed] [Google Scholar]
- 12.Marie JC, Liggitt D, Rudensky AY. Cellular mechanisms of fatal early-onset autoimmunity in mice with the T cell-specific targeting of transforming growth factor-beta receptor. Immunity. 2006;25(3):441–454. doi: 10.1016/j.immuni.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 13.Fahlen L, Read S, Gorelik L, Hurst SD, Coffman RL, Flavell RA, Powrie F. T cells that cannot respond to TGF-beta escape control by CD4(+)CD25(+) regulatory T cells. J Exp Med. 2005;201(5):737–746. doi: 10.1084/jem.20040685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorelik L, Flavell RA. Immune-mediated eradication of tumors through the blockade of transforming growth factor-beta signaling in T cells. Nat Med. 2001;7(10):1118–1122. doi: 10.1038/nm1001-1118. [DOI] [PubMed] [Google Scholar]
- 15.Dulin NO, Sorokin A, Reed E, Elliott S, Kehrl JH, Dunn MJ. RGS3 inhibits G protein-mediated signaling via translocation to the membrane and binding to Galpha11. Mol Cell Biol. 1999;19(1):714–723. doi: 10.1128/mcb.19.1.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zloza A, Jagoda MC, Lyons GE, Graves MC, Kohlhapp FJ, O’Sullivan JA, Lacek AT, Nishimura MI, Guevara-Patino JA (2011) CD8 Co-receptor promotes susceptibility of CD8(+) T cells to transforming growth factor-beta (TGF-beta)-mediated suppression. Cancer Immunol Immunother. doi:10.1007/s00262-010-0962-6 [DOI] [PMC free article] [PubMed]
- 17.Beveridge NE, Price DA, Casazza JP, Pathan AA, Sander CR, Asher TE, Ambrozak DR, Precopio ML, Scheinberg P, Alder NC, Roederer M, Koup RA, Douek DC, Hill AV, McShane H. Immunisation with BCG and recombinant MVA85A induces long-lasting, polyfunctional mycobacterium tuberculosis-specific CD4+ memory T lymphocyte populations. Eur J Immunol. 2007;37(11):3089–3100. doi: 10.1002/eji.200737504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Precopio ML, Betts MR, Parrino J, Price DA, Gostick E, Ambrozak DR, Asher TE, Douek DC, Harari A, Pantaleo G, Bailer R, Graham BS, Roederer M, Koup RA. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J Exp Med. 2007;204(6):1405–1416. doi: 10.1084/jem.20062363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107(12):4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Betts MR, Price DA, Brenchley JM, Lore K, Guenaga FJ, Smed-Sorensen A, Ambrozak DR, Migueles SA, Connors M, Roederer M, Douek DC, Koup RA. The functional profile of primary human antiviral CD8+ T cell effector activity is dictated by cognate peptide concentration. J Immunol. 2004;172(10):6407–6417. doi: 10.4049/jimmunol.172.10.6407. [DOI] [PubMed] [Google Scholar]
- 21.Kroger CJ, Alexander-Miller MA. Dose-dependent modulation of CD8 and functional avidity as a result of peptide encounter. Immunology. 2007;122(2):167–178. doi: 10.1111/j.1365-2567.2007.02622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kroger CJ, Alexander-Miller MA. Cutting edge: CD8+ T cell clones possess the potential to differentiate into both high- and low-avidity effector cells. J Immunol. 2007;179(2):748–751. doi: 10.4049/jimmunol.179.2.748. [DOI] [PubMed] [Google Scholar]
- 23.Desser L, Holomanova D, Zavadova E, Pavelka K, Mohr T, Herbacek I. Oral therapy with proteolytic enzymes decreases excessive TGF-beta levels in human blood. Cancer Chemother Pharmacol. 2001;47(Suppl):10–15. doi: 10.1007/s002800170003. [DOI] [PubMed] [Google Scholar]
- 24.Pircher H, Rohrer UH, Moskophidis D, Zinkernagel RM, Hengartner H. Lower receptor avidity required for thymic clonal deletion than for effector T-cell function. Nature. 1991;351(6326):482–485. doi: 10.1038/351482a0. [DOI] [PubMed] [Google Scholar]
- 25.Sant’Angelo DB, Janeway CA., Jr Negative selection of thymocytes expressing the D10 TCR. Proc Natl Acad Sci USA. 2002;99(10):6931–6936. doi: 10.1073/pnas.102182499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bellavance EC, Kohlhapp FJ, Zloza A, O’Sullivan JA, McCracken J, Jagoda MC, Lacek AT, Posner MC, Guevara-Patino JA. Development of tumor-infiltrating CD8+ T cell memory precursor effector cells and antimelanoma memory responses are the result of vaccination and TGF-beta blockade during the perioperative period of tumor resection. J Immunol. 2011;186(6):3309–3316. doi: 10.4049/jimmunol.1002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanjabi S, Mosaheb MM, Flavell RA. Opposing effects of TGF-beta and IL-15 cytokines control the number of short-lived effector CD8+ T cells. Immunity. 2009;31(1):131–144. doi: 10.1016/j.immuni.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leignadier J, Labrecque N. Epitope density influences CD8+ memory T cell differentiation. PLoS One. 2010;5(10):e13740. doi: 10.1371/journal.pone.0013740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obar JJ, Lefrancois L. Early signals during CD8 T cell priming regulate the generation of central memory cells. J Immunol. 2010;185(1):263–272. doi: 10.4049/jimmunol.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi GX, Harrison K, Han SB, Moratz C, Kehrl JH. Toll-like receptor signaling alters the expression of regulator of G protein signaling proteins in dendritic cells: implications for G protein-coupled receptor signaling. J Immunol. 2004;172(9):5175–5184. doi: 10.4049/jimmunol.172.9.5175. [DOI] [PubMed] [Google Scholar]
- 31.Reif K, Cyster JG. RGS molecule expression in murine B lymphocytes and ability to down-regulate chemotaxis to lymphoid chemokines. J Immunol. 2000;164(9):4720–4729. doi: 10.4049/jimmunol.164.9.4720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of CD3 and CD8 is not affected by priming ligand affinity. OT-I cells were primed with the various ligands and incubated as described in Fig. 1. At day 5 after priming, the cells were analyzed for surface expression of CD3 and CD8. Expression levels of these markers are shown in offset histograms. Data shown are representative of at least three individual experiments with similar results (TIFF 120 kb)
Neither ligand affinity nor TGFβ signaling affects CD8+ T-cell activation status markers. OT-I cells were primed with the various ligands and incubated as shown in Fig. 1. At day 5, the cells were analyzed for surface expression of CD44, CD62L, KLRG1, and CD127 and intracellular expression of the transcription factors T-bet and Eomes. Each histogram indicates the expression level of the marker in CD3+CD8+ cells incubated in the presence or absence of 20 ng/ml TGFβ. Data shown are representative of at least three individual experiments with similar results (TIFF 338 kb)
Initial responses to OVA APLs. OT-I splenocytes were stimulated in vitro for 8 h with decreasing concentrations of OVA257 or the APLs. Following stimulation, cells were analyzed for polycytokine production. The data were fit with sigmoidal dose–response curves. Separate curves were accepted for each peptide as the extra sum-of-squares F test yielded P values less than 0.0001 for each plot. Data shown are representative of at least three individual experiments with similar results (TIFF 130 kb)
No affinity-dependent phenotypic changes are observed before the addition of TGFβ. OT-I cells were primed with the various ligands and incubated as described for the first 2 days. The cells were then analyzed by flow cytometry. Expression levels of CD3, CD44, CD62L, RGS3, TGFβRI, TGFβRII, KLRG1, CD127, and T-bet are shown in offset histograms. Data shown are representative of at least three individual experiments with similar results (TIFF 390 kb)